Abstract
Malaria during the first pregnancy causes a high rate of fetal and neonatal death. The decreasing susceptibility during subsequent pregnancies correlates with acquisition of antibodies that block binding of infected red cells to chondroitin sulfate A (CSA), a receptor for parasites in the placenta. Here we identify a domain within a particular Plasmodium falciparum erythrocyte membrane protein 1 that binds CSA. We cloned a var gene expressed in CSA-binding parasitized red blood cells (PRBCs). The gene had eight receptor-like domains, each of which was expressed on the surface of Chinese hamster ovary cells and was tested for CSA binding. CSA linked to biotin used as a probe demonstrated that two Duffy-binding-like (DBL) domains (DBL3 and DBL7) bound CSA. DBL7, but not DBL3, also bound chondroitin sulfate C (CSC) linked to biotin, a negatively charged sugar that does not support PRBC adhesion. Furthermore, CSA, but not CSC, blocked the interaction with DBL3; both CSA and CSC blocked binding to DBL7. Thus, only the DBL3 domain displays the same binding specificity as PRBCs. Because protective antibodies present after pregnancy block binding to CSA of parasites from different parts of the world, DBL-3, although variant, may induce cross-reactive immunity that will protect pregnant women and their fetuses.
Plasmodium falciparum malaria is more severe in pregnant women, especially during the first pregnancy (primigravida), and causes disease in the mother and fetal death even in those women who were previously immune (1). In the primigravida, massive numbers of parasitized red blood cells (PRBCs) sequester in the maternal circulation of the placenta, binding to chondroitin sulfate A (CSA) (2). Antibodies that develop after multiple pregnancies are associated with reduced PRBCs in the placenta and block CSA binding of PRBCs from different parts of the world (3). This suggests that the parasite ligand for binding CSA may be a vaccine candidate for protecting pregnant women and their fetuses against malaria.
Members of the recently described var gene family and their expressed proteins, P. falciparum erythrocyte membrane protein 1 (PfEMP-1), mediate PRBC binding to several adhesion receptors such as CD36 and intercellular adhesion molecule 1 (ICAM-1) (4, 5) and are involved in antigenic variation (4–6). Recent work on var gene switching showed transcription of a particular var gene (FCR3.varCSA) in parasites selected for binding to CSA but not in parasites selected for adhesion to CD36 or ICAM-1 (7). Thus, var genes adhere dichotomously either to CD36 and other receptors on endothelium or to CSA in placenta and not to CD36. Potential receptor domains in var genes include Duffy binding-like (DBL) domains, named for their homology to the Duffy binding domain of Plasmodium vivax (6), and cysteine-rich interdomain regions (CIDR). The CIDR1 domain, located after the first DBL, was demonstrated to mediate PRBC adhesion to CD36 (4, 8). DBL1 has been identified as a receptor for binding PRBCs to uninfected RBCs in var genes from PRBCs that rosette normal RBCs (9, 10). The ligands for binding CSA and other endothelial receptors are yet to be defined. Antibodies to two different domains of a var gene (different from the one described here) expressed in CSA-binding parasites reduced binding to CSA (11), indicating the possible involvement of var genes in adhesion to CSA, although direct identification of the domains that bind CSA was lacking. To identify the domain of the parasite ligand that mediates adhesion to CSA, we cloned the expressed var gene of FCR3-CSA-PRBCs. Here we identify the DBL domain within a particular PfEMP-1 that binds CSA.
Materials and Methods
Parasites and Cell Lines.
P. falciparum FCR3 parasites were cultured and selected on the adhesion receptors CD36 and CSA as described (7). A stable transfectant of Chinese hamster ovary (CHO)–745 cells (CSA negative) (12) permanently expressing cDNA of CD36 (13), ICAM-1 (14), vascular cell adhesion molecule 1 (VCAM-1) (15), and E-selectin (16) was constructed by using Fugene 6 transfection reagent (Roche Diagnostics). A stably transformed human umbilical vein endothelial cell line was kindly provided by D. Paulin and P. Vicart (Institut Pasteur) (17). Surface expression of platelet endothelial cell adhesion molecule 1, ICAM-1, E-selectin, and VCAM-1 was analyzed by using specific mAbs (R & D Systems). Anti-CD36 mAb was a gift of L. Edelman (Institut Pasteur).
