Abstract
The mitochondrial outer membrane-anchored monoamine oxidase (MAO) is a biochemically important flavoenzyme that catalyzes the deamination of biogenic and xenobiotic amines. Its two subtypes, MAOA and MAOB, are linked to several psychiatric disorders and therefore are interesting targets for drug design. To understand the relationship between structure and function of this enzyme, we extended our previous low-resolution rat MAOA structure to the high-resolution wild-type and G110A mutant human MAOA structures at 2.2 and 2.17 Å, respectively. The high-resolution MAOA structures are similar to those of rat MAOA and human MAOB, but different from the known structure of human MAOA [De Colibus L, et al. (2005) Proc Natl Acad Sci USA 102:12684–12689], specifically regarding residues 108–118 and 210–216, which surround the substrate/inhibitor cavity. The results confirm that the inhibitor selectivity of MAOA and MAOB is caused by the structural differences arising from Ile-335 in MAOA vs. Tyr-326 in MAOB. The structures exhibit a C-terminal transmembrane helix with clear electron density, as is also seen in rat MAOA. Mutations on one residue of loop 108–118, G110, which is far from the active center but close to the membrane surface, cause the solubilized enzyme to undergo a dramatic drop in activity, but have less effect when the enzyme is anchored in the membrane. These results suggest that the flexibility of loop 108–118, facilitated by anchoring the enzyme into the membrane, is essential for controlling substrate access to the active site. We report on the observation of the structure–function relationship between a transmembrane helical anchor and an extra-membrane domain.
Keywords: single transmembrane helix, transmembrane helical anchor, harmine, x-ray structure
Monoamine oxidase (MAO) is an enzyme localized to the mitochondrial outer membrane and catalyzes the deamination of biogenic and xenobiotic amines, such as neuroactive serotonin, norepinephrine, and dopamine. MAO contains a flavin adenine dinucleotide (FAD) covalently bound to a cysteine residue by an 8α-(S-cysteinyl)-riboflavin linkage (1). MAO plays a decisive role in some psychiatric and neurological disorders, including depression and Parkinson's disease. Because inhibition of MAO increases the level of neurotransmitters in the central nervous system, searching for the effective inhibitors represents one important approach to developing novel drugs to treat such illnesses. MAO has two subtypes, MAOA and MAOB, whose amino acid sequences are up to 70% identical, although each enzyme has unique substrate and inhibitor specificities (2): MAOA oxidizes serotonin, whereas MAOB does not; MAOA is selectively inhibited by clorgyline, whereas MAOB is highly inhibited by deprenyl. In addition, the oxidative deamination produces harmful hydrogen peroxide that may further generate free radicals (3). Development of selective and reversible MAO inhibitors is important not only from the standpoint of treating symptoms (i.e., by increasing the biological half-life of monoamine neurotransmitters), but also with regard to the neuroprotective effects (i.e., prevention or delay of neurodegeneration itself) (4). To develop more effective and specific inhibitors, it is important to understand the inhibition and catalytic mechanism, based on 3D protein structures. Binda et al. (5) first determined the x-ray structures of human MAOB at 3.0 Å and later improved the resolution to 1.7 Å, including cocrystals with various inhibitors (6). On the other hand, the structure of MAOA was determined only at lower resolution: we determined the structure of rat MAOA at 3.2 Å (7), and De Colibus et al. (8) solved the human MAOA structure at 3.0 Å. Despite the limited resolution, we were still able to use these structures to obtain information that was useful in understanding the distinct substrate and inhibitor specificities of MAOA and MAOB (7).
Interestingly, the x-ray structure of monoclinic human MAOA (8) at 3.0 Å differs from those of rat MAOA and human MAOB in the loop conformations of residues 108–118 and 210–216, both important components of the active site. These components affect the substrate/inhibitor specificities of human MAOA, as reported (8, 9). From our earlier result, the backbone structure of rat MAOA, including residues 108–118 and 210–216, is nearly identical to that of human MAOB. Because the amino acid sequences of the two enzymes are 70% identical, we had anticipated this similarity. The sequence identity between human MAOA and rat MAOA is as high as 87% over the whole molecule and 90% in residues 108–118 and 210–216; therefore, in light of their high sequence identity, the structural differences between the two enzymes in these regions are exceptional. Because these regions take part in the composition of active center, it is therefore important to understand whether the differences between rat and human MAOA in these regions truly exist, or whether they are any artifacts, which is critical for the guidance of drug design.
