Abstract
Accurate profiling of lipidomes relies upon the quantitative and unbiased recovery of lipid species from analyzed cells, fluids, or tissues and is usually achieved by two-phase extraction with chloroform. We demonstrated that methyl-tert-butyl ether (MTBE) extraction allows faster and cleaner lipid recovery and is well suited for automated shotgun profiling. Because of MTBE's low density, lipid-containing organic phase forms the upper layer during phase separation, which simplifies its collection and minimizes dripping losses. Nonextractable matrix forms a dense pellet at the bottom of the extraction tube and is easily removed by centrifugation. Rigorous testing demonstrated that the MTBE protocol delivers similar or better recoveries of species of most all major lipid classes compared with the “gold-standard” Folch or Bligh and Dyer recipes.
Keywords: mass spectrometry, sample preparation, direct infusion, nano-ESI-MS
Recent developments in mass spectrometric technology enabled the comprehensive characterization of eukaryotic lipidomes, fostering the molecular biology of lipids and metabolism-related disorders (reviewed in Refs. 1–4). Typically, lipidome profiling by mass spectrometry proceeds along LC-MS or shotgun approaches. The former identifies and quantifies lipid species preseparated by normal or reversed-phase chromatography coupled online to a mass spectrometer, which is capable of fast acquisition of MS or MS/MS spectra (5–8). In contrast, in shotgun lipidomics, total lipid extracts are infused directly into a mass spectrometer, and the molecular characterization of lipid species relies either on the accurately determined m/z of precursor ions (9) or on the detection of specific fragment ions or neutral losses in tandem mass spectrometric experiments (1, 9–12).
Regardless of the analytical approach used, its success depends on the completeness of the extraction of lipids from corresponding cells, fluids, or tissues. Lipids of all major classes could be recovered via chloroform/methanol extraction, typically according to the Folch, Lees, and Sloane Stanley (13) or Bligh and Dyer (14) recipes (15), in which they are mostly enriched in the chloroform phase.
Electrospray mass spectrometry, a major tool for analyzing complex lipidomes, is particularly sensitive towards the quality of lipid extracts. Coextracted components of biological matrix and salts (often, without further definition, termed background) affect both the sensitivity and specificity of lipid analysis. Often, abundant background ions obscure lipid precursors, and their MS/MS spectra are densely populated with “ghost” peaks and abundant chemical noise. Adducts with common background cations (sodium, potassium) and anions (chloride) increase the ambiguity of molecular species assignment and affect the accuracy of quantitative determination.
Because of the higher density of chloroform compared with a water/methanol mixture, it forms the lower phase of the two-phase partitioning system. While collecting the chloroform fraction, a glass pipette or a needle of the pipetting robot reaches it through a voluminous layer of nonextractable insoluble matrix, usually residing at the interface of the water/methanol and chloroform phases. However, even a minute amount of insoluble precipitate accidentally grabbed together with the chloroform fraction clogs the electrospray ion source or LC system, because of the micrometer size of the spraying orifice and/or connecting tubing. We note that, because of high density and the viscosity of chloroform, centrifugation is usually of little help. Although mass spectrometry enables lipid profiling at the low femtomole level, much higher amounts are usually required to circumvent the difficulties in handling microvolumes of total extracts and ensure the sufficient stability of the analytical pipeline.
Additionally, the known carcinogenicity of chloroform involves considerable health risk for laboratory personnel (16). Also, chloroform decomposition yields phosgene and hydrochloric acid, inflicting chemical modification of labile lipid species (17).
Here, we report an extraction protocol specifically developed for shotgun profiling of complex lipidomes from samples with excessive amounts of biological matrices. Lipid extraction by methyl-tert-butyl ether (MTBE)/methanol (18, 19) greatly simplifies sample handling and enables automated processing of minute amounts of biological samples. Rigorous testing established that the recovery of lipid species of almost all major classes is the same or better than was typically achieved by the Folch recipe (13), which is generally regarded the “gold standard” in lipid biochemistry.
MATERIALS AND METHODS
Materials and lipid standards
Synthetic lipid standards were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL); MTBE and 2-propanol were from Sigma-Aldrich Chemie GmbH (Munich, Germany). Chloroform, methanol, and ammonium acetate were LC grade; water with 0.1% ammonium acetate was LC-MS grade and purchased from Fluka (Buchs, Switzerland). LC-MS-grade water was purchased from Fisher Scientific (Loughborough, UK).
Sample preparation
Escherichia coli (NA-22 strain) were grown on Luria-Bertani medium, collected by centrifugation, washed three times by M9 solution (22 mM KH2PO4, 22 mM Na2HPO4, 85 mM NaCl, and 1 mM MgSO4) followed by rinsing with 0.1% ammonium acetate, and frozen.
A sample of mouse brain tissue was dissected from adult mouse of NMRI strain. Brain hemispheres were separated and minced into small pieces in ice-cold 0.1% ammonium acetate followed by homogenization in a Potter homogenizer.
The daf-22 strain of Caenorhabditis elegans was grown on NGM agar plates with E. coli (NA-22 strain) as a food source (20). To collect eggs, worms were bleached with basic hypochlorite solution as described (21). To remove worm debris, egg suspension was filtered through 80 μm nylon mesh, rinsed with LC-MS-grade water, and frozen in liquid nitrogen in 200 μl aliquots.
Lipid profiling by quadrupole time-of-flight mass spectrometry
Mass spectrometric analysis was performed on a modified QSTAR Pulsar i quadrupole time-of-flight mass spectrometer (MDS Sciex, Concord, Canada) equipped with a robotic nanoflow ion source NanoMate HD (Advion BioSciences, Ltd., Ithaca, NY). Ionization voltage was set to 1.05 kV, gas pressure to 1.25 p.s.i., and the source was controlled by Chipsoft 6.3.2 software (Advion BioSciences). All lipid samples were diluted in MS-mix buffer [7.5 mM ammonium acetate in chloroform-methanol-2-propanol (1:2:4, v/v/v)] and infused at the flow rate of ∼250 nl/min. A typical sample volume of 10 μl allowed 35 min of stable electrospray time. Data-dependent acquisition (DDA) experiments were performed as described previously (22). The analytical quadrupole Q1 was operated under the unit resolution settings, and fragments were detected within the m/z range 100–1,000. Each MS/MS spectrum was acquired for 10–30 s under the fixed collision energy offset of 40 eV. Lipid species were identified and quantified using LipidInspector software (22).
Mass spectrometric analysis of lipids from human plasma extracts
Plasma lipid extracts were analyzed by direct flow injection on a triple quadrupole mass spectrometer (Quattro Ultima; Micromass, Manchester, UK) equipped with an electrospray ion source as described (23, 24). Briefly, 20 μl of total lipid extracts was injected for each lipid class-specific analysis using a HTS PAL autosampler (Zwingen, Switzerland) and an Agilent 1100 binary pump (Waldbronn, Germany). A mixture of methanol-chloroform (3:1, v/v) containing 