Adhesion Assays.
PRBC adhesion assays on transfected CHO cells and fresh cryosections of human placenta were performed as described (18). Adhesion of plasmagel-enriched PRBCs to various receptors coated on plastic was performed as follows: 10 μl of receptor in PBS was immobilized directly on Petri dishes (Falcon 1001) overnight at 4°C. Biotinylated CSA (Biot-CSA), biotinylated CSC (Biot-CSC), or soluble CD36 was immobilized via mouse mAbs (5 μg/ml) directed either against biotin (Sigma) or against an epitope tag incorporated into a recombinant CD36 molecule (mAb 179) (19). [Receptor concentration: recombinant human thrombomodulinCSA (hTM), 5 μg/ml; CSA, 10 μg/ml (Sigma); CSC, 10 μg/ml (Sigma); Biot-CSA and Biot-CSC, 100 μg/ml]. The coated dots were blocked with 1% BSA and were incubated with 10 μl of trophozoites (0.5% hematocrit) in binding medium (BM) [RPMI medium1640 with 25 mM Hepes (pH 6.8)] for 20 min at 37°C. Unfixed cells were removed by washes in BM, were fixed with 2% glutaraldehyde, and were stained in Giemsa for microscopic examination.
Cloning DNA Sequence Analysis and Reverse Transcription–PCR and Northern Blot Analysis of the FCR3.varCSA Gene.
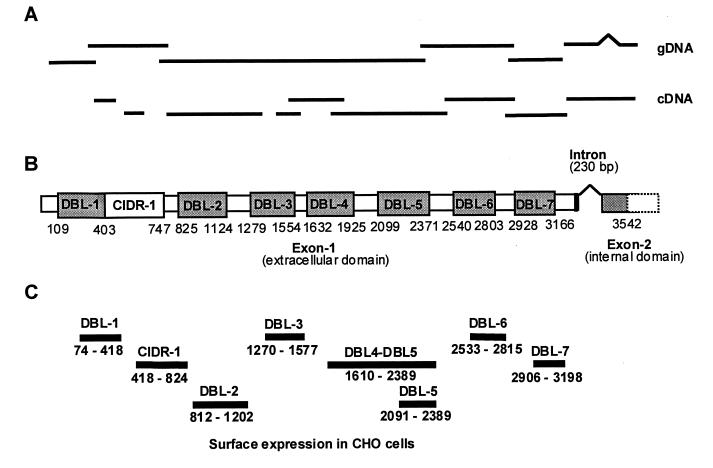
The Vectorette technique (Genosys, The Woodlands, TX) was used to extend a short sequence tag corresponding to the DBL-1 domain transcribed in CSA binding FCR3 parasites (7). We confirmed the linear order of the DNA sequence obtained from genomic FCR3-CSA DNA by using overlapping PCR fragments from cDNA of FCR3.varCSA trophozoites (Fig. 1A) and a YAC clone (gift of M. Lanzer, University of Heidelberg) spanning most of the exon I of the FCR3.varCSA gene (data not shown). DNA fragments from DBL1, DBL3/4, and DBL6/7 hybridized to the same 13-kilobase transcript (Fig. 2A; data not shown). Reverse transcription–PCR and Northern blot analyses were performed as described (7) from total parasite RNA prepared by using the TRIZOL (Life Technologies, Gaithersburg, MD) extraction method (19).
Figure 1.
Cloning and expression of the FCR3.varCSA gene. (A) Overlapping clones of the varCSA gene were isolated from genomic FCR3-CSA parasites and were sequenced. Regions amplified by reverse transcription–PCR from FCR3-CSA trophozoites mRNA confirm that the genomic varCSA gene sequence is contiguous with the exception of the intron region. (B) Schematic domain organization of the FCR3.varCSA gene. An unusually small intron of 230 bp separates exon I and exon II of the varCSA gene. The amino acid boundaries of the different DBL and CIDR-1 domains are indicated. (C) Domain regions that have been expressed on the surface of CHO-745 cells with their amino acid boundaries.