The functional role of the C-terminal transmembrane helix has also been of biological interest. The significance of the binding of MAO to the mitochondrial outer membrane remains unclear. The x-ray structure of rat MAOA revealed that the C terminus maintains a transmembrane structure (7), whereas the present available structures of both human MAOB and monoclinic human MAOA have resolved only a few residues in this helical region (6, 8). Studies showed that the C-terminal 29-aa residues in MAOB are responsible for targeting and anchoring the protein to the mitochondrial outer membrane (10). A C-terminal truncation leads to a significant decrease in MAOB catalytic activity, but does not produce any significant change in inhibitor specificity (11). Therefore, C-terminal anchoring for this enzyme must be important for its biological functions. Here, we report the x-ray structure of human MAOA complexed with a reversible MAOA-specific inhibitor, harmine, at 2.2-Å resolution. The high-resolution structure provides greater insight into the enzymatic details of MAOA, especially in terms of substrate/inhibitor binding specificities. We also show human MAOA structure with the full transmembrane helix. We measured activities of wild-type protein and mutants with mutations at residue G110 in loop 108–118, both in the solubilized and membrane-bound forms, to better understand the role of the transmembrane anchor.
Results and Discussion
The Overall Structure of Human MAOA.
Using the molecular replacement method, with rat MAOA lacking the C-terminal helix as a search model, the structure of wild-type human MAOA was determined and refined to R = 0.201 and Rfree = 0.255 at 2.2-Å resolution. Because the G110A mutant was isomorphous with the wild type, the mutant structural model was prepared by replacing residue 110 with alanine; this model refined to R = 0.193 and Rfree = 0.244 at 2.17-Å resolution. The human MAOAs were monomers rather than dimers, as in the case of rat MAOA. This monomeric state in crystals was consistent with that of the monoclinic human MAOA (8). The C-terminal helix was built by iterations of structural refinements and calculations of composite-omit maps. The statistics of the structural determinations of the wild-type and G110A mutant are shown in Table 1.
Table 1.
Data collection and refinement statistics
| Statistic | Wild type | Mutant (G110A) |
|---|---|---|
| Wavelength, Å | 0.9 | |
| Exposure time, s | 10.0 | |
| Oscillation angle, ° | 0.5 | |
| Space group | C222 | |
| Cell dimensions, Å | ||
| a | 135.3 | 135.5 |
| b | 218.7 | 217.4 |
| c | 54.4 | 54.8 |
| Resolution range, Å | 60.28∼2.20 (2.32∼2.20) | 49.57∼2.17 (2.25∼2.17) |
| Completeness, % | 99.8 (99.7) | 71.4 (21.5) |
| No. of unique reflections | 41,775 (6,020) | 31,614 (1,063) |
| Redundancy | 4.9 (4.9) | 2.8 (1.4) |
| Rmerge | 0.138 (0.670) | 0.079 (0.310) |
| I/σ | 6.7 (1.6) | 7.1 (1.5) |
| Refinement | ||
| Rcryst | 0.201 | 0.193 |
| Rfree | 0.255 | 0.244 |
| rmsd | ||
| Bonds, Å | 0.023 | 0.019 |
| Angle, ο | 2.125 | 1.812 |
| Ramachandran plot | ||
| Allowed region, % | 90.8 | 90.2 |
| Additionally allowed, % | 8.8 | 9.4 |
| Generously allowed, % | 0.2 | 0.2 |
| Disallowed region, % | 0.2 | 0.2 |
Completeness (%) is a percentage of independent reflections observed and is 90.5 (76.8) at 2.55 Å and 94.1 (84.4) at 2.76 Å. Redundancy is the number of observed reflections for each independent reflection. I/σ is the average of intensity signal-to-noise ratio. Rmerge, is ΣhΣI|I(h,i) − <I(h)> /ΣhΣII(h, i), where I(h, i) is the intensity value of the ith measurement of h and <I(h)> is the corresponding mean value of I(h) for all I measurements. The summation is over the reflections with I/σ (I) >1.0. Rcryst is the conventional crystallographic R factor, Σ|FO − FC|/Σ|FO|, where FO and FC are the observed and calculated structure factors, respectively. Rfree is a free R factor in the program CNS (21) evaluated for the 5% of reflections that are excluded from the refinement.
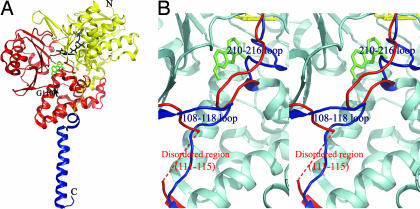
Fig. 1A shows the overall structure of human MAOA. It is nearly identical to that of rat MAOA, which has 90% sequence identity to human MAOA. Residues 108–118 and 210–216 of the wild-type human MAOA were clearly assigned in the MR/DM map. The MR/DM and composite-omit maps and structural features, including B factors, were reasonable for residues 108–118 and 210–216 and for other regions. Residues 108–118 and 210–216 of human MAOA superposed well with the homologous regions of rat MAOA (7), and with the corresponding regions of human MAOB (6), but differently from those of the early reported monoclinic human MAOA (8), as shown in Fig. 1B. To double-check the validity of the structure at these regions, we used the monoclinic human MAOA as a search model for the molecular replacement. The resultant maps at the two loop regions did not fit those of the monoclinic human MAOA, but fit our structure well. A Fo − Fc difference Fourier map had no significant residual density at residues 108–118 and 210–216 of our structure. These results further confirmed our structure.
