Abstract
Overexpression of membrane proteins in Escherichia coli frequently leads to the formation of aggregates or inclusion bodies, which is undesirable for most studies. Ideally, one would like to optimize the expression conditions by monitoring simultaneously and rapidly both the amounts of properly folded and aggregated membrane protein, a requirement not met by any of the currently available methods. Here, we describe a simple gel-based approach with green fluorescent protein as folding indicator to detect well folded and aggregated proteins simultaneously. The method allows for rapid screening and, importantly, pinpointing the most likely bottlenecks in protein production.
Keywords: folding indicator, in gel GFP fluorescence, optimization of overexpression, inclusion bodies
Protein overexpression in a functional state is one of the first hurdles encountered for the structural analysis of membrane proteins. The biogenesis of integral membrane proteins involves many steps such as targeting of the nascent polypeptide chain to the membrane and insertion into and assembly in the membrane, and each of these steps requires distinct components (1, 2). In Escherichia coli, the most commonly used expression host (3), exceeding the capacity of the cell to process the nascent membrane protein correctly, may result in the production of aggregated material in inclusion bodies. Refolding of membrane proteins from these inclusion bodies is challenging and in most cases possible only for β-barrel type membrane proteins. Consequently, functional overexpression of membrane proteins in the cytoplasmic membrane is preferred.
The amount of well folded protein produced in the membrane can often be increased by systematic optimization of several parameters such as the expression strain, induction temperature, growth medium, and promoter strength (4). Evaluation of conditions may be done by determining the amount of protein present in isolated membrane vesicles or by monitoring the activity of the protein, provided suitable activity assays are available. However, these techniques are laborious and therefore unsuitable for the routine analysis of many expression conditions. The use of green fluorescent protein (GFP) fusions has enabled a quicker determination of the amount of folded protein (5). Because proper folding of GFP fused to the C terminus of a target protein depends on the correct folding of the latter, only folded fusion protein will become fluorescent. This approach has been shown to be applicable to membrane proteins and soluble proteins alike (5, 6).
All of the above-mentioned methods quantify the amount of folded protein only. Information on the absolute expression levels or the amount of aggregated protein produced is not obtained. Consequently, the use of these techniques alone does not suffice to assess whether overexpression of folded membrane proteins is limited at the level of the transcription/translation or the trajectory beyond. For example, low fluorescence of GFP fusion proteins could indicate low expression levels but also a large aggregated fraction of a highly overexpressed protein. Whereas the first might be overcome by supplementing the expression strain with a plasmid encoding for rare tRNAs, the latter could possibly be relieved by a decrease in temperature during induction. Obviously, quantification of both aggregated and well folded protein would be helpful to accelerate the optimization process. Ideally, the quantification should avoid extra experimental steps, which would prevent high-throughput analysis, and should not involve relating different types of data, which would make comparisons cumbersome.
Here, we present an application based on the use of GFP as protein folding indicator, in which not only the folded protein is quantified, but also the nonfolded protein. The method is based on differential migration of folded and aggregated GFP fusion proteins during SDS/PAGE. Subsequent immunodetection of both species allows simultaneous determination of the levels of folded and aggregated protein present. This additional information greatly accelerates the optimization of the functional overexpression of both membrane and soluble proteins.
Results
In Gel Mobility Shift of GFP Fusion Proteins.
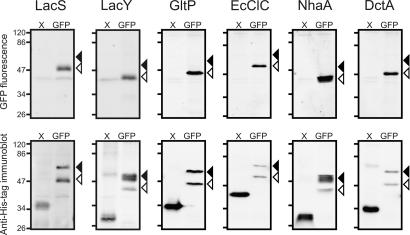
We initially expressed six membrane proteins in E. coli with a GFP-His10 moiety fused to their C termini. The proteins represent members of four evolutionary unrelated families and are all polytopic integral membrane transporters [supporting information (SI) Table S1]. After cell disruption, the total cellular protein extract of the different cultures was analyzed by SDS/PAGE and in gel detection of GFP fluorescence. For all GFP fusion proteins, a single prominent fluorescent band was observed (Fig. 1 Upper, right lanes). The gel was also analyzed by Western blotting and immunodetection with an antibody directed against the His tag to detect the expressed proteins. The blot revealed a band at the same position as the fluorescent signal but, surprisingly, also an additional band of an apparent molecular mass that was ≈10 kDa higher (Fig. 1 Lower, right lanes). Two bands were observed for all GFP fusion proteins tested, but the ratios of the intensities varied between different proteins.
Fig. 1.