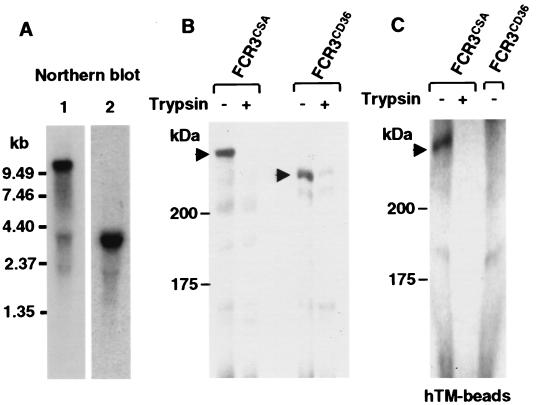
Figure 2.
(A) A large var transcript is detected in total RNA of FCR3-CSA trophozoites. Lanes: 1, FCR3.varCSA DBL-1 sequence probe; 2, Hsp70-specific probe hybridizing to the P. falciparum heat shock gene transcript of ≈3 kilobases. (B) Identification of a high molecular weight 125I-labeled and trypsin-sensitive erythrocyte surface molecule. Shown is the separation of SDS extracts of surface iodinated FCR3-CSA (first lane) and FCR3-CD36 (third lane) trophozoites. The labeled high molecular mass proteins of ≈400 and 250 kDa are sensitive to trypsinization (second lane and fourth lane). (C) CSA carrying human thrombomodulin (hTM)-coated Dynabeads affinity-purifies a high molecular weight molecule in SDS extracts from FCR3-CSA but not from FCR3-CD36 trophozoites. The purified molecule is sensitive to trypsin treatment. 125I-PfEMP-1 molecules are indicated by arrows.
Surface Iodination, Trypsinization, and Affinity Purification of the CSA Ligand.
Mature intact PRBC were selected by the receptor panning procedure, were grown for 1–2 cycles, and were enriched to >75% by the plasmagel technique (20). Surface iodination, sequential extraction with 1% Triton X-100 followed by 2% SDS, and trypsinization [l-1-tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Sigma)] of PRBC were performed as described (21). Samples derived from iodination were separated on a 0.5% agarose/4% acrylamide composite gel (22), were dried, and were exposed to Kodak X-Omat XAR-5 film. Prestained protein markers were purchased from Life Technologies and New England BioLabs. For affinity purification of the CSA ligand, 30 μg of recombinant human thrombomodulin was incubated with 1 × 108 tosyl-activated M-450 Dynabeads (4.5 μm diameter) (Dynal, Oslo, Norway) in 1 ml of 0.1 M phosphate buffer (pH 7.4) according to the protocol provided by the manufacturer. Beads were incubated overnight at 4°C with 15 μl of iodinated SDS extracts diluted in 500 μl of BM (pH 6.8) containing 1% BSA. Beads were washed by using a magnet (Dynal MPC) and were processed as described (21).
CHO Surface Expression of FCR3.varCSA Domains.
PCR products spanning each single domain were cloned into the expression vector pSRα5 (19). Stable transfectants that express FCR3.varCSA regions on the surface of CHO-745 cells were selected on a fluorescent-activated cell sorter, were cloned, and were used for receptor binding studies as described (19).
Conjugation of Biotin to Chondroitin Sulfate.