Fig. 1.
The structure of human MAOA and the comparison with the early known structure. (A) The overall structure drawn in ribbon mode. N, N terminus; C, C terminus. The structure can be divided into two domains, extra-membrane domain (shown in yellow and red) and membrane binding domain (shown in blue). The extra-membrane domain was further divided as two regions, FAD binding region (yellow) and substrate/inhibitor binding region (red). FAD (black) and harmine (green) molecules are shown as stick models. The black arrow indicates the position of G110, a residue at which we introduced mutations. (B) Stereoview of the superposed structures of human MAOA and the early published human MAOA. The identical parts between the two structures are shown in cyan. The different folds at loops 108–118 and 210–216 in the two structures are shown in blue (our structure) and red (early known structure). The fragment of A111-V115 in the early structure is disordered and not visible. Pictures were generated by PyMOL (26).
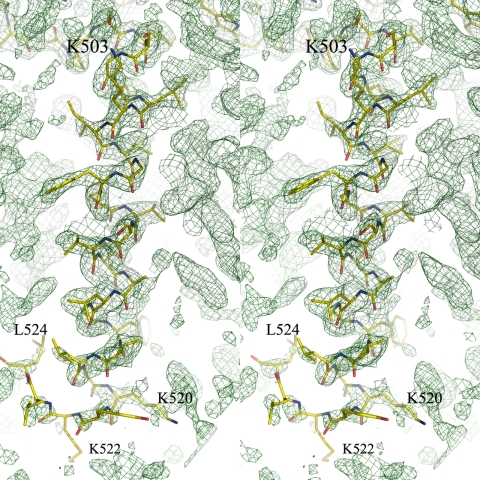
In the single transmembrane helix from residues 498–524, residues 498–521 were fitted uniquely on the composite-omit map as in Fig. 2 and converged well to a helical conformation after structural refinement. Although the map at the end of C-terminal region was poor in electron density, residues 522–524 were able to successfully built into the model, and their structures converged to a nonhelical conformation after refinement. The very end three residues, 525–527, were not visible. The side chains of the transmembrane helix were located in the map, although the B factors were higher than those of the extra-membrane domain. The averaged B factor of backbone atoms for the C-terminal helix was 64.2 Å2, whereas that for the extra-membrane domain was 34.3 Å2.
Fig. 2.
Stereoview of the C-terminal transmembrane helical structure. The composite-omit map for the C-terminal domain is contoured at the 1.0-σ level at 2.2-Å resolution. A structural model of the transmembrane helix from residues 498 to 524 is superposed on the composite-omit map. The other parts of the protein structure are not shown for clarity.
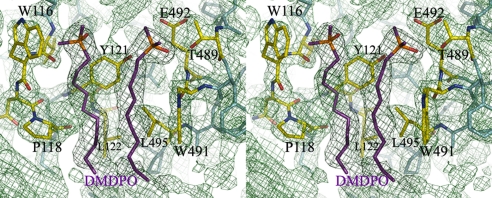
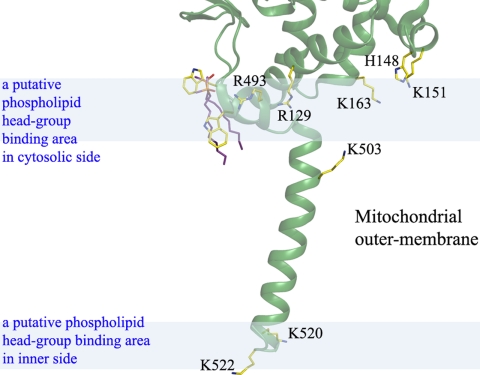
We also detected two molecules of dimethyldecylphosphine oxide (DMDPO) in a Fo − Fc difference Fourier map and composite-omit map (Fig. 3). DMDPO is the detergent that was used in crystallizing the protein. The molecules were surrounded by three aromatic residues and a proline: Trp-116, Trp-491, Tyr-121, and Pro-118. In vivo, the DMDPO site is likely occupied by phospholipid in the mitochondrial outer membrane. In addition, the horizontal arrangement of the positively charged residues (Arg-129, His-148, Lys-151, Lys-163, Arg-493, Lys-503, Lys-520, and Lys-522) that interact with the phospholipid hydrophilic head groups indicates the location of the outer-membrane surface, as shown in Fig. 4. A one-turn helix parallel to the membrane surface was supposed to be buried in the membrane, the single transmembrane helix, as seen in rat MAOA (7).
Fig. 3.
