Abstract
We report a group of TRIMs (terminal-repeat retrotransposons in miniature), which are small nonautonomous retrotransposons. These elements, named Cassandra, universally carry conserved 5S RNA sequences and associated RNA polymerase (pol) III promoters and terminators in their long terminal repeats (LTRs). They were found in all vascular plants investigated. Uniquely for LTR retrotransposons, Cassandra produces noncapped, polyadenylated transcripts from the 5S pol III promoter. Capped, read-through transcripts containing Cassandra sequences can also be detected in RNA and in EST databases. The predicted Cassandra RNA 5S secondary structures resemble those for cellular 5S rRNA, with high information content specifically in the pol III promoter region. Genic integration sites are common for Cassandra, an unusual feature for abundant retrotransposons. The 5S in each LTR produces a tandem 5S arrangement with an inter-5S spacing resembling that of cellular 5S. The distribution of 5S genes is very variable in flowering plants and may be partially explained by Cassandra activity. Cassandra thus appears both to have adapted a ubiquitous cellular gene for ribosomal RNA for use as a promoter and to parasitize an as-yet-unidentified group of retrotransposons for the proteins needed in its lifecycle.
Keywords: pol III, genome evolution, transcription, transposable element
Retrotransposons, excepting SINEs (short interspersed nuclear elements) and LINEs (long interspersed nuclear elements), resemble retroviruses in their structure and intracellular life cycle. They are ubiquitous in the genomes of plants, animals, and fungi and account for >50% of large plant genomes (1, 2). Their life cycle comprises transcription of genomic copies, translation of their encoded proteins, packaging of the transcripts into virus-like particles, reverse transcription, and targeting of the cDNA copy to the nucleus for integration into the genome (3, 4). The transcriptional signals for RNA polymerase II (pol II) are found in the long terminal repeats (LTRs) at either end of the element, flanking the priming sites for reverse transcription and the coding domain specifying the proteins needed for replication and integration [supporting information (SI) Fig. S1].
In addition to the classical retrotransposons, several well conserved nonautonomous groups have been discovered that lack all or part of their coding capacity (5). The BARE2 elements cannot express the capsid protein GAG (6), and Morgane lacks most of its coding capacity (7). The TRIM (terminal repeat retrotransposon in miniature) and LARD (large retrotransposon derivative) elements (Fig. S1) entirely lack reading frames for retrotransposon proteins (8–12). The TRIM elements are composed of 100- to 250-bp LTRs, priming sites for reverse transcriptase, and a small intervening segment. Evidence for past mobility suggests that they are activated by transcomplementation (10). These have been found in at least 13 species from four plant families (9, 10).
Here, we describe a group of TRIM elements, which we refer to as Cassandra, that carry 5S RNA sequences having well conserved RNA polymerase III promoters as part of their LTRs. 5S rRNAs are universal 120-nt components of ribosomes (13). We present the structure, distribution, transcription, and insertional polymorphism of Cassandra elements, as well as features of the 5S sequences they contain, and discuss their possible function.
Results
Isolation of Cassandra Elements.
To rapidly isolate uncharacterized retrotransposons, we exploited the general presence, in LTR-containing retrotransposons, of the primer binding site (PBS) for (−)-strand cDNA synthesis by reverse transcriptase (3, 14). The PBS is positioned just internal to the left LTR (Fig. 1A and Fig. S1). Generally, tRNA genes are not clustered sufficiently to produce a PCR product from tRNA amplification primers. However, retrotransposons in plants are frequently clustered or nested (15, 16). Hence, most of the PCR products amplified and isolated are derived from retrotransposons. The 3′ end of the LTR is adjacent to the PBS and can thus be identified for the design of LTR primers. Here, we amplified genomic sequences between PBS motifs using primers matching the methionyl-initiator tRNA, which is the most common retrotransposon PBS (3). The identified LTR termini were then used to design primers for inter-LTR amplification to clone entire retrotransposons.
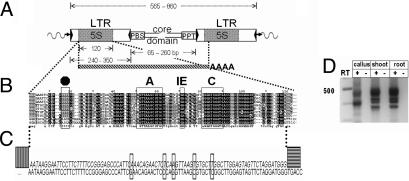
Fig. 1.
Cassandra structure and transcription. (A) Structure of a Cassandra element. Flanking genomic DNA is indicated as a wavy line with the target site duplications (TSDs) as arrowheads. The element components, including the reverse-transcriptase priming sites PBS and PPT, are shown as boxes. The terminal inverted repeats (TIRs) of the long terminal repeats (LTRs) are shown as black triangles, and the 5S domain is hatched. The size ranges in base pairs for the sequenced elements and segments therein are shown above and below the diagram. The predicted pol III-mediated transcript is shown below as a hatched bar with a poly(A) tail. (B) Alignment of cellular Cassandra 5S RNA from cereals. The A-, IE-, and C-Boxes of pol III promoters are marked, as is the predicted pol III terminator (black octagon). The sequences are, from top to bottom: Cassandra 5S of Triticum durum, Secale cereale, Hordeum vulgare, Avena sativa, Zea mays, Sorghum bicolor, and Oryza sativa and cellular 5S of T. aestivum and Z. mays. The complete alignment is shown in Fig. S2. (C) Alignment of Cassandra 5S RNA transcripts with the Cassandra genomic sequence. The sequenced product generated by RLM-RACE is shown in italics. The RACE adapter for the 5′ end of the transcript is shown as a vertically hatched block, and the nested 3′ primer as a horizontally hatched block. Mismatches are boxed. Because the 3′ primer is nested, the 3′ terminus of the 5S RNA transcript is not found in the sequence. (D) RT-PCR amplification. Lanes show products from three tissues, with controls lacking reverse transcriptase in the cDNA protocol. The smaller product (430 nt) represents either a deletion or strand-jumping resulting from secondary structure.