Conjugation of biotin to chondroitin sulfate A (bovine trachea, Sigma) and chondroitin sulfate C (shark cartilage, Sigma) was performed by modification of a previous procedure (23). In brief, biotinyl-(aminocaproyl)3-hydraside was synthesized by Fmoc-based solid phase peptide synthesis. A portion (0.71 mmol) of P-alokoxybenzylalcohol-Wang resin (0.71 mmol/g 100–200 mesh, Watanabe Chemical Co., Osaka) was treated with p-nitrophenylchloroformate (3.55 mmol, 5 eq) and pyridine (7.1 mmol, 10 eq) in CHCl3 overnight at room temperature. The resin was washed with CHCl3 (six times) and with dimethylformamide (DMF) (six times). The resin in DMF was treated with NH2NH2 hydrate (7.1 mmol, 10 eq) by shaking for 3 hr at room temperature and was washed with DMF six times. The resulting hydrasinated resin in DMF was acylated with Fmoc-aminocaproic acid (3 eq) 1-hydroxybenzotriazole (3 eq), and diisopropylcarbodiimide (3 eq) for coupling. Twenty percent piperidine-DMF was used for deprotection, and the reaction was repeated twice more with aminocaproic acid, followed by (+)-Biotin (Wako Pure Chemical, Osaka). The protected peptide resin was treated with TFA-m-cresol-ethanedithiol (9:0.5:0.5 cocktail) for deprotection and cleavage, was purified by HPLC in 0.1% MeCNaq, and was characterized by mass-spectrometry. Conjugation between the biotinyl-(aminocaproyl)3-hydraside and chondroitin sulfates was carried out by reductive hydrazination of chondroitin sulfate via terminal aldhyde with NaBH3(CN) in 1 M AcOH at room temperature. This procedure gave a stable covalent conjugation. The conjugates were purified by gel filtration on Sephadex G-15 in 0.1 M AcOH and were analyzed by HPLC, and the incorporation ratio was determined by combustive amino acid analysis as follows. Biotinyl-(aminocaproyl)3-NHNH-chondroitin sulfate was hydrolyzed with 6 M HCL containing 0.2% phenol at 110°C for 24 hr, and an aliquot of the resulting hydrolysates was subjected to amino acid analyzer [Hitachi (Tokyo) 835 A] equipped with Chromato-Integrator (Hitachi D-2500). An unknown peak corresponding to the same retention time of aminocaproic acid was observed with CSA. Thus, the molar ratio of conjugation was calculated from the difference between the biotin-CSA to CSA alone. The incorporation was only 15–25%.
Binding Assay of Chondroitin Sulfate Linked to Biotin.
Sheep anti-mouse IgG M-450 Dynabeads (2 × 106) were incubated overnight at 5°C with 2 μg of mouse anti-biotin mAb (Jackson Immunoresearch) in PBS with continuous agitation. The beads were washed three times with BM (pH7.2) and 1% BSA (BMB) and were resuspended with 45 μl of BMB to 4 × 107 beads/ml. Transfected CHO-745 cells (100,000) were grown for 48 hr on four glass coverslips in a six-well plate. Coverslips were transferred into a 12-well plate containing 1 ml of BMB and 50 μg of Biot-CSA or Biot-CSC (Sigma) and were incubated 1 hr. For inhibition assays, the cells were incubated for 1 hr with 200 μg/ml of CSA or CSC (Sigma) before addition of the biotin-conjugated carbohydrates. The coverslips were washed three times in a basin containing BMB, were transferred to a humidified chamber, and were incubated for 1 hr at room temperature with the coated Dynal beads (45 μl of 4 × 107 beads/ml). The coverslips then were flipped cell-side down onto a stand and were incubated for 3 min to allow unbound beads to settle by gravity. Coverslips then were washed three times with BM and were fixed with 2% paraformaldehyde (Polysciences) in PBS, and the degree of bead association with cells was examined. In some experiments, chondrotinase ABC (Fluka) at 1 unit/ml was added to the cells for 1 hr at room temperature before addition of the beads.
Results
Adhesive Phenotype of Parasites Selected for CSA Binding.
The binding characteristics of the CSA-selected PRBCs (Table 1) resemble the adhesive phenotype observed in PRBCs isolated from placentas of malaria-infected women, i.e., binding to CSA but not to CD36 (2). These adhesion properties are very different from those of CD36-selected PRBCs, which bound to several receptors but not to CSA (Table 1). Furthermore, sera from multigravid women from Cameroon and Senegal efficiently block adhesion of FCR3-CSA PRBCs to CSA (J.G. and A.S., unpublished data).
Table 1.