The detergent molecules bound at the one-turn helix next to the transmembrane helix. The composite-omit map (1.0-σ level) was generated by CNSsolve (22). The pocket is surrounded by hydrophobic amino acid residues: W116, P118, Y121, L122, and W491. These models were generated by PyMOL (26).
Fig. 4.
Binding model of MAOA into the mitochondrial outer membrane. The positively charged residues Arg-129, His-148, Lys-151, Lys-163, Arg-493, Lys-503, Lys-520, and Lys-522 are shown. These residues are presumed to interact with the phospholipid hydrophilic head group at the membrane surface shown as blue semitransparent areas. The upper area represents the cytosolic side.
Structure of the Substrate/Inhibitor Cavity with a Specific Reversible Inhibitor.
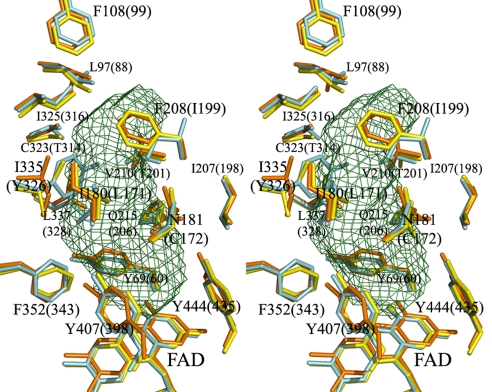
The superposed residues surrounding the substrate/inhibitor cavity of human MAOA, rat MAOA, and human MAOB are shown in Fig. 5. All 16 residues surrounding the substrate/inhibitor cavity are conserved between human and rat MAOA, and six of the 16 residues differ between MAOA and MAOB.
Fig. 5.
Stereoview of the substrate/inhibitor binding sites of human MAOA, human MAOB, and rat MAOA. Residues of human MAOA are shown in yellow, rat MAOA in orange, and human MAOB in cyan. The residues that are important in forming the substrate/inhibitor cavity are labeled. The residue numbering is according to the residue positions in human MAOA, which are the same as in rat MAOA. The residue numbers of human MAOB are shown in parentheses. Two residues, I199 of human MAOB and I335 of human or rat MAOA, are present as different rotamers in different complexes. The cavity was calculated by VOIDOO (13) with a 1.57-Å radius probe. These models were generated in PyMOL (26) (rmsd was 0.545 Å for human MAOB and 0.612 Å for rat MAOA).
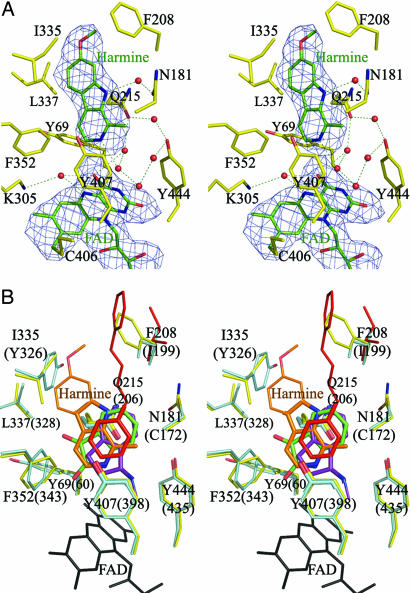
Harmine, a reversible inhibitor, is located in the active center cavity of the enzyme. It interacts with Tyr-69, Asn-181, Phe-208, Val-210, Gln-215, Cys-323, Ile-325, Ile-335, Leu-337, Phe-352, Tyr-407, Tyr-444, and FAD (Fig. 6A). Seven water molecules occupy the space between the inhibitor and these groups. The inhibitor and the FAD are bridged through two water molecules by hydrogen bonds. The amide group of the Gln-215 side chain interacts tightly with harmine by a π–π interaction, with an interplane distance of 3.4 Å. We superposed the structures of human MAOA with harmine and other inhibitors including isatin [Protein Data Bank (PDB) ID code 1OJA], rasagiline analogue (PDB ID code 2C67), and 1,4-diphenyl-2-butene (PDB ID code 1OJ9) in human MAOB. The positions of the aromatic rings of these reversible inhibitors are highly conserved, as shown in Fig. 6B. Coplanar aromatic rings make π–π interactions with Gln-215 of human MAOA or Gln-206 of MAOB (Gln-215/206). The aromatic rings interact with Phe-352/343 and Tyr-407/398 at the opposite side of Gln-215/206. The structure of the harmine molecule in human MAOA could not be accommodated into human MAOB because of its structural overlap with Tyr-326 of MAOB. Thus, Ile-335 in MAOA and Tyr-326 in MAOB play a crucial role in substrate/inhibitor selectivity. These results are consistent with our previous structural analysis of rat MAOA and site-directed mutagenesis studies (7, 12). The structure of 1,4-diphenyl-2-butene in MAOB would collide with Phe-208 of MAOA, which corresponds to Ile-199 of MAOB. The selectivity of the reversible inhibitors is caused by the different size and shape of the substrate/inhibitor cavity, restricted by Ile-335 and Phe-208 in MAOA, which corresponds to Tyr-326 and Ile-199 of MAOB. When structures of human MAOA with harmine and rat MAOA with clogyline are compared with each other, the side chains of Ile-335 have different conformations. The different inhibitors are accommodated by the induced fit of Ile-335, as observed for Ile-199 of human MAOB (6). The bond between the flavin-substituted Cys-406 and Tyr-407 is in the cis-conformation, as is that of human MAOB determined at 1.7-Å resolution.