Overall Organization of Cassandra Elements.
We isolated Cassandra retrotransposons from 50 species across the plant kingdom, including ferns and both monocotyledonous and dicotyledonous angiosperms (Table S1). Cassandra elements are 565–860 bp, with LTRs varying in length by species, from 240 to 350 bp (Table S2 and SI Text). The LTRs of the sequenced Cassandra contain conserved termini with a universal 5′ TG… CA 3′ structure and terminal inverted repeats (TIRs), varying from 6 to 12 bp, typical of LTR retrotransposons. The canonical TIR pair for Cassandra is 5′-TGTrABA–GTkACA-3′, except for ferns, where 5′-TGTTGGG–AyyTACA-3′ is found. The internal domains comprise a highly conserved ≈18-nt PBS for reverse transcriptase, complementary to methionyl initiator tRNA, and an ≈13-nt (+)-strand priming site (PPT), separated by intervening sequences as short as 34 nt. This internal domain is considerably smaller than previously reported for other TRIMs (9, 10).
5S Sequences in Cassandra LTRs.
The most singular feature of Cassandra is the presence of 5S sequences 42–205 bp in from the LTR termini (Table S2), with a length mirroring the cellular 5S rRNA consensus of 120 nt. Cellular 5S rRNA genes are universally transcribed by pol III (13). The A-, IE-, and C-Boxes, which constitute the pol III internal promoter (13, 17), are highly conserved in Cassandra between nucleotides 40 and 120 of the 5S (Fig. 1B and Fig. S2). This segment is 78–91% identical to the 5S rRNA gene of its corresponding species (Table S3).
The beginning of the 5S region, nucleotides 1–40, diverges from the cellular 5S genes and is less conserved overall (Fig. 2 and Fig. S2). Phylogenetic analyses of Cassandra 5S sequences show that they form a clade distinct from cellular 5S. (Fig. 2). Both the TIRs and the PPT showed conservation consonant with the plant family from which they derived (Tables S1 and S2).
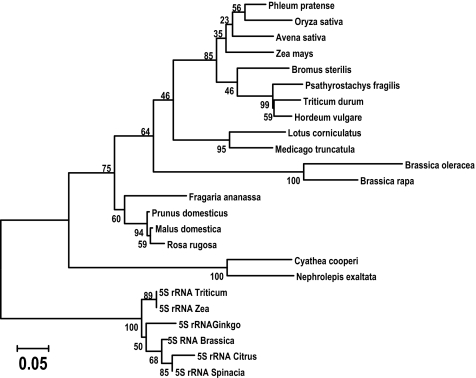
Fig. 2.
Phylogenetic relationships among selected Cassandra 5S domains and cellular 5S rRNA genes. A minimum evolution tree was produced from aligned 5S rRNAs and Cassandra 5S RNA regions. Bootstrap values from 500 tests are indicated at the nodes. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The neighbor-joining tree of the same data (data not shown) is topologically identical, except that Gingko 5S rRNA is basal to the other 5S sequences.
Cassandra 5S Domains Are Transcriptionally Functional.
The presence of a pol III promoter in the 5S region raised the possibility that Cassandra replicates via pol III transcription rather than by pol II, which is generally used by LTR retrotransposons. Pol II generates capped and polyadenylated transcripts, whereas pol III produces uncapped transcripts usually without poly(A) tails. Full-length cDNA libraries are prepared by selecting for the cap (18); BLAST searches of full-length rice cDNA thus prepared (http://red.dna.affrc.go.jp/cDNA/) found accessions containing complete Cassandra elements within longer cellular transcripts (data not shown). Matches in plant EST databases (Table S4) also indicate pol II-driven read-through transcription of Cassandra.
Several lines of evidence nevertheless indicate that Cassandra itself is transcribed by the pol III promoter in its 5S region. First, uncapped barley Cassandra transcripts, initiated specifically at the beginning of the 5S in the LTR, can be detected by PCR amplification using RNA adapters ligated to the RNA 5′ ends (Fig. 1C and SI Text). Second, 3′ ends of Cassandra transcripts that were amplified from polyadenylated barley leaf mRNA by nested 3′ RACE (19) terminated in the 3′ LTR just beyond a putative pol III termination signal (20), TTTT (Fig. 1B). The terminator is found in all Cassandra 5S but in no cellular 5S (Fig. S2). Cellular 5S terminators are located in the intergenic spacer just beyond the 5S (21).
Polyadenylated, read-through transcripts that contain Cassandra solo LTRs do not terminate at this signal (data not shown); it is apparently not recognized by pol II. The predicted size of the Cassandra transcript from the beginning of the 5S sequence in the 5′ LTR to the pol III terminator in the 3′ LTR is 480 nt. Consistent with this, isolated total RNA from barley callus, shoots, and roots, amplified with primers located in the Cassandra-specific first 40 nt of the 5S region, displays the LTR-to-LTR transcripts typical of retrotransposons (Fig. 1D).