Binding characteristics of FCR3-CSA and FCR3-CD36 parasites to various host receptors
| Adhesion receptor | FCR3-CSA | FCR3-CD36 |
|---|---|---|
| hTMCSA* | 8910 ± 352 | 34 ± 24 |
| CSA | 3545 ± 278 | 68 ± 26 |
| Biot-CSA | 2866 ± 156 | 22 ± 15 |
| Biot-CSC | 32 ± 12 | Not determined |
| Placenta† | 850 ± 230 | 58 ± 46 |
| CHOCSA‡ | 3450 ± 234 | 23 ± 34 |
| CHOCD36 | 45 ± 32 | 2035 ± 143 |
| CHOICAM-1 | 24 ± 21 | 679 ± 64 |
| CHOVCAM-1 | 46 ± 56 | 456 ± 69 |
| CHOE-selectin | 82 ± 34 | 235 ± 36 |
| Human thrombospondin§ | 45 ± 34 | 78 ± 53 |
| HUVECPECAM-1, ICAM-1, VCAM¶ | 124 ± 67 | 1879 ± 98 |
Recombinant hTM carrying chondroitin sulfate A (hTM) produced in CHO cells (D.P. and T.F., unpublished work). Binding of PRBC to receptors bound to plastic is expressed as number of PRBC/mm2 ± SD of two independent experiments.
Adhesion of PRBC to serial 7-μm cryosections of snap-frozen placenta tissue was performed as described elsewhere (18). Only PRBC adhesion on syncytiotrophoblasts and syncytial bridges were counted and expressed as the mean number of PRBC ± SE per 20 high-power microscopic fields (1,000x Leitz Diaplan microscope).
CHO-745 cells (CSA negative) stably transfected with the human adhesion receptors CD36, ICAM-1, VCAM-1, and E-selectin are described in this work. Cytoadherence on confluent cells is expressed as number of PRBC/mm2 ± SD of two independent experiments.
Cytoadhesion of PRBC was performed on purified human thrombospondin at 50 μg/ml in 20 mM Tris⋅HCl (pH 7.2), 150 mM NaCl, and 2 mM CaCl spotted onto Petri dishes. Shown is the number of PRBC/mm2 ± SD of two independent experiments.
Nonactivated human umbilical vein endothelial cells express PCAM-1, ICAM-1, and VCAM-1 as detected on by those cells immunofluorescence using mAb9G11, 11C81, and 4B2.
The FCR3.varCSA Gene Has Eight Receptor-Like Domains.
A specific sequence tag of the FCR3.varCSA gene corresponding to the DBL-1 of a previous publication (7) was used to extend the gene sequence in the 5′ and 3′ directions. We obtained 10,628 bp that contain the entire extracellular region encoded by exon I (9,931 bp) and 698 bp of exon II, the intracellular domain (Fig. 1). An unusually short intron of 230 bp separates the two exons. The DNA sequence predicts an ORF of 3,542 amino acids and an overall structure homologous to the published var sequences, with seven DBL domains and a CIDR1 domain found after DBL1 (Fig. 1B). FCR3.varCSA sequence probes corresponding to DBL-1, DBL3/4, and DBL6/7 hybridize to a large transcript of ≈13 kilobases in total RNA of FCR3-CSA trophozoites (Fig. 2A; data not shown). Reverse transcription–PCR of mRNA and PCR of genomic DNA proved that the sequence was contiguous (Fig. 1A).
FCR3.varCSA Codes for a Large Trypsin-Sensitive Erythrocyte Surface Molecule That Binds to CSA.
Surface iodination of FCR3-CSA trophozoite-infected RBCs identified a single molecule of ≈400 kDa (Fig. 2B) with the characteristics of the var gene product PfEMP-1 (21). First, it was degraded by trypsin treatment of intact PRBCs (Fig. 2B). Second, it was efficiently extracted only in denaturing detergent (2% SDS). Third, it was variant [absent in CD36-selected PRBCs (FCR3-CD36), which instead express an iodinated molecule of 250 kDa] (Fig. 2B). The mild trypsinization removed the iodinated part of the molecule but did not ablat binding to CSA. Adhesion was interrupted only at very high concentrations of trypsin (data not shown). This is in agreement with the trypsin-resistant phenotype of PfEMP1 from other CSA-adherent PRBCs (12).