Fig. 6.
Stereoviews of the substrate/inhibitor binding site. (A) The Fo − Fc difference Fourier map contoured at 2.0 σ was generated at 2.2-Å resolution for the inhibitor (harmine) and FAD. Amino acid residues are shown in yellow, and FAD and harmine are shown in green. Dotted lines indicate hydrogen bonds. (B) The structure of the substrate/inhibitor binding sites in human MAOA and MAOB complexed with specific inhibitors. The residues are numbered according to human MAOA, and the numbers in parentheses are for human MAOB. MAOA and MAOB residues are shown in yellow and light blue, respectively. Inhibitors are colored as follows: orange, harmine; green, isatin (PDB ID code 1OJA); purple, rasagiline analogue (PDB ID code 2C67); and red, 1,4-diphenyl-2-butene (PDB ID code 1OJ9). FAD is shown in black. Nitrogen and oxygen atoms are shown in blue and red, respectively. Residue I199 of MAOB is present as different rotamers in different complexes. The rotamer of this residue, in MAOB with 1,4-diphenyl-2-butene, is shown in red. The residues Q215 and Y407 that form important hydrophobic interactions to the inhibitors are shown as thick stick models.
The Loop Structures at the Substrate Entrance and Its Function in Enzyme Activity.
When the structure of the substrate/inhibitor cavity was calculated by using the VOIDOO program (13) with a radius of 1.57 Å, the cavity was completely closed, as shown in Fig. 5. When we calculated with a radius of 1.40 Å, however, a narrow path was detected between the cavity and the outside of molecule [supporting information (SI) Fig. S1]. The entrance for substrate/inhibitor is surrounded by residues V93-E95, Y109-P112, and F208-N212, which lie in three different loops. The steady-state width of the entrance is too narrow for such compounds as harmine to pass through. We infer that structural fluctuations that enlarge the entrance are essential for the cavity to accept substrates. To understand the relationship between the structural fluctuations and the enzymatic reactions, we made a mutation at Gly-110 (in the loop 109–112 beside the entrance) in rat MAOA and human MAOA. Although this site is far from the active center, the activity of the purified G110A mutants dropped significantly. To further analyze the structure–function relationship at this site, we determined the structure of human G110A. The Cα trace of human G110A was almost identical compared with wild-type MAOA, both in the loop structures and in other parts of the protein, implying that the activity change was derived from a dynamic alteration of the structure of the enzyme. To further confirm the role of the flexibility of G110, a G110P mutant was made, which gave this region a highly rigid quality. As expected, the G110P mutant showed an increase of Km of 19-fold (Table 2). Together with the structural characteristics of the substrate/inhibitor binding site, these results suggest that the loop flexibility is critical for opening the entry for substrates/inhibitors.
Table 2.