Structural Prediction for Cassandra 5S RNA.
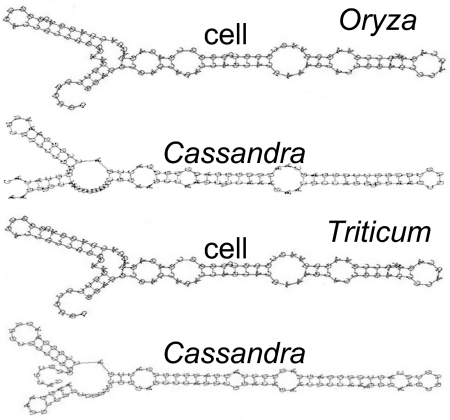
We modeled the folding of the predicted Cassandra 5S and compared these with modeled cellular 5S rRNAs. As shown (23, 24), not all cellular 5S rRNAs fold into the canonical structure derived from x-ray crystallography (13). The predicted Cassandra 5S RNA folds varied, but at least some resembled the canonical structure of cellular 5S rRNA (Fig. 3 and SI Text), whereas other Cassandra and cellular 5S formed noncanonical folds. All Cassandra 5S RNA folds display structural conservation and thermodynamic stability, unlike reversed sequences sharing the same degree of sequence conservation. Tests for neutrality (25, 26) rejected the null hypothesis, indicating that selection is acting to maintain the secondary structure of Cassandra 5S RNA. Analyses of the information content in the Cassandra 5S RNA fold compared with cellular 5S rRNAs (Fig. S3 and SI Text) were made. Information content is a measure of the nonrandomness or conservation of a sequence or structure at a particular alignment position (27, 28). These show peaks in information content for both Cassandra and cellular 5S RNA folds between positions 62 and 114, overlapping the pol III promoter.
Fig. 3.
Structural predictions for Cassandra 5S RNA compared with cellular 5S rRNA.
Cassandra Retrotransposons Are Abundant and Insertionally Polymorphic.
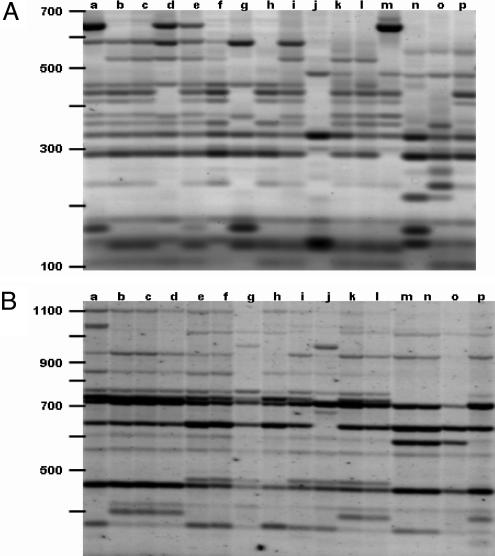
In addition to transcription, evidence for competence in retrotransposition includes conservation of replication and packaging signals as well as integration of replicated copies. Polymorphisms in retrotransposon genomic distribution, visualized by transposon display methods, serve as evidence for integration. Furthermore, because of the role of replication in transposition, the prevalence of a particular group of retrotransposons is evidence for past propagation. Application of the IRAP and REMAP methods (29, 30) with Cassandra primers indicates that these elements are polymorphic in their integration sites in barley germplasm accessions (Fig. 4A). We have applied these methods as well to various members of the Rosaceae (12) including apple (Fig. 4B) and to bread wheat (T. aestivum), timothy (Phleum pretense), cultivars of turnip rape (Brassica rapa), and canola (B. napus) (data not shown) and observed levels of polymorphism that are generally higher than those obtained with families of protein-coding, autonomous retrotransposons.
Fig. 4.
Insertional polymorphism of Cassandra elements by transposon display. (A) Polymorphism of Cassandra insertion sites by IRAP for barley. The template DNA was from cultivars (left to right): a, Tammi; b, Hankija 673; c, Otra; d, Vega; e, Edda; f, Paavo; g, OA.C.21; h, Gull; i, Pomo; j, Djau Kabutak; k, Marinka; l, Borwinia; m, Gaulois; n, Rondo; o, Krona; p, Union. (B) REMAP for Cassandra elements in apple. Cultivars, including their sports, from left to right: a, Antonovka; b, Melba; c, Melba, red Plats; d, Melba, red Pate; e, Bergius; f, Sävstaholm; g, White Astrakan; h, Red Astrakan; i, Gyllenkroks Astrakan; j, Big transparent Astrakan; k, Åkerö, Tarko; l, Åkerö, Rajalin; m, Yellow Cinnamon apple; n, Red Cinnamon apple; o, Brown Cinnamon apple; p, Transparente Blanche. The positions of 100-kb size markers are shown at the left.
Integrase, encoded by retrotransposons, creates target site duplications (TSDs) as it inserts new elements (31). Hence, detection of TSDs flanking genomic copies provides evidence for retrotransposition. Public genomic sequences containing Cassandra elements from a variety of species display 5-bp TSDs, many of which have not yet accumulated mismatches due to mutation after insertion (Table S5). Taken together, the data suggest that Cassandra is, or recently has been, transpositionally active.