Surface-iodinated PfEMP-1 extracted from FCR3-CSA PRBCs before trypsinization bound to human thrombomodulin-coated Dynabeads (Fig. 2C) whereas the FCR3-CD36 PfEMP-1 did not bind thrombomodulin (Fig. 2C). Thus, the CSA-containing thrombomodulin that bound FCR3-CSA PRBCs (Table 1) affinity-purified a red cell surface molecule with the characteristics of PfEMP-1.
DBL-3 of the FCR3.varCSA Gene Binds Specifically to CSA.
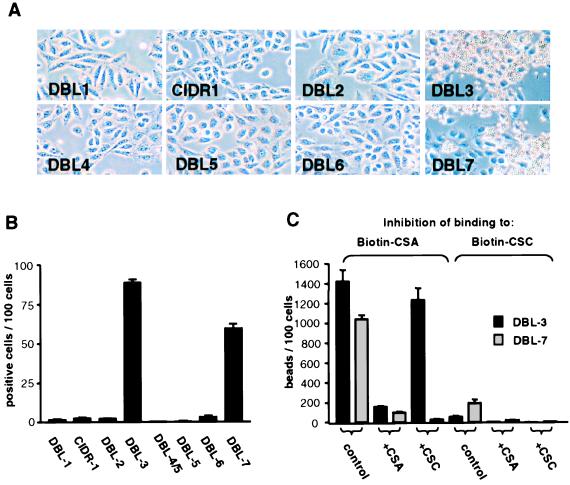
Biot-CSA and Biot-CSC were developed as specific reagents to measure CSA binding to the FCR3.varCSA domains. The activity of the biotinylated compounds was identical to that of the nonbiotinylated material (Table 1), with PRBCs binding only to Biot-CSA. Of the eight receptor-like domains expressed on the surface of CHO-745 cells (mutants not expressing CSA) (Fig. 1C), only DBL3 and DBL7 transfectants bound Biot-CSA (Fig. 3 A and B). No binding was observed if cells were incubated with biotin alone or if the cells were treated with chondroitinase ABC after incubation with Biot-CSA (data not shown). Addition of CSA blocked the binding of Biot-CSA to both DBLs; however, addition of CSC, a molecule that does not block CSA-mediated PRBC adhesion (12, 24), had no effect on binding to DBL3 but blocked binding to DBL7 (Fig. 3C). In addition, only DBL7 expressed on CHO cells bound Biot-CSC, and this binding was inhibited by addition of CSA or CSC (Fig. 3C). Thus, the binding properties of DBL3, but not of DBL7, are compatible with the adhesion properties of CSA-adherent PRBCs (Table 1).
Figure 3.
Binding of Biot-CSA to transfected CHO-745 cells. (A) Binding of anti-biotin-coated Dynabeads to CHO-745 cells expressing domains of FCR3varCSA incubated with CSA-biotin. (B) Percentage of transfected cells that bound four or more beads. (C) Inhibition of binding of CSA and CSC to DBL-3 and DBL-7 transfectant. Transfected cells were incubated with Biot-CSA or Biot-CSC without (control) or after preincubation with 200 μg/ml CSA (+CSA) or CSC (+CSC). Binding is given as number of positive cells (B) or number of beads (C) per 100 cells. Error bars represent the standard deviation from three different experiments.
Discussion
A previous study demonstrated that antibodies to two domains of a var gene (varCS2) from CSA PRBCs reduced binding to CSA (11); however, the identity of the parasite CSA-binding ligand molecule remained ambiguous. We determined by direct binding studies that the var gene domain DBL-3 binds CSA. Specific studies of domain binding are necessary to determine whether the DBL-3 domain of varCS2 mediates adhesion to CSA and whether other domains, such as CIDR1, are involved with this interaction in varCS2. The two DBL3 sequences (ref. 11 and present study) show no specific homology other than the homology found among all DBL3 domains, including those from PRBCs that do not bind CSA (19). The same is true for the CIDR1 domain. The CIDR1 domain of the FCR3-CSA var did not bind CD36 (data not shown), which is in full agreement with the failure of the PRBCs to bind CD36. This is distinct from other CIDR1 domains from CD36-binding PRBCs that bind CD36 (8, 19). Thus, we cannot predict the adhesion properties of a particular var domain from its primary sequence.