Electrophoretic mobility of membrane proteins fused or not fused to GFP. All proteins and GFP fusion proteins were expressed in E. coli MC1061, except for GltP and DctA, which were expressed in E. coli TOP10. Whole-cell samples of cultures induced with 1 × 10−2% (wt/vol) l-arabinose were disrupted and analyzed as described in Materials and Methods. (Upper) In gel GFP fluorescence. (Lower) Immunoblots of the same gels decorated with anti-His tag antibody. X and GFP indicate the absence or presence of a GFP moiety at the C terminus, respectively. Black and white arrows indicate the positions of nonfluorescent and fluorescent species of the GFP fusion proteins, respectively. Molecular masses (in kilodaltons) of the marker proteins are indicated on the left of each panel. Molecular mass of the nonmodified proteins are mentioned in Table S1. The increase in molecular mass caused by the GFP fusion is ≈27 kDa. The faint doublet bands at ≈40 kDa (Upper) represent (an) endogenous fluorescent protein(s) from E. coli.
To analyze the origin of the dual migration, we compared the electrophoretic mobility of the GFP fusion proteins with that of the same proteins without GFP fusion (Fig. 1, left lanes). Nonfused proteins did not show the dual electrophoretic mobility and migrated as single bands. With these nonfused proteins as a reference, the apparent molecular mass of the upper band of the GFP fusions was in agreement with the predicted mass increase from the fusion with GFP (+27 kDa), whereas the lower fluorescent band migrated only 10–15 kDa higher than the band of the nonfused protein. These observations suggest that the GFP moiety of the fusion protein in the upper band was fully denatured, consistent with the predictable migration in SDS/polyacrylamide gels and the absence of fluorescence. The anomalous migration of the lower band can be explained by the preservation of the structure of the GFP moiety, which is consistent with the observed fluorescence.
Because the correct folding of the GFP moiety in GFP fusion proteins is known to depend on the productive folding of the preceding protein domain (5, 6), the observed dual in gel migration of the GFP fusion proteins could be representative of the folded and aggregated protein populations produced under these expression conditions. Clearly, methodology allowing such rapid and simultaneous quantification of both folded and aggregated protein would be of great value during, for instance, optimization of protein expression and purification conditions. We thus set out to establish whether the dual in gel migration of the GFP fusion proteins is a suitable indicator for the amounts of folded and aggregated protein present in a sample.
The Intensity Ratios of the Two Bands Depend on Expression Levels.
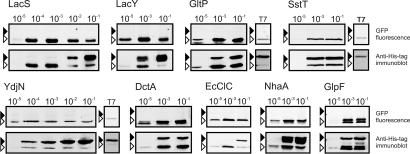
To determine whether the double bands of the expressed GFP fusion proteins were not a mere artifact but indeed originated from different folding states of the expressed proteins, we investigated whether the ratio between the intensities of the two bands changed when the expression conditions were altered to produce more or less aggregated protein. High expression levels generally produce more aggregated protein in inclusion bodies than lower expression levels. In our experiments, the expression of the GFP fusions was controlled by the PBAD promoter, and therefore variation in the expression levels was achieved by altering the concentration of the inducer l-arabinose in the medium during induction. A wide range of inducer concentrations, spanning 4 orders of magnitude, was used. We analyzed the electrophoretic mobility of nine membrane protein GFP fusions (22) (Table S1) by in gel GFP fluorescence and immunoblotting (Fig. 2, Upper and Lower, respectively). For each protein, the combined signal of the two bands on the immunoblots increased with the inducer concentration, indicating higher overall expression levels. However, the distribution of the intensities over the two bands varied markedly. At high inducer concentrations, the relative contribution of the upper band was highest, but at low inducer concentrations, for several proteins the higher band was absent, and only the lower (fluorescent) protein could be detected, consistent with a high fraction of properly folded protein.
Fig. 2.
Electrophoretic mobility of GFP fusion proteins expressed to different levels. Expression of all GFP fusion proteins was controlled by the AraC/PBAD system, except for panels labeled T7, for which the T7 expression system was used. All proteins are polytopic membrane proteins (Table S1). Cultures were induced with 1 × 10−5%, 1 × 10−4%, 1 × 10−3%, 1 × 10−2%, or 1 × 10−1% (wt/vol) l-arabinose as indicated above the lanes. For the T7 expression system, 0.4 mM IPTG was used. (Upper) In gel GFP fluorescence. (Lower) Immunoblots of the same gels decorated with anti-His tag antibody. Black and white arrows indicate the positions of nonfluorescent and fluorescent species of the GFP fusion proteins, respectively.
To exclude that the dual migration was specific for the combination of the expression host E. coli MC1061 and the AraC/PBAD expression system (22), we also studied the expression of the GFP fusions of GltP, YdjN, and SstT in E. coli BL21(DE3) controlled by the T7 system. Again, dual migration of the GFP fusion proteins was observed (Fig. 2) (7). Thus, the dual migration is not specific for the combination of the expression host E. coli MC1061 and the AraC/PBAD expression system. Noticeably, the intensities of the upper bands of the proteins expressed with the T7 system were high compared with the lower bands. The ratios between the two equaled or exceeded those of proteins expressed by using the AraC/PBAD system with high inducer concentrations [1 × 10−1% (wt/vol) arabinose]. Apparently, the expression conditions with the T7 system disfavored the formation of fluorescent proteins.