Kinetic parameters for wild-type and mutant human MAOA
| Enzyme | Solubilized form |
Membrane-bound form |
||||
|---|---|---|---|---|---|---|
| Km, mM | kcat, min−1 | kcat/Km, min−1·mM−1 | Km, mM | kcat, min−1 | kcat/Km, min−1·mM−1 | |
| Rat WT | 0.096 ± 0.016 | 146 ± 21 | 1,530 ± 143 | 0.026 ± 0.001 | 152 ± 5 | 5,792 ± 434 |
| Rat G110A | 0.913 ± 0.096 | 122 ± 10 | 134 ± 14 | 0.039 ± 0.003 | 145 ± 3 | 3,757 ± 268 |
| Rat G110P | 1.834 ± 0.178 | 44 ± 3 | 24 ± 4 | 0.184 ± 0.039 | 142 ± 29 | 773 ± 14 |
| Human WT | 0.117 ± 0.004 | 181 ± 31 | 1,544 ± 263 | 0.044 ± 0.004 | 165 ± 2 | 3,803 ± 349 |
| Human G110A | 0.526 ± 0.083 | 116 ± 13 | 221 ± 20 | 0.093 ± 0.003 | 131 ± 4 | 1,407 ± 41 |
Interestingly, the mutant enzymes showed more significant changes when they were solubilized from the membrane, as compared with wild-type enzymes. For rat G110A, the Km for the substrate kynuramine increased nearly 10-fold (0.096 to 0.913 mM) compared with the wild-type in the detergent-solubilized form. However, there was very little change in the membrane-bound form (0.026 to 0.039 mM). The human mutant also showed similar results. The kcat values for the wild-type and all mutants of rat MAOA or human MAOA are comparable, ranging from 116 to 181 min−1 for both the solubilized and membrane-bound forms, except for solubilized rat G110P mutant, which has a kcat of 44 min−1. Because solubilized G110P has a very high Km value, it is difficult to accurately estimate the maximum activity, because we observed some substrate-inhibition phenomena at very high concentrations of substrate (i.e., the activity decreases while the substrate concentration increases too high). Therefore, the low kcat may not reflect a true quality of G110P. These results suggest that the mutation on G110 does not affect enzyme catalytic activity, but rather affects substrate binding, especially in the solubilized form. These differences in Km between the membrane-bound and solubilized forms were previously thought to be caused by the inactivating effect of the detergent. Although the detergent may have some effects on enzyme activity, it cannot fully explain why the mutants showed significantly bigger change than the wild type. Therefore, we conclude that it is the membrane anchoring, but not detergent, that affects enzyme catalytic efficiency (Km) in MAOA. The decrease of the activity in C-terminal truncated MAOB (11) also supports the idea that membrane anchoring is essential for MAO's functions. There is also a possibility that the different Km values between mutant and wild type are caused by the larger residues in the mutants, which may block the entrance. If the size plays a major role in blocking the entrance, the mutants should show similar decreases in substrate affinity in both the membrane-anchored and solubilized forms. However, our results showed that the solubilized form, but not the membrane-anchored form, had a dramatic change in Km, indicating that the residue size at G110 is not a major factor that affects the Km. Instead, flexibility at this site, together with the membrane anchoring, plays the major role. Because the mutation site is far from the active center, and the crystal structure of G110A shows little change in the detailed structure, the only possible change would be in the flexibility of the loop of 109–112, as supported by the above data. From the membrane binding model shown in Fig. 4, the C-terminal helix and a small part of the enzyme are buried in the membrane, leaving large parts of enzyme in the cytosol. Therefore, it is possible that fluctuations of the extra-membrane domain (e.g., because of Brownian motion) of the MAO enzyme are not synchronized with its transmembrane domain buried in the membrane. This discrepancy between the motions of the extra-membrane and transmembrane domain may therefore cause conformational changes in or around the entrance loops near the active site, which in turn open the entrance for the substrate/inhibitor. Obviously, the flexible G110 is important for inducing this conformational change. Substitution of G110 with alanine produces a small amount of resistance to this change, but it becomes more obvious when enzyme is solubilized. When G110 is replaced with the rigid residue proline, the resistance to conformational change at the loop becomes more significant, both in membrane-bound and solubilized forms. This model partially explains the role of the C-terminal anchoring of MAOA and is a novel concept in membrane protein function.
Materials and Methods
Construction of Expression Plasmids for Wild-Type and Mutant MAOA.
cDNA of human MAOA was amplified by PCR with a 6× His tag at the N terminus. The resulting cDNA was inserted into a yeast expression vector, YEp51, as described for the expression of rat MAOA (7, 14, 15). A G110A expression plasmid was constructed by using a QuikChange XL site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. The primers (synthesized by Gene Design) used were 5′-GGGGAAAACATATCCATTTCGGGCCGCCTTTCCACCAGTATGG-3′ and 5′-CCATACTGGTGGAAAGGCGGCCCGAAATGGATATGTTTTCCCC-3′ for human G110A; 5′-CCCATTCCGTGCTGCATTCCCACC-3′ and 5′-GGTGGGAATGCAGCACGGAATGGG-3′ for rat G110A; and 5′-CTTACCCATTCCGTCCTGCATTCCCACC-3′ and 5′-GGTGGGAATGCAGGACGGAATGGGTAAG-3′ for rat G110P. All constructs were confirmed by DNA sequencing with an ABI PRISM 3100 Genetic Analyzer.
Protein Expression and Purification.
His-tagged wild-type and mutant MAOA proteins were expressed in Saccharomyces cerevisiae strain BJ2168 (a prc1–407 prb1122 prp4–3 leu2 trp1 ura3–52) (Wako Pure Chemical) and purified by using a published method (14). Briefly, cells transformed with YEp51 vector carrying His-MAOA cDNA were cultured in leucine-free synthesized medium, and expression was induced by galactose for 36 h. The cells were collected and treated with yeast lytic enzyme (Zymolyase; Seikagaku) for 3 h at 30°C and then disrupted by sonication. After removing cellular debris by low-speed centrifugation, crude membrane fractions were collected by ultracentrifugation at 40,000 rpm in a Beckman 45-Ti rotor for 25 min. MAOA proteins were solubilized from the crude membrane fraction by using N-dodecylphosphocholine (Anatrace) and purified by two repeats of affinity chromatography on a Ni column. The purified protein was eluted with buffer containing 500 mM imidazole and dialyzed against buffer containing 10 mM sodium phosphate (pH 7.6), 100 mM sodium chloride, 10% (vol/vol) glycerol, 5 mM β-mercaptoethanol, and 0.05% (wt/vol) N-dodecylphosphocholine. The final concentration of protein before crystallization was 10 mg/ml.