Plant cellular 5S RNAs are found in large clusters (32). In barley, we have estimated the number of Cassandras and their associated 5S RNA domains by slot blot, in four varieties (winter barley varieties Tu Dam Mai 1, China; Han 85–222, China; Casbon, USA; Tennessee Winter, USA; data not shown). Using a probe that includes most of the Cassandra element except the 5S domains, and hence does not detect cellular 5S, we found 6,697 ± 588 copies. Searches of the full-length rice genome found 352 elements with alignments ≈100 nt in length, 84 complete elements, and 268 solo LTRs, corresponding to 436 Cassandra 5S RNA sequences. (Table S6 and SI Text). A similar number of cellular 5S genes, 384, have been identified in rice, although the latter may be an underestimate (33). We estimate Cassandra to number in the thousands in the ferns (data not shown). The primer annealing sites and BamHI restriction site used to systematically define, amplify, and clone 5S rRNA genes in barley (32) are not found in the Cassandra 5S RNA domains; hence, these were not previously recorded as 5S rRNA gene variants.
Analyses of the rice genome sequence revealed that 15% of Cassandra LTRs and 21% of complete elements are inserted into genes, although only 1% of the total is in coding sequences (Table S6). By comparison, retrotransposon Tos17, distinctive in its preference for genic insertions, displays a similar distribution in the rice genome but approximately half the genic insertions are into exons (34). Unlike Cassandra, Tos17 is generally silent and rare, being found in one to five copies (34). The EST data (above) are consistent with many Cassandra elements being inserted in transcribed genes.
Discussion
Cassandra retrotransposons have two salient features. First, as TRIMs, they are nonautonomous and must rely on the proteins of autonomous retrotransposons for replication (5). The autonomous partner(s) of Cassandra remains to be identified. Nevertheless, they are a fairly abundant family conserved in structure and sequence. The occurrence of Cassandra in the ferns, tree ferns, and all angiosperms investigated places their origin at least in the Permian, 250 MYA (35). Their widespread distribution supports evolutionary radiation rather than horizontal transfer.
The second notable feature is the presence of 5S domains with conserved RNA polymerase III promoters in the LTRs of all cloned Cassandra elements. This distinguishes them from all previously described Class I retrotransposons (3). In addition to read-through transcripts containing Cassandra elements, Cassandra specifically produces the LTR-to-LTR transcripts typical of retroelements at least in barley. Transcripts initiate from the internal RNA polymerase III promoter found in the 5S RNA domain of the 5′ LTR and terminate in the 3′ LTR at a canonical pol III terminator that is universal in Cassandra but absent from within cellular 5S genes. An R region, needed for LTR retrotransposon reverse transcription, would thus be formed from the 5′ end of the 5S region and comprises a relatively short 18 nt.
Polyadenylation of pol III transcripts is rare except in quality-control surveillance (36). However, many Cassandra 5S, but not cellular 5S genes, possess a putative polyadenylation signal, CAA(T/C)AA, located 17 nt before the pol III terminator at the beginning of the 5S domain (Fig. S2). Although the signal differs from the canonical AATAAA, it resembles other noncanonical signals and its distance from the terminator is quite typical (22). Hence, Cassandra polyadenylation more likely represents RNA maturation than turnover. Furthermore, polyadenylated cellular 5S has recently been reported (21).
The presence of pol III promoters nested within pol II read-through transcripts is not unique to Cassandra. A well known example is the Alu SINE elements of the human genome. Both independent copies transcribed by pol III and nested copies transcribed by pol II contribute to the RNA pool and have roles in gene regulation (37). Another SINE, B2 of mouse, carries both a pol III and a pol II promoter, which function independently (38).
We speculate that Cassandra may have originated from the retroposition of a SINE element derived from 5S rRNA (39, 40) into an LTR, which was then copied into the other LTR by standard retrotransposon reverse transcription. In phylogenetic trees (Fig. 2), Cassandra 5S sequences are completely separated from cellular 5S at 100% bootstrap values, suggesting a single origin for Cassandra rather than multiple independent acquisitions of the 5S domain.
The maintenance of the 5S RNA domain begs a functional explanation. It may aid Cassandra replication. Secondary structural models of the Cassandra 5S region show conservation of a single nucleotide bulge associated with transcription factor IIIA (TF IIIA) binding (13); the ability of TF IIIA to bind both RNA and DNA and the role of TF IIIA in mediating 5S nuclear transport may offer selective advantages to Cassandra. Alternatively, the ability of the 5S pol III promoter to evade silencing by methylation alone (41, 42) may be important in Cassandra propagation. Information-content analyses suggest that the structure for the pol III promoter is functional and under selection. The role of the 5S domain and its promoter in the Cassandra life cycle remains to be elucidated.
In the plants (32, 43–47) and fungi (40), evidence has accumulated both for the lack of concerted evolution (48) and for variability and rapid rearrangements in 5S rRNA loci. An uncharacterized transpositional process even has been suggested to explain these phenomena (40, 43, 47). We believe that at least part of the apparent 5S gene dynamism may result from the activity of Cassandra retrotransposons. Strikingly, the presence of a 5S RNA region in each LTR interspersed with the LTR termini and internal domain of the Cassandra is reminiscent of the arrangement of cellular 5S genes in plants (32). In plants, the nontranscribed spacers (NTS) of cellular 5S genes vary between 100 and 700 nt, the barley NTS varying from 171 to 388 bp (32). In barley, for example, the two 5S RNA regions of a Cassandra are separated by 340 bp within an element of 724 bp, similar in length (but not sequence) to the NTS spacing of “long class” 5S rRNA genes.