In conclusion, the intensities of both bands of the GFP fusion protein were affected by the relative promoter strength. Overall, low expression levels increased the relative contribution of the lower, fluorescent band, whereas high expression levels increased the proportion of the upper band.
Characterization of the Fluorescent GFP Fusion Species.
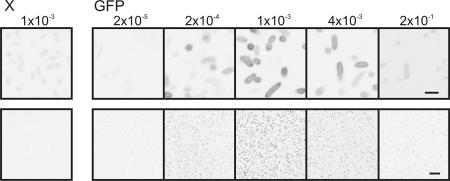
Initial demonstration of the suitability of GFP as an indicator for the productive folding of (soluble) proteins was done by relating whole-cell GFP fluorescence to protein solubility (5). Accordingly, we verified whether the GFP fluorescence measured in gel was in correspondence to the fluorescence as determined in vivo in nondisrupted cells. E. coli cells expressing LacS-GFP were imaged by confocal microscopy. The cells displayed fluorescence at their periphery as expected for a membrane protein GFP fusion (Fig. 3, Upper). Cells exposed to increasing concentrations of inducer showed an optimum in fluorescence at ≈1 × 10−3% (wt/vol) l-arabinose (67 μM; Fig. 3). This result is in perfect agreement with the fluorescence of the lower band of LacS-GFP as detected in gel (Fig. 2).
Fig. 3.
Confocal microscopy images of cells expressing LacS-GFP to different levels. E. coli MC1061 cells were induced with the percentages of l-arabinose indicated above the panels. X and GFP indicate the absence or presence of a GFP moiety at the C terminus of LacS, respectively. The fluorescence shown is inverted. For each sample, similar numbers of cells were imaged. (Upper) Close-up of cells to indicate the distribution of the fluorescence. (Scale bar, 2 μm.) (Lower) Overview of the culture. (Scale bar, 10 μm.)
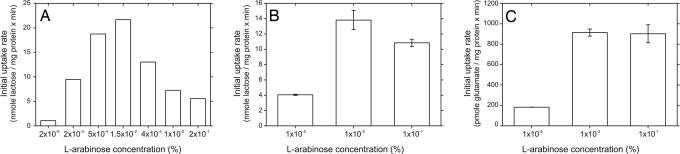
Next, we related the intensity of the fluorescent signal of the GFP fusion protein to the activity of the lactose transporters LacS and LacY and the glutamate transporter GltP. The initial rate of transport, which is indicative of the amount of functionally expressed protein, was determined in cells expressing these proteins to different levels (Fig. 4). Initial rates of lactose counterflow transport by LacS increased up to an inducer concentration of 1.5 × 10−3% (wt/vol) arabinose (Fig. 4A). Above this concentration, the transport rate decreased and leveled off to ≈30% of the maximal activity observed. Similar results were obtained for LacS-GFP (data not shown). In cells expressing LacY-GFP, transport activity increased up to inducer concentrations of 1 × 10−3% (wt/vol) arabinose but decreased slightly toward higher inducer concentrations (Fig. 4B). The initial rate of glutamate transport of GltP-GFP leveled off above 1 × 10−3% (wt/vol) arabinose (Fig. 4C). In all cases, the relation between the transport activity and the inducer concentration matched the course of the intensity of the GFP fluorescence in gel (for LacS, LacY, and GltP) and in vivo (for LacS), suggesting that the fluorescent species observed in gel represents the fraction active protein.
Fig. 4.
Transport activities of membrane proteins expressed at different levels. Initial rates of transport were determined in whole E. coli MC1061 cells for LacS (A) and LacY-GFP (B), or membrane vesicles for GltP-GFP (C). Arabinose concentrations during induction were varied from 1 × 10−5% to 1 × 10−1% (wt/vol). E. coli MC1061 is devoid of the endogenous E. coli lactose transporter LacY, hence background lactose transport activity was absent. The GltP-GFP transport rates were corrected for background glutamate transport activity.
Characterization of the Nonfluorescent GFP Fusion Species.
The previous observations suggest that the fluorescent species is indicative for folded protein and, consequently, that the nonfluorescent species represents aggregated or unfolded protein. In E. coli, incorrectly folded protein often leads to the formation of inclusion bodies. We determined whether insoluble aggregates were present in cells induced at two extremes of the arabinose concentrations, which are conditions at which the upper band for LacS-GFP was either absent (1 × 10−3%) or prominently visible (2 × 10−1%). Indeed, we could isolate inclusion bodies containing LacS from cells induced with the high arabinose concentration, but not from cells induced with the intermediate arabinose concentration (1 × 10−3%) (Fig. S1). This observation confirms that the nonfluorescent GFP species is indicative of aggregated protein.