Identification of the domain that binds CSA allows the extension of the Fried and Duffy model of maternal malaria to the molecular level (2, 3). At the age of first pregnancy, most residents of endemic areas are clinically immune and develop a repertoire of anti-PfEMP-1 antibodies against endothelial adherent PRBCs (CD36-binding PRBCs) but not against the CSA-binding placental adherent PRBCs. Primigravid women who do not yet display antibodies against the CSA binding ligand offer a new niche for sequestration and proliferation of those parasites. Our data support the view that antigenic variation of PfEMP-1, in addition to its role in immune evasion, can lead to drastic changes in parasite tropism. A switch to a PfEMP-1 that mediates CSA adhesion is probably the key molecular event involved in the disease process observed during the first pregnancy. The data published by Fried and Duffy (2, 3) and from J.G. and A.S. (unpublished data) demonstrated that antibodies from multigravid women block binding of PRBCs from placenta to CSA. This blockade of adhesion was not specific for a particular clone, as sera from multigravid females blocked PRBCs not only from Africa but also from other parts of the world. Although CSA-binding PfEMP-1s are variant in primary sequence (ref. 11 and present study), the adhesion inhibition results suggest a conserved structure or conserved antigenic determinants among various CSA-adherent strains. It is conceivable that the blocking antibodies are directed at the CSA-adhesion domain, DBL3; however, it is also possible that a crossreactive immune response to the more N-terminal regions of the varCSA molecule, such as the CIDR1 domain, might provide the adhesion blocking activity. In this case, steric hindrance or conformational change promoted by antibody binding will result in reduced binding to CSA. If these antibodies are reactive with conserved determinants of the binding domain, then immunization with recombinant DBL3 may induce antibodies that will protect pregnant women from this complication. Thus, the DBL3 domain becomes a candidate as a vaccine for pregnant women in Africa. Today, protection of pregnant women depends on prophylaxis with drugs. With the increasing drug resistance in Africa and other countries, vaccines would be a welcome alternative for prevention of disease in pregnancy.
Acknowledgments
We thank Dr. Hiroto Nishioka (University of Kyoto Japan) for technical assistance and Affymax Research Institute for providing the expression vector and mAb 179. This work has been supported by a grant from the Commission of the European Communities for research and technical development (Contracts CT96-0071 and CT98-0362), GDR, and a French Army grant (Contract DSP/STTC-97/070). P.B. was supported by Fondation Mérieux and Fond d’Etudes de l’Assistance Publique/Hopitaux de Paris.
Abbreviations
- CSA
chondroitin sulfate A
- CSC
chondroitin sulfate C
- Biot-CSA
biotinylated CSA
- Biot-CSC
biotinylated CSC
- CIDR
cysteine-rich interdomain region
- DBL
Duffy binding-like
- PfEMP-1
Plasmodium falciparum erythrocyte membrane protein 1
- hTM
human thrombomodulin
- ICAM-1
intercellular adhesion molecule 1
- VCAM-1
vascular cell adhesion molecule 1
- PRBCs
parasitized red blood cells
- CHO
Chinese hamster ovary
- BM
binding medium
- DMF
dimethylformamide
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ133811).