In conclusion, Cassandra is thus a striking example of adaptation by transposable elements of cellular genes. The reciprocal phenomenon, recruitment of transposable elements by cellular genes, is well known. The L1 LINE element provides promoters for human genes (49) and contributes to gene remodeling by exon shuffling (50, 51). Among Class II transposons, Pack-MULEs (52) and Helitrons (53, 54) can move cellular genes or fragments and likewise contribute to both genic and genome remodeling. In addition, the RAG1 and RAG2 proteins essential for V(D)J recombination in the immune system originate from transposase (55, 56). In addition to Cassandra, one finds very few examples of the recruitment of a cellular component by a transposable element; at least chromodomains appear to have been borrowed early in evolution by a clade of retrotransposon integrases (57). Cassandra, in contrast, appears both to have coopted a ubiquitous ribosomal RNA that continues to be transcribed as its component and to parasitize another group of retrotransposons for the proteins needed in its lifecycle.
Materials and Methods
Plant DNA Preparation.
DNA was prepared as described in ref. 58.
Isolation of Cassandra Elements.
The Cassandra elements were first isolated with PCR primers corresponding to the (−)-strand priming site (PBS). Later, additional Cassandra elements were specifically isolated by PCR using nested primers that match the pol III promoter region. For PBS–PBS amplification, the primers matched initiator-methionyl tRNA: 5′-ACTTGGATGCTGATACCA-3′. Amplifications were carried out in 20-μl reaction volumes containing: 1× buffer [75 mM Tris·HCl (pH 8.8), 20 mM (NH4)2SO4, 2 mM MgCl2, 0.01% Tween-20], 20–100 ng of DNA, 600 nM primer, 200 μM dNTP, 1 unit of TaqDNA polymerase and 0.04 units of Pfu DNA polymerase. PCR was performed with an initial denaturation at 95°C for 3 min, followed by 32 cycles of 95°C for 15 sec, 55°C for 60 sec, 72°C for 90 sec, and a final elongation at 72°C for 5 min. The PCR products were cloned and sequenced. To screen for Cassandra sequences, PCRs were carried out on these cloned PCR products by using two primers, one matching the vector, the other complementary to the A-Box of the pol III promoter belonging to the 5S RNA sequence expected in the Cassandra LTRs (primer 1,033, 5′-CATCGGAACTCCGAAGTTAAGCGAG-3′). Clones containing Cassandra segments yield amplification products between 220 and 300 bp.
Alternatively, once Cassandra was identified, we carried out amplification between a PBS (primer 5′-TAGGTCGGAACAGGCTCTGATACCA-3′) and the 5S RNA region of the adjacent LTR (using either of several primers: 621, 5′-CTGGAGCAATTTTAGGATGGGTGACC-3′; 623 5′-TGATGGGTGACCTCCTGGGAAG-3′; 625, 5′-ACTCCATGGTTAAGTGTGCTTG-3′). Amplification conditions were as above, except 200 nM primers were used and reactions consisted of an initial denaturation at 94°C for 4 min; 30 cycles of 94°C for 40 sec, 55°C for 40 sec, 68°C for 10 sec, and a final elongation at 68°C for 10 min. Products were cloned and sequenced, and the sequences corresponding to the 3′ ends (with respect to transcriptional direction) of LTRs lying between the 5S domain and the PBS used for the design of adjacent, outward-facing PCR primers. These amplified the region between the 3′ end of the 5′ LTR and the 3′ end of the 3′ LTR.
LTR–LTR Amplification.
To amplify entire Cassandra elements the 3′ termini of the Cassandra 5′ LTRs were identified, from the products described immediately above, by the final 5′ CA 3′ motif and its position several base pairs from the end of the PBS primer. These were used to design primers at the LTR termini facing toward each other. Both full-length and LTR products are amplified. For some plant families, the LTRs were sufficiently conserved that specific, overlapping, inverted primers could be used across the family. These were: Poaceae, primers 977 5′-TTGTCCTCACTCATGCGCACC-3′ and 784 5′-CGAGTGAGGACAAAGTGCGCAG-3′; Rosaceae, primers 1,129 5′-AGGATGTGACGATTTGGTATCAGAGC-3′ and 1,130 5′-GGGCTTCACTACATCCTGGGATCG-3′; Pteridophyta (ferns), primers 1,119 5′-TGGATGGCTAGACCAGTTTATGCAAC-3′ and 1,120 5′-TAAGGTGTTAGGAACCTCCGGTCTAGC-3′. Amplifications were carried out as above, with 20–100 ng of template DNA and 200 nM concentrations of each primer with PCR programs of: 95°C for 3 min; 20 to 27 times a cycle of 95°C for 15 sec, 55°C for 40 sec, 72°C for 20 sec, and a final elongation step at 72°C for 10 min.
Cloning of RNA Polymerase III Transcripts by RT-PCR.