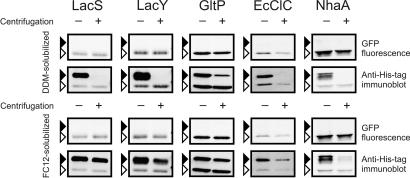
To validate this observation further, we analyzed whether the two GFP fusion species could be solubilized by mild detergents. Detergent-solubilized and properly folded membrane proteins are expected to sediment slower than their insoluble aggregates. If the upper band would consist of improperly folded and aggregated GFP fusion proteins, differential centrifugation should separate them from properly folded proteins. Cells, induced with high arabinose concentrations [1 × 10−1% (wt/vol)] to induce the formation of the upper band, were disrupted and solubilized with 1% (wt/vol) n-dodecyl-β-d-maltoside (DDM) or Fos-choline-12 (FC-12). Both DDM and FC-12 are highly effective in solubilizing membrane proteins (8–10), and DDM has been used successfully in obtaining structures of many membrane proteins (3). Solubilized cells expressing GFP fusions of LacS, LacY, GltP, EcClC, or NhaA were analyzed by in gel fluorescence and immunoblotting before and after ultracentrifugation (Fig. 5). The electrophoretic mobility and the intensities of the bands were not affected by the addition of the detergents (data not shown). However, after centrifugation, the band pattern changed significantly. For LacS, LacY, GltP, and NhaA, the lower, fluorescent band was almost quantitatively recovered for all fusion proteins solubilized in DDM (between 70% and 100% based on GFP fluorescence). For EcClC, the recovery of this band was ≈50% based on GFP fluorescence. For the DDM-solubilized proteins, centrifugation resulted in complete removal (LacS, LacY, EcClC, and NhaA) or a significant reduction (GltP) of the upper nonfluorescent band. These results indicate that the upper nonfluorescent band indeed represents the aggregated fraction.
Fig. 5.
Differential sedimentation of the two species of GFP fusion proteins. E. coli MC1061 cells were induced with 1 × 10−1% (wt/vol) l-arabinose. The GFP fusion proteins expressed are indicated above the panels. After disruption by bead beating, cells were solubilized by the addition of 1% DDM (Upper) or 1% FC-12 (Lower). Samples were taken before (−) and after (+) ultracentrifugation and analyzed by in gel fluorescence (indicated by GFP fluorescence) and immunodetection with an anti-His tag antibody (indicated by anti-His tag immunoblot). Black and white arrows indicate the positions of nonfluorescent and fluorescent species of the GFP fusion proteins, respectively.
Although qualitatively similar results were obtained with FC-12-solubilized material, the absolute decrease of the intensity of the upper band after centrifugation was less pronounced. To analyze this observation in more detail, we performed size-exclusion chromatography (SEC) on the supernatant of disrupted and FC-12-solubilized cells expressing GFP fusions of LacS, LacY, and GltP (Fig. S2). For all proteins, the lower (fluorescent) band peaked near the expected elution volume for the monomeric (LacS and LacY) or trimeric (GltP) species in a detergent and lipid micelle. For LacS and LacY, the upper band eluted as a very broad peak and was more abundant in the initial fractions, which is indicative of (partially) unfolded, less compact proteins. For GltP-GFP, a large fraction of the upper nonfluorescent band was lost during SEC. Whereas the intensities of the two bands were almost identical in the sample that was loaded onto the column, the intensity of the upper band in the elution fractions was severely reduced, possibly caused by retention of this species on the column or on the filter preceding the column. Overall, the results indicate that the fraction of the FC-12-solubilized GFP fusion that is represented by the upper band consists of misfolded protein, which surprisingly could not be pelleted easily. Possibly, this observation reflects the ability of FC-12 to solubilize aggregates of membrane proteins partially, preventing their complete sedimentation.
A final characterization was done to establish whether all of the aggregated protein was present in dense inclusion bodies or whether some of the misfolded material was membrane-associated. After cell rupturing, the dense and large inclusion bodies (and unbroken cells) should sediment in a low-speed centrifugation step and thereby separate from the membrane vesicles. We disrupted cells expressing LacS-GFP and analyzed the pellet and supernatant fractions obtained after low-speed centrifugation (Fig. S3). As expected, the pellet fraction showed a relative increase in the signal corresponding to the aggregated protein (upper band), indicating that part of the aggregated material was present as dense inclusion bodies (in agreement with Fig. S1). The pellet also contained a small amount of the lower, fluorescent band most likely because of the presence of intact cells that escaped disruption. In contrast, the supernatant fraction showed a relative increase in the signal of the lower fluorescent band, representing the well folded protein. But surprisingly, the supernatant also contained some aggregated protein. The supernatant was further analyzed by sucrose density centrifugation (Fig. S3). Again, the lower-density fraction containing the membrane vesicles also contained some of the aggregated protein that migrated in the upper band, confirming that the aggregated material is present not only in dense inclusion bodies but also in a form that cofractionates with the membranes, thereby preventing removal by centrifugation. Similar observations were made for cells expressing LacY-GFP (data not shown).