References
- 1.Steketee R W, Wirima J J, Slutsker L, Heymann D L, Breman J G. Am J Trop Med Hyg. 1996;55:2–7. doi: 10.4269/ajtmh.1996.55.2. [DOI] [PubMed] [Google Scholar]
- 2.Fried M, Duffy P. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 3.Fried M, Nosten F, Brockman A, Brabin B J, Duffy P E. Nature (London) 1998;395:851–852. doi: 10.1038/27570. [DOI] [PubMed] [Google Scholar]
- 4.Baruch D I, Pasloske B L, Singh H B, Bi X, Ma X C, Feldman M, Taraschi T F, Howard R J. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 5.Smith J D, Chitnis C E, Craig A G, Roberts D J, Hudson-Taylor D E, Peterson D S, Pinches R, Newbold C I, Miller L H. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su X Z, Heatwole V M, Wertheimer S P, Guinet F, Herrfeldt J A, Peterson D S, Ravetch J A, Wellems T E. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 7.Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baruch D I, Ma X C, Singh H B, Bi X, Pasloske B L, Howard R J. Blood. 1997;90:3766–3775. [PubMed] [Google Scholar]
- 9.Rowe J A, Moulds J M, Newbold C I, Miller L H. Nature (London) 1997;388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- 10.Chen Q, Barragan A, Fernandez V, Sundstrom A, Schlichtherle M, Sahlen A, Carlson J, Datta S, Wahlgren M. J Exp Med. 1998;187:15–23. doi: 10.1084/jem.187.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeder J C, Cowman A F, Davern K M, Beeson J G, Thompson J K, Rogerson S J, Brown G V. Proc Natl Acad Sci USA. 1999;96:5198–5202. doi: 10.1073/pnas.96.9.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogerson S J, Chaiyaroj S C, Ng K, Reeder J C, Brown G V. J Exp Med. 1995;182:15–20. doi: 10.1084/jem.182.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berendt A R, Simmons D L, Tansey J, Newbold C I, Marsh K. Nature (London) 1989;341:57–59. doi: 10.1038/341057a0. [DOI] [PubMed] [Google Scholar]
- 14.Simmons D, Makgoba M W, Seed B. Nature (London) 1988;331:624–627. doi: 10.1038/331624a0. [DOI] [PubMed] [Google Scholar]
- 15.Osborn L, Hession C, Tizard R, Vassallo C, Luhowskyj S, Chi-Rosso G, Lobb R. Cell. 1989;59:1203–1211. doi: 10.1016/0092-8674(89)90775-7. [DOI] [PubMed] [Google Scholar]
- 16.Bevilacqua M P, Stengelin S, Gimbrone M A, Jr, Seed B. Science. 1989;243:1160–1165. doi: 10.1126/science.2466335. [DOI] [PubMed] [Google Scholar]
- 17.Vicart P, Testut P, Schwartz B, Llorens-Cortes C, Perdomo J J, Paulin D. J Cell Physiol. 1993;157:41–51. doi: 10.1002/jcp.1041570106. [DOI] [PubMed] [Google Scholar]
- 18.Gysin J, Pouvelle B, Le Tonqueze M, Edelman L, Boffa M C. Mol Biochem Parasitol. 1997;88:267–271. doi: 10.1016/s0166-6851(97)00082-0. [DOI] [PubMed] [Google Scholar]
- 19.Smith J D, Kyes S, Craig A G, Fagan T, Hudson-Taylor D, Miller L H, Baruch D I, Newbold C I. Mol Biochem Parasitol. 1998;97:133–148. doi: 10.1016/s0166-6851(98)00145-5. [DOI] [PubMed] [Google Scholar]
- 20.Pasvol G, Wilson R J, Smalley M E, Brown J. Ann Trop Med Parasitol. 1978;72:87–88. doi: 10.1080/00034983.1978.11719283. [DOI] [PubMed] [Google Scholar]
- 21.Baruch D I, Gormely J A, Ma C, Howard R J, Pasloske B L. Proc Natl Acad Sci USA. 1996;93:3497–3502. doi: 10.1073/pnas.93.8.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiesner J, Mattei D, Scherf A, Lanzer M. Parasitol Today. 1998;14:38–40. doi: 10.1016/s0169-4758(97)01155-1. [DOI] [PubMed] [Google Scholar]
- 23.Shinohara Y, Sota H, Kim F, Shimizu M, Gotoh M, Tosu M, Hasegawa Y. J Biochem (Tokyo) 1995;117:1076–1082. [PubMed] [Google Scholar]
- 24.Robert C, Pouvelle B, Meyer P, Muanza K, Fujioka H, Aikawa M, Scherf A, Gysin J. Res Immunol. 1995;146:383–393. doi: 10.1016/0923-2494(96)81042-x. [DOI] [PubMed] [Google Scholar]