The 5′ ends of transcripts were amplified by ligation of an RNA adapter, followed by RT-PCR (59). The method was carried out with the aid of a kit (FirstChoice RLM-RACE, product 1700; Ambion). To determine whether the transcripts were uncapped, amplifications were preceded by dephosphorylation, which blocks RNA ligation to an uncapped RNA. The details are described in SI Text.
To determine the sequence of the 3′ ends, mRNA was extracted from barley leaves and DNase-treated (Ambion kit AM1906). The first-strand cDNA was synthesized with a tagged oligo(dT) primer (E1820; 5′-AAGC A G T G G T AACAACGCAGAGTACT30NA). Amplifications were carried out by nested PCR, using a forward primer matching the PBS (5′-TGGTATCAGAGCCGACCCTC-3′) and a reverse primer (E2146) matching the tag of E1820. The program used denaturation at 94°C for 30 sec, annealing at 56°C for 30sec, amplification at 72°C for 1 min, and 34 cycles of repetition. The second PCR was carried out on 0.2 μl of the first PCR product as template, with a forward primer matching the beginning of the LTR (E1160; 5′-CCTGGCTTATTAGGGATGATAGACTAC-3′), E2146 as the reverse primer, annealing at 53°C, and 24 cycles. Products were cloned into the PGEMTe vector and sequenced.
Transposon Display Methods.
IRAP (interretrotransposon amplified polymorphism) and REMAP (retrotransposon-microsatellite amplified polymorphism) were carried out essentially as before (30), except that for barley IRAP, two nested primers were used: 978, 5′-GGTGTGTCCGGGGCGTTACA-3′; 979, 5′-CCGGGAGCCCATTCGAAC-3′. The REMAP reactions were carried out on apple DNA samples by using the protocol described above for IRAP. The Cassandra primer was 879, 5′-TGATCCACTCCCCTGGGCGATGTGG-3′, used together with a microsatellite primer anchored by 1 nt at its 3′ end, primer 439, 5′-AGAGAGAGAGAGAGAGAGC-3′.
Copy Number Estimation.
The Cassandra copy number was estimated by slot blot essentially as described (60). Blots were probed with a PCR fragment amplified from barley cv. Bomi with primers 975, 5′-AGTTCTGTTCGAATGGGCTCC-3′ and 784, 5′-CGAGTGAGGACAAAGTGCGCAG-3′. This generated a 388-bp fragment, which extends from the 5′ LTR beyond the 5S RNA promoter through the internal region to the 3′ LTR and terminates before the 5S RNA promoter of the 3′ LTR. Thus, the part of the Cassandra 5S most conserved with cellular 5S was not part of the probe, avoiding cross-hybridization.
Sequence Analysis, Searches, and Alignment.
Sequence analyses using the tools of EMBOSS and ClustalW were run in the BioBox of the CSC–Scientific Computing Ltd. (www.csc.fi). Alignments were also made with the MULTALIN (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page = npsa_multalin.html) and GeneDoc (www.nrbsc.org/gfx/genedoc/index.html) (61) tools. The cellular 5S sequences were retrieved from a dedicated database (http://rose.man.poznan.pl/5SData/). We aligned the Cassandra 5S domains first within plant families and then realigned each set with the aligned cellular 5S rRNA set. Finally, a global alignment was carried out. Based on the alignments, PCR primers were designed by FastPCR software (www.biocenter.helsinki.fi/bi/programs/fastpcr.htm). The BLAST searches for sequence similarity were made online at the National Center for Biotechnology Information web site (www.ncbi.nlm.nih.gov/blast/). Searches for Arabidopsis transcripts, however, were made on the BLAST server and At_transcripts database maintained at the TAIR site (www.arabidopsis.org/Blast/) and against the GenBank collection.
Searches for Cassandra copies were made within the available pseudomolecules for the rice genome from TIGR (www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml). The query strings were consensus sequences for the isolated Cassandra copies from rice. Cassandra (or the LTR and internal domain segments thereof) was queried against the corresponding genome by using either BLAT (62) or BLASTN (63, 64), each with default parameters. The entire Cassandra consensus and each of its parts were also searched against the various sections of the rice genome (CDS, intergenic, introns, UTR) by using BLAT and BLAST. The results were parsed, cutoffs were applied, and remaining hits were checked and counted.
Phylogenetic Analyses and Tree Building.
Evolutionary history was inferred by using the minimum evolution method (65). The bootstrap consensus tree inferred from 500 replicates (66) was taken to represent the evolutionary history of the sequences (66). The evolutionary distances were computed by using the maximum composite likelihood method (67); the units represent the number of base substitutions per site. The tree was searched by using the close-neighbor-interchange (CNI) algorithm (68) at a search level of 1. The neighbor-joining algorithm (69) was used to generate the initial tree. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (pairwise deletion option). There were a total of 141 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (70).
Modeling of Secondary Structure.
RNA fold prediction was carried out with the ViennaRNA package version 1.6 (www.tbi.univie.ac.at/∼ivo/RNA/) (72), at a folding temperature of 17°C. This was chosen to reflect ambient conditions for plants. Information content was determined as described (27). Further details for secondary structure modeling and information content determination can be found in the SI Text.
Supplementary Material
Acknowledgments.