Taken together, our observations indicate that the intensity of the upper band represents aggregated protein. Consistently, this fraction of the GFP fusion protein is less prone to solubilization by detergents. In contrast, the intensity of the lower, fluorescent band corresponds to the GFP fluorescence observed in whole cells and the amount of active protein present. It represents the fraction of the GFP fusion proteins that is functionally expressed and readily solubilized upon exposure to detergents.
Discussion
Optimization of the expression of membrane proteins in a functional state often is an iterative process in which parameters such as the vector design (e.g., type of promoter and affinity tags) and cultivation conditions (e.g., host, inducer concentration, and temperature) are varied while monitoring the quantity and quality of the protein produced (4). Available methods for evaluation are based on the quantification of either the amount of properly folded protein (fluorescence of GFP fusions and activity assays) or the amount of aggregated species (inclusion body isolations), whereas data on both species would provide information on the expression bottlenecks, which is essential to guide the optimization process. Here, we describe a generic method for the rapid simultaneous quantification of folded and aggregated membrane protein.
The method is based on the use of GFP as indicator for stably folded protein (5, 6) and exploits the ability of folded GFP to maintain its tertiary structure during analysis by SDS/PAGE (this work and refs. 11 and 12). The approach takes advantage of the differential electrophoretic mobility of folded and misfolded GFP fusions. Immunodetection allows determination of the ratio of both species. Dual migration of GFP fusions has been observed (11), but an explanation was not provided. All complementary techniques clearly indicate that the upper band represents the fraction of aggregated protein, whereas the lower, fluorescent band represents properly folded, membrane-inserted, and active protein.
The two protein species could be separated by differential centrifugation after solubilization with the detergent DDM. Aggregated proteins sediment faster than detergent-solubilized membrane proteins and pelleted at appropriate centrifugation velocities while the properly folded proteins stayed in the supernatant. Removal of the aggregates from solubilized membranes by centrifugation was less complete for the detergent FC-12 than for DDM. SEC of FC-12-solubilized GFP fusions indicated that FC-12 could partially solubilize the aggregated material. In favor of this interpretation are the recurrent observations that FC-12 is one of the most effective detergents for the solubilization of membrane proteins (8–10). FC-12 has not been very successful in the crystallographic analysis of membrane proteins. The only membrane protein thus far crystallized in the FC-12-solubilized state, the rat monoamine oxidase A, contains only one isolated transmembrane segment (13).
The reliability of the method presented depends to a large extend on the quality of GFP as folding indicator. Previously, this application was validated by relating the GFP fluorescence to the amount of fusion protein present in the appropriate cell fraction (inclusion bodies, soluble or membrane-bound) (5, 6). Here, we extend this validation by relating the GFP fluorescence to the activity of the fusion partner. We observed excellent correlation between the activity of the transporter proteins and GFP fluorescence.
Our observations on several unrelated membrane proteins are in line with those of others (4), claiming that high expression levels often lead to aggregated protein, whereas reduction of the protein production rate increases the fraction of properly folded protein (Fig. 2). Importantly, the production rates at which misfolding and aggregation emerged varied per protein and did not relate to their absolute expression levels. Additionally, the fraction of protein that was properly folded diminished for some proteins toward higher expression levels (e.g., LacS, EcClC, and NhaA; Fig. 2) but stayed constant for others (e.g., GltP, DctA and GlpF; Fig. 2). Together, these observations emphasize that the optimization of protein expression needs to be done on an individual basis. The methodology presented here will facilitate this process.
The use of GFP fusions to optimize membrane protein overproduction can be integrated seamlessly with other downstream applications of GFP; e.g., a protocol to select the most effective detergents for solubilization of membrane proteins has been described (12). Also, by using fluorescent size-exclusion chromatography (FSEC), the stability of GFP fusions can be rapidly characterized without purification (14). To assure that purification procedures are sufficiently selective to remove the aggregated protein from the properly folded fraction, the in gel mobility shift assay presented here can be applied. Apart from these analytical applications, removal of the His-tagged GFP moiety by digestion with the tobacco etch virus (TEV) protease on a preparative scale allows further processing of the nonfused membrane protein for functional assays or crystallization trials.
In summary, we present a generic method for the simultaneous quantification of folded and aggregated membrane protein using GFP fusions. The method requires only standard equipment and small culture samples; it is not labor-intensive and can greatly facilitate the optimization of the overexpression of both membrane and soluble proteins.
Materials and Methods
Plasmid Constructions.