Crystallization.
The enzyme was crystallized into an orthorhombic space group by the sitting drop vapor diffusion method. Before crystallization, DMDPO (Anatrace) was added to the sample to a final concentration of 0.8% (wt/vol). Harmine hydrochloride (Wako Pure Chemical), a reversible inhibitor, was cocrystallized with human MAOA. Harmine hydrochloride was added at a protein/harmine ratio of 1:5. The best crystals were obtained by equilibrating with reservoir solution containing 1.6 M ammonium sulfate and 100 mM citric acid (pH 5.6) at 277 K. The crystals were successfully frozen in a nitrogen gas stream at 100 K using 30% glycerol as the cryoprotectant. The crystal belonged to an orthorhombic space group C222. It diffracted at 2.2 Å better than both rat MAOA with P43212 (7) and monoclinic human MAOA (8).
X-Ray Experiments.
The diffraction data were collected at 100 K by using an imaging plate detector (DIP6040) at beamline BL44XU at SPring-8 (Hyogo, Japan). Data were indexed and integrated with the MOSFLM program and scaled with the SCALA program in the CCP4 suite (16, 17). Five wild-type and two mutant crystals were used for data collection. Each image was taken with 10 s of exposure and an oscillation angle of 0.5°. Statistics of the intensity data and structure determination are summarized in Table 1. Wild-type crystals diffracted X-rays isotropically, whereas mutant crystals exhibited anisotropy in intensity distribution of their diffractions. Therefore, after merging 232 images of the mutant crystals, the effective resolution of each image was determined by inspecting its R factor, Σ|I(h) − <I(h)>|/ΣhI(h), where I(h) is the intensity value of h and <I(h)> is the corresponding mean value of I(h) for all images. The resolutions of 232 images were different from each other and were in the wide range from 2.17 to 3.38 Å.
Structure Determination.
The structure of the wild-type protein was determined by the molecular replacement method using the MOLREP program (18). A reference model for molecular replacement was prepared by truncating residues 108–118, 210–216, and 501–527 from rat MAOA (PDB ID code 1O5W). The initial phase was calculated at 3.0-Å resolution by using the reference model. Because the solvent content of the human MAOA crystal was as high as 63%, phase extension was performed from 3.0 to 2.2 Å by the solvent flattening method (19) with the program DM (20). The resulting map was referred to as the MR/DM map. The initial model of human MAOA was built on the MR/DM map. The Coot program (21) was used for modeling, and the CNSsolve program (22) and REFMAC5 program (23) were used for structural refinement. Electron density maps were generated by CNSsolve.
The structure of the mutant was determined by the molecular replacement method with the wild-type structure as a reference model. The MR/DM maps at several resolutions were compared to inspect the effective resolution of the intensity data set. Because the electron density map at 2.17-Å resolution exhibited finer structure than those of the lower resolution, the intensity data up to 2.17 Å were used for the structural refinement of the mutant.
Determination of Kinetic Properties.
The activity of MAOA was measured spectrophotometrically by monitoring the increase in absorbance at 314 nm upon oxidation of kynuramine and the formation of 4-hydroxyquinoline (24) in assay buffer [25 mM Tris (pH 7.5), 150 mM NaCl]. The concentration of kynuramine ranged from 0.0625 to 5.0000 mM. The extinction coefficient of 4-hydroxyquinoline at 314 nm was determined to be 12,300 M−1·cm−1. The kinetic constants were calculated according to the method of Ma and Ito (14). The concentration of purified MAOA was determined by the absorbance of FAD. The oxidized form of FAD has an absorption peak at 456 nm and an extinction coefficient of 11,800 M−1·cm−1 (25). The concentration of MAOA protein in the mitochondrial outer membrane was determined by titration with an irreversible inhibitor, clorgyline. Briefly, 10 μl of prepared crude membranes from yeast cells expressing MAOA was incubated with 0, 10, 20, 30, 40, 50, 60, 80, or 100 pmol of clorgyline in assay buffer at room temperature for 2 h. The remaining enzymatic activity was measured by using kynuramine as a substrate, as described above. The minimum concentration of clorgyline that completely inhibited MAOA was considered equivalent to the concentration of the enzyme.
Supplementary Material
Acknowledgments.