The authors thank Ursula Lönnqvist and Anne-Mari Narvanto for excellent technical assistance, Jean-Marc Deragon for discussions on 5S, and Alexander Bolshoy for discussions on information content. This work was supported by Academy of Finland Grants 106949 and 207485.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF538603–AF538610, AF538605–AF538618, AY164585, AY271957–AY271963, AY359471, AY603364–AY603377, AY860307–AY860317, AY923749, DQ094839–DQ094843, DQ673669, DQ767972, DQ788719, and EF125870–EF125877).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709698105/DCSupplemental.
References
- 1.Vitte C, Panaud O. LTR retrotransposons and flowering plant genome size: Emergence of the increase/decrease model. Cytogenet Genome Res. 2005;110:91–107. doi: 10.1159/000084941. [DOI] [PubMed] [Google Scholar]
- 2.Liu R, et al. A GeneTrek analysis of the maize genome. Proc Natl Acad Sci USA. 2007;104:11844–11849. doi: 10.1073/pnas.0704258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 4.Kumar A, Bennetzen J. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 5.Sabot F, Schulman AH. Parasitism and the retrotransposon life cycle in plants: A hitchhiker's guide to the genome. Heredity. 2006;97:381–388. doi: 10.1038/sj.hdy.6800903. [DOI] [PubMed] [Google Scholar]
- 6.Tanskanen JA, Sabot F, Vicient C, Schulman AH. Life without GAG: The BARE-2 retrotransposon as a parasite's parasite. Gene. 2006;390:166–174. doi: 10.1016/j.gene.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Sabot F, Sourdille P, Chantret N, Bernard M. Morgane, a new LTR retrotransposon group, and its subfamilies in wheats. Genetica. 2006;128:439–447. doi: 10.1007/s10709-006-7725-5. [DOI] [PubMed] [Google Scholar]
- 8.Kalendar R, et al. LARD retroelements: Conserved, non-autonomous components of barley and related genomes. Genetics. 2004;166:1437–1450. doi: 10.1534/genetics.166.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang TJ, et al. Characterization of terminal-repeat retrotransposon in miniature (TRIM) in Brassica relatives. Theor Appl Genet. 2007;114:627–636. doi: 10.1007/s00122-006-0463-3. [DOI] [PubMed] [Google Scholar]
- 10.Witte CP, Le QH, Bureau T, Kumar A. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci USA. 2001;98:13778–13783. doi: 10.1073/pnas.241341898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang N, Jordan IK, Wessler SR. Dasheng and RIRE2. A nonautonomous long terminal repeat element and its putative autonomous partner in the rice genome. Plant Physiol. 2002;130:1697–1705. doi: 10.1104/pp.015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonius-Klemola K, Kalendar R, Schulman AH. TRIM retrotransposons occur in apple and are polymorphic between varieties but not sports. Theor Appl Genet. 2006;112:999–1008. doi: 10.1007/s00122-005-0203-0. [DOI] [PubMed] [Google Scholar]
- 13.Szyman′ski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5 S rRNA: structure and interactions. Biochem J. 2003;371(Pt 3):641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marquet R, Isel C, Ehresmann C, Ehresmann B. tRNAs as primer of reverse transcriptases. Biochemie. 1995;77:113–124. doi: 10.1016/0300-9084(96)88114-4. [DOI] [PubMed] [Google Scholar]
- 15.Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P. A contiguous 66 kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SanMiguel P, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 17.Cloix C, et al. In vitro analysis of the sequences required for transcription of the Arabidopsis thaliana 5S rRNA genes. Plant J. 2003;35:251–261. doi: 10.1046/j.1365-313x.2003.01793.x. [DOI] [PubMed] [Google Scholar]
- 18.Seki M, et al. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 1998;15:707–720. doi: 10.1046/j.1365-313x.1998.00237.x. [DOI] [PubMed] [Google Scholar]
- 19.Borson ND, Salo WL, Drewes LR. A lock-docking oligo(dT) primer for 5′ and 3′ RACE PCR. PCR Methods Appl. 1992;2:144–148. doi: 10.1101/gr.2.2.144. [DOI] [PubMed] [Google Scholar]
- 20.Cozzarelli NR, et al. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 21.Fulnecek J, Kovarik A. Low abundant spacer 5S rRNA transcripts are frequently polyadenylated in Nicotiana. Mol Genet Gen. 2007;278:565–573. doi: 10.1007/s00438-007-0273-6. [DOI] [PubMed] [Google Scholar]
- 22.Loke JC, et al. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 2005;138:1457–1468. doi: 10.1104/pp.105.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathews DH. Predicting a set of minimal free energy RNA secondary structures common to two sequences. Bioinformatics. 2005;21:2246–2253. doi: 10.1093/bioinformatics/bti349. [DOI] [PubMed] [Google Scholar]
- 24.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Y-X, Li W-H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peleg O, et al. RNA secondary structure and sequence conservation in C1 region of human immunodeficiency virus type 1 env gene. AIDS Res Hum Retroviruses. 2002;18:867–878. doi: 10.1089/08892220260190353. [DOI] [PubMed] [Google Scholar]
- 28.Peleg O, Trifonov EN, Bolshoy A. Hidden messages in the nef gene of human immunodeficiency virus type 1 suggest a novel RNA secondary structure. Nucleic Acids Res. 2003;31:4192–4200. doi: 10.1093/nar/gkg454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalendar R, Schulman A. IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nat Protoc. 2006;1:2478–2484. doi: 10.1038/nprot.2006.377. [DOI] [PubMed] [Google Scholar]
- 30.Schulman AH, Flavell AJ, Ellis THN. The application of LTR retrotransposons as molecular markers in plants. Methods Mol Biol. 2004;260:145–173. doi: 10.1385/1-59259-755-6:145. [DOI] [PubMed] [Google Scholar]