E. coli MC1061 (15) was used as the cloning host. DNA manipulations were done according to standard protocols. All vectors used in this work were derivatives of pBADMycHisB (Invitrogen). The construction of pBADLacSC320AΔIIA, pBADLacSC320AΔIIA-GFP, and pBADcLIC has been described (16, 17). Plasmid pBADcLIC-GFP was constructed from a 4,045-bp SwaI–XbaI fragment of pBADcLIC and an XbaI-digested PCR product (784 bp) holding an NcoI-free sequence coding for EGFP. The sequence coding for EGFP was obtained from pET28aGltP-TEV-GFPHis8 (18), a derivative of pWaldo (5). By using ligation-independent cloning, genes cloned in pBADcLIC were expressed with a small N-terminal extension (MGGGFA; resulting from cloning-related sequences) and a C-terminal TEV protease cleavage site (ENLYFQG) followed by a His10 tag. Genes cloned in pBADcLIC-GFP have similar modifications but additionally have the gene coding for the EGFP protein inserted between the sequences for the TEV protease cleavage site and the His10 tag. Plasmid pBADcLIC_LacSΔIIA and pBADcLIC-GFP_LacSΔIIA, containing the lacSΔIIA gene, were constructed by ligation-independent cloning as described in ref. 17. LacSΔIIA is a truncation mutant of LacS devoid of the regulatory soluble IIA domain (16); for convenience, in the main text and figures we refer to LaSDIIA as LacS. The construction of derivatives of pBADcLIC and pBADcLIC-GFP, containing gltP, dctA, ydjN, and sstT, will be described elsewhere. Derivatives of pBADcLIC and pBADcLIC-GFP, containing lacY, ecClC, nhaA, and glpF, were a kind gift from G. B. Erkens (University of Groningen). Derivatives of pWaldo, containing gltP, dctA, and ydjN, were a kind gift from D. O. Daley (Stockholm University).
Transport Assays.
LacS.
Lactose counterflow transport was assayed in E. coli MC1061/pBADLacSC320AΔIIA as described in ref. 16.
LacY.
Lactose transport driven by the proton motive force was assayed in E. coli MC1061/pBADcLIC-LacY-GFP as described for LacS but at 23°C (19).
GltP.
Glutamate transport driven by the proton motive force was assayed in membrane vesicles of E. coli MC1061/pBADcLIC-GltP-GFP as described in ref. 20.
Cultivation.
E. coli MC1061 (15), TOP10 (21), and BL21(DE3) were cultivated at 37°C on Luria broth supplemented with 100 μg/ml ampicillin under vigorous aeration. Cultivation of E. coli MC1061 and E. coli TOP10 containing derivatives of pBADMycHisB (Invitrogen) was started with a 1–2% (vol/vol) inoculum of an overnight culture. Cells were grown until OD660 = ≈0.6 was reached and induced with the appropriate amount of l-arabinose. Cultivation was continued for 2 h. Cultivation of E. coli BL21(DE3) containing derivatives of pWaldo was done as described above for E. coli MC1061, but cells were induced with 0.4 mM IPTG.
Purification of Inclusion Bodies.
Whole-cell pellets of E. coli MC1061/pBADLacSC320AΔIIA, corresponding to ≈7 mg of protein, were resuspended in 1.25 ml of 50 mM Tris·HCl, 1 mM EDTA (pH 8.0), 0.73 M sucrose plus 2 mg/ml lysozyme and incubated on ice for 30 min. Subsequently, MnCl2, MgCl2, and DNase were added to final concentrations of 1.8 mM, 18 mM, and 0.8 mg/ml, respectively, and the sample was vortexed and incubated on ice for 30 min. After the addition of 3 ml of detergent buffer [20 mM Tris·HCl (pH 7.5), 200 mM NaCl, 10 mg/ml deoxycholate, 10 mg/ml Nonidet P-40, 2 mM EDTA, and 10 mM 2-mercaptoethanol], the sample was vortexed thoroughly and centrifuged for 5 min at 10,000 ×g at 4°C. Next, the pellet was resuspended two times in 3 ml of Tris-Triton buffer [20 mM Tris·HCl (pH 7.5), 0.5% (wt/vol) Triton X-100, 1 mM EDTA plus 10 mM 2-mercaptoethanol] and centrifuged for 5 min at 10,000 × g at 4°C, followed by two rounds of resuspension in 3 ml of Tris–DTT buffer [50 mM Tris·HCl (pH 8.0), 1 mM EDTA plus 10 mM DTT] and centrifugation at 10,000 × g for 5 min at 4°C. Pellets were resuspended in 200 μl of 7 M urea and stored at −80°C until use.
In Gel Fluorescence and Immunodetection.
Whole-cell samples corresponding to ≈1 mg of protein were resuspended in 400 μl of ice-cold 50 mM KPi (pH 7.2), 1 mM MgSO4, 10% (wt/vol) glycerol, 1 mM PMSF, and trace amounts of DNase. Glass beads (300 mg, 0.1-mm diameter) were added, and samples were shaken in a FastPrep device (Bio101) for 20 s at force 6. This procedure was repeated once after cooling the samples for 5 min on ice. Aliquots (40 μl, 100 μg of protein) were taken, and 10 μl of 5× protein sample buffer [120 mM Tris·HCl (pH 6.8), 50% glycerol, 100 mM DTT, 2% (wt/vol) SDS, and 0.1% (wt/vol) bromophenol blue] was added. Samples were stored on ice until use.