We thank all members of beamline BL44XU of SPring-8 and Ms. Tomoko Nagasue for providing technical assistance with this research. This work was supported in part by Grant-in-Aid for Scientific Research on Priority Areas 16087206 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.T.), the Japan Biological Informatics Consortium (T.T.), and the Strategic Japan-U.K. Cooperative Program of the Japan Science and Technology Agency (T.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and the structural factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2Z5X and 2Z5Y).
This article contains supporting information online at www.pnas.org/cgi/content/full/0710626105/DCSupplemental.
References
- 1.Walker WH, Kearney EB, Seng RL, Singer TP. The covalently bound flavin of hepatic monoamine oxidase. 2. Identification and properties of cysteinyl riboflavin. Eur J Biochem. 1971;24:328–331. doi: 10.1111/j.1432-1033.1971.tb19690.x. [DOI] [PubMed] [Google Scholar]
- 2.Bach AW, et al. cDNA cloning of human liver monoamine oxidase A and B: Molecular basis of differences in enzymatic properties. Proc Natl Acad Sci USA. 1988;85:4934–4938. doi: 10.1073/pnas.85.13.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youdim MBH, Riederer P. The relevance of glial monoamine oxidase B and polyamines to the action of selegiline in Parkinson's disease. Movement Disorders. 1993;8:S8–S13. doi: 10.1002/mds.870080504. [DOI] [PubMed] [Google Scholar]
- 4.Saura J, et al. Localization of monoamine oxidases in human peripheral tissues. Life Sci. 1996;59:1341–1349. doi: 10.1016/0024-3205(96)00459-6. [DOI] [PubMed] [Google Scholar]
- 5.Binda C, Newton-Vinson P, Hubalek F, Edmondson DE, Mattevi A. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat Struct Biol. 2002;9:22–26. doi: 10.1038/nsb732. [DOI] [PubMed] [Google Scholar]
- 6.Binda C, et al. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proc Natl Acad Sci USA. 2003;100:9750–9755. doi: 10.1073/pnas.1633804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma J, et al. Structure of rat monoamine oxidase A and its specific recognitions for substrates and inhibitors. J Mol Biol. 2004;338:103–114. doi: 10.1016/j.jmb.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 8.De Colibus L, et al. Three-dimensional structure of human monoamine oxidase A (MAOA): Relation to the structures of rat MAOA and human MAOB. Proc Natl Acad Sci USA. 2005;102:12684–12689. doi: 10.1073/pnas.0505975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edmondson DE, De Colibus L, Binda C, Li M, Mattevi A. New insights into the structures and functions of human monoamine oxidases A and B. J Neural Transm. 2007;114:703–705. doi: 10.1007/s00702-007-0674-z. [DOI] [PubMed] [Google Scholar]
- 10.Mitoma J, Ito A. Mitochondrial targeting signal of rat liver monoamine oxidase B is located at its carboxyl terminus. J Biochem (Tokyo) 1992;111:20–24. doi: 10.1093/oxfordjournals.jbchem.a123712. [DOI] [PubMed] [Google Scholar]
- 11.Rebrin I, Geha RM, Chen K, Shih JC. Effects of carboxyl-terminal truncations on the activity and solubility of human monoamine oxidase B. J Biol Chem. 2001;276:29499–29506. doi: 10.1074/jbc.M100431200. [DOI] [PubMed] [Google Scholar]
- 12.Geha RM, Rebrin I, Chen K, Shih JC. Substrate and inhibitor specificities of human monoamine oxidase A and B are influenced by a single amino acid. J Biol Chem. 2001;276:9877–9882. doi: 10.1074/jbc.M006972200. [DOI] [PubMed] [Google Scholar]
- 13.Kleywegt GJ, Jones TA. Detection, delineation, and display of cavities in macromolecular structures. Acta Crystallogr D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 14.Ma J, Ito A. Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable and active conformation of rat monoamine oxidase A. J Biochem (Tokyo) 2002;131:107–111. doi: 10.1093/oxfordjournals.jbchem.a003064. [DOI] [PubMed] [Google Scholar]
- 15.Ma J, et al. Crystallization and preliminary crystallographic analysis of rat monoamine oxidase A complexed with clorgyline. Acta Crystallogr D. 2004;60:317–319. doi: 10.1107/S0907444903025770. [DOI] [PubMed] [Google Scholar]
- 16.Leslie AG. Integration of macromolecular diffraction data. Acta Crystallogr D. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 17.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 18.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 19.Wang BC. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- 20.Cowtan K. DM: An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Weissbach H, Smith TE, Daly JW, Witkop B, Udenfriend S. A rapid spectrophotometric assay of monoamine oxidase based on the rate of disappearance of kynuramine. J Biol Chem. 1960;235:1160–1163. [PubMed] [Google Scholar]
- 25.Weyler W, Salach JI. Purification and properties of mitochondrial monoamine oxidase type A from human placenta. J Biol Chem. 1985;260:13199–13207. [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.