- 31.Katzman M, Katz RA. Substrate recognition by retroviral integrases. Adv Virus Res. 1999;52:371–395. doi: 10.1016/s0065-3527(08)60307-3. [DOI] [PubMed] [Google Scholar]
- 32.Baum BR, Johnson DA. The molecular diversity of the 5s rRNA gene in barley (Hordeum vulgare) Genome. 1994;37:992–998. doi: 10.1139/g94-140. [DOI] [PubMed] [Google Scholar]
- 33.Cloix C, et al. Analysis of 5S rDNA arrays in Arabidopsis thaliana: physical mapping and chromosome-specific polymorphisms. Genome Res. 2000;10:679–690. doi: 10.1101/gr.10.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyao A, et al. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pryer KM, et al. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature. 2001;409:618–622. doi: 10.1038/35054555. [DOI] [PubMed] [Google Scholar]
- 36.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Häsler J, Samuelsson T, Strub K. Useful ‘junk’: Alu RNAs in the human transcriptome. Cell Mol Life Sci. 2007;64:1793–1800. doi: 10.1007/s00018-007-7084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrigno O, et al. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nat Genet. 2001;28:77–81. doi: 10.1038/ng0501-77. [DOI] [PubMed] [Google Scholar]
- 39.Kapitonov VV, Jurka J. A novel class of SINE elements derived from 5S rRNA. Mol Biol Evol. 2003;20:694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- 40.Rooney AP, Ward TJ. Evolution of a large ribosomal RNA multigene family in filamentous fungi: Birth and death of a concerted evolution paradigm. Proc Natl Acad Sci USA. 2005;102:5084–5089. doi: 10.1073/pnas.0409689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Besser D, et al. DNA methylation inhibits transcription by RNA polymerase III of a tRNA gene, but not of a 5S rRNA gene. FEBS Lett. 1990;269:358–362. doi: 10.1016/0014-5793(90)81193-r. [DOI] [PubMed] [Google Scholar]
- 42.Vaillant I, et al. Regulation of Arabidopsis thaliana 5S rRNA genes. Plant Cell Physiol. 2007;48:745–752. doi: 10.1093/pcp/pcm043. [DOI] [PubMed] [Google Scholar]
- 43.Raskina O, Belyayev A, Nevo E. Quantum speciation in Aegilops: Molecular cytogenetic evidence from rDNA cluster variability in natural populations. Proc Natl Acad Sci USA. 2004;101:14818–14823. doi: 10.1073/pnas.0405817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shishido R, Sano Y, Fukui K. Ribosomal DNAs: An exception to the conservation of gene order in rice genomes. Mol Gen Genet. 2000;263:586–591. doi: 10.1007/s004380051205. [DOI] [PubMed] [Google Scholar]
- 45.Pontes O, et al. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc Natl Acad Sci USA. 2004;101:18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davison J, Tyagi A, Comai L. Large-scale polymorphism of heterochromatic repeats in the DNA of Arabidopsis thaliana. BMC Plant Biol. 2007;7:44. doi: 10.1186/1471-2229-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Datson PM, Murray BG. Ribosomal DNA locus evolution in Nemesia: Transposition rather than structural rearrangement as the key mechanism? Chrom Res. 2006;14:845–857. doi: 10.1007/s10577-006-1092-z. [DOI] [PubMed] [Google Scholar]
- 48.Zimmer EA, et al. Rapid duplication and loss of genes coding for the chains of hemoglobin. Proc Natl Acad Sci USA. 1980;77:2158–2162. doi: 10.1073/pnas.77.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. 2006;2006:71753. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 51.Xing J, et al. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci USA. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang N, et al. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 53.Lai J, Li Y, Messing J, Dooner HK. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA. 2005;102:9068–9073. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgante M, et al. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 55.Jones JM, Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev. 2004;200:233–248. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 56.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kordiš D. A genomic perspective on the chromodomain-containing retrotransposons: Chromoviruses. Gene. 2005;347:161–173. doi: 10.1016/j.gene.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 58.Vicient CM, et al. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X, Gorovsky MA. Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE) Nucleic Acids Res. 1993;21:4954–4960. doi: 10.1093/nar/21.21.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalendar R, et al. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicholas KB, Nicholas HB., Jr . GeneDoc: A Tool for Editing and Annotating Multiple Sequence Alignments. 1997. ( www.psc.edu/biomed/genedoc) [Google Scholar]
- 62.Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rzhetsky A, Nei M. A simple method for estimating and testing minimum evolution trees. Mol Biol Evol. 1992;9:945–967. [Google Scholar]
- 66.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 67.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nei M, Kumar A. Molecular Evolution and Phylogenetics. New York: Oxford Univ Press; 2000. [Google Scholar]
- 69.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 70.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 71.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 72.Hofacker IL, et al. Fast folding and comparison of RNA secondary structures. Monatshefte Chemie. 1994;125:167–188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.