Protein samples were analyzed by 10% SDS/PAGE, and in gel GFP fluorescence was immediately visualized with an LAS-3000 imaging system (Fujifilm) and AIDA software (Raytest). Subsequently, gels were submitted to semidry electroblotting and immunodetection with a primary antibody raised against a His6 tag (Amersham Pharmacia Biotech). Chemiluminescence detection was done by using the Western light kit (Tropix, Inc.) and the Fujifilm LAS-3000 imaging system.
Differential Sedimentation of Folded and Aggregated Proteins.
Whole-cell samples were prepared as described for in gel fluorescence and immunodetection. After bead beating, aliquots (100 μl, 250 μg) were transferred to TLA 100.1 centrifuge tubes. Samples were solubilized for 45 min at 4°C in the presence of 1% (wt/vol) DDM or FC-12 and occasionally mixed by brief vortexing. A small aliquot (20 μl, 45 μg) was removed to represent the sample before ultracentrifugation. Next, the remaining sample was centrifuged at 355,000 × g for 10 min at 4°C in a TLA 100.1 rotor. A small aliquot (20 μl, 45 μg) was removed from the supernatant to represent the sample after ultracentrifugation. Both aliquots were mixed with 5 μl of 5× sample buffer and analyzed by in gel fluorescence and immunodetection.
SEC of Disrupted, FC-12-Solubilized Cells.
Whole-cell samples were prepared as described for in gel fluorescence and immunodetection, except that each sample contained ≈7.2 mg of protein and 600 μl of ice-cold 50 mM KPi (pH 7.2), 1 mM MgSO4, 10% (wt/vol) glycerol, 1 mM PMSF, and trace amounts of DNase. Two preparations were processed for each sample, and after bead beating, 400 μl was recovered from each. These fractions were pooled. Aliquots of 540 μl were mixed with 60 μl of 10% (wt/vol) FC-12 and incubated on ice for 1 h with occasional mixing. Next, the solubilized material was centrifuged at 355,000 × g for 10 min at 4°C in a TLA 100.1 rotor (Beckmann). Samples were taken before and after centrifugation to monitor the band pattern by in gel fluorescence and immunodetection. A 200-μl fraction of the supernatant was analyzed by SEC with a Superdex 200 column (10/300 GL; Amersham Biosciences) preequilibrated with 50 mM KPi (pH 7), 200 mM NaCl, and 0.04% (wt/vol) DDM, similar to that described in refs. 10 and 14. Fractions obtained from the SEC were analyzed as described for in gel fluorescence and immunodetection.
Isolation of Membrane Vesicles.
Whole cells corresponding to ≈160 mg of protein were resuspended in 40 ml of ice-cold 50 mM KPi (pH 7.2), 1 mM MgSO4, 10% (wt/vol) glycerol, 1 mM PMSF, and a few milligrams of DNase. Cells were passed twice through a French pressure cell at 10,000 psi, supplemented with 2 mM EDTA (pH 7.5) (the resulting sample was labeled “disrupted cells”), followed by centrifugation at 22,500 × g for 10 min at 4°C in a JA 25.5 rotor. Without disturbing the pellet, 25 ml of the supernatant was removed (the resulting fraction was labeled “LS supernatant”). The remaining supernatant was disposed of, and the pellet was resuspended in ice-cold 50 mM KPi (pH 7.2), to the original volume (40 ml; fraction labeled “LS pellet”). Aliquots (10 μl) of the fractions were analyzed by in gel fluorescence and immunoblotting. Subsequently, 1 ml of the LS supernatant fraction was supplemented with 115 μl of 87% (wt/vol) glycerol and layered on top of a step sucrose gradient consisting of 1 ml of 55%, 2 ml of 51%, 1 ml of 45%, and 1 ml of 36% (wt/vol) sucrose. All sucrose solutions contained 50 mM KPi (pH 7.5). The gradient was centrifuged at 250,802 × g for 40 min at 4°C in an MLA 80 rotor. The top 5.1 ml of the gradient was removed and homogenized by vortexing (fraction labeled “top”). The remaining 1 ml was adjusted to 6.1 ml with 50 mM KPi (pH 7.5) and homogenized (fraction labeled “bottom”). Aliquots (31 μl) of the fractions were analyzed by in gel fluorescence and immunoblotting.
Microscopic Imaging of LacSΔIIA-GFP.
Cells were pelleted, washed twice with ice-cold 50 mM KPi (pH 7.2), plus 2 mM MgSO4 (KPM) to remove adhering medium components and l-arabinose, and resuspended to ≈OD660 = 120 (≈36 mg of protein per ml). After overnight incubation, cells were deenergized by incubation with 50 μM SF6847 plus 30 mM NaN3 for 2 h. Aliquots of a cell suspension at OD660 of 0.6 were placed on a coverslip that held an imaging chamber gasket (Molecular Probes); the chamber was sealed by a microscope slide. The cells were allowed to sediment for at least 30 min before imaging. The fluorescence of LacS and LacS-GFP was examined by confocal laser microscopy, by using a commercial ConfoCor2 combination (Zeiss) with a Plan-Apochromat ×63, NA 1.4 oil-immersion objective. Samples were excited at 488 nm, and emission was detected at 500–550 nm. Parallel with confocal images of fluorescence (8 bit, 512 × 511 pixels), cells were imaged by differential interference contrast with Normarski optics.
Supplementary Material
Acknowledgments.
We thank G. van den Bogaart, D. M. Veltman, and A. M. Krikken for technical assistance with the confocal microscopy. We thank G. B. Erkens and D. O. Daley for the kind gift of several plasmids used in this work. This work was funded by European Membrane Protein Consortium (E-MeP, 504601), The Netherlands Organisation for Scientific Research (NWO, Vidi fellowship to D.-J.S.) and the Netherlands Proteomics Centre.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802190105/DCSupplemental.
References
- 1.Drew D, Froderberg L, Baars L, de Gier JW. Assembly and overexpression of membrane proteins in Escherichia coli. Biochim Biophys Acta. 2003;1610:3–10. doi: 10.1016/s0005-2736(02)00707-1. [DOI] [PubMed] [Google Scholar]
- 2.Dalbey RE, Chen M. Sec-translocase-mediated membrane protein biogenesis. Biochim Biophys Acta. 2004;1694:37–53. doi: 10.1016/j.bbamcr.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Raman P, Cherezov V, Caffrey M. The Membrane Protein Data Bank. Cell Mol Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang DN, et al. Practical aspects of overexpressing bacterial secondary membrane transporters for structural studies. Biochim Biophys Acta. 2003;1610:23–36. doi: 10.1016/s0005-2736(02)00709-5. [DOI] [PubMed] [Google Scholar]
- 5.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 6.Drew DE, von Heijne G, Nordlund P, de Gier JW. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 2001;507:220–224. doi: 10.1016/s0014-5793(01)02980-5. [DOI] [PubMed] [Google Scholar]
- 7.Terpe K. Overview of bacterial expression systems for heterologous protein production: From molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 8.Eshaghi S, et al. An efficient strategy for high-throughput expression screening of recombinant integral membrane proteins. Protein Sci. 2005;14:676–683. doi: 10.1110/ps.041127005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White MA, Clark KM, Grayhack EJ, Dumont ME. Characteristics affecting expression and solubilization of yeast membrane proteins. J Mol Biol. 2007;365:621–636. doi: 10.1016/j.jmb.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newstead S, Kim H, von Heijne G, Iwata S, Drew D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:13936–13941. doi: 10.1073/pnas.0704546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki T, Takahashi Y, Koch KS, Leffert HL, Watabe H. Construction of a fusion protein between protein A and green fluorescent protein and its application to Western blotting. FEBS Lett. 1996;384:193–197. doi: 10.1016/0014-5793(96)00289-x. [DOI] [PubMed] [Google Scholar]
- 12.Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. Optimization of membrane protein overexpression and purification using GFP fusions. Nat Methods. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- 13.Ma J, et al. Structure of rat monoamine oxidase A and its specific recognitions for substrates and inhibitors. J Mol Biol. 2004;338:103–114. doi: 10.1016/j.jmb.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 14.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Wertman KF, Wyman AR, Botstein D. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene. 1986;49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]
- 16.Geertsma ER, Duurkens RH, Poolman B. The activity of the lactose transporter from Streptococcus thermophilus is increased by phosphorylated IIA and the action of β-galactosidase. Biochemistry. 2005;44:15889–15897. doi: 10.1021/bi051638w. [DOI] [PubMed] [Google Scholar]
- 17.Geertsma ER, Poolman B. High-throughput cloning and expression in recalcitrant bacteria. Nat Methods. 2007;4:705–707. doi: 10.1038/nmeth1073. [DOI] [PubMed] [Google Scholar]
- 18.Drew D, et al. A scalable, GFP-based pipeline for membrane protein overexpression screening and purification. Protein Sci. 2005;14:2011–2017. doi: 10.1110/ps.051466205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geertsma ER, Duurkens RH, Poolman B. Identification of the dimer interface of the lactose transport protein from Streptococcus thermophilus. J Mol Biol. 2003;332:1165–1174. doi: 10.1016/j.jmb.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Groeneveld M, Slotboom DJ. Rigidity of the subunit interfaces of the trimeric glutamate transporter GltT during translocation. J Mol Biol. 2007;372:565–570. doi: 10.1016/j.jmb.2007.06.067. [DOI] [PubMed] [Google Scholar]
- 21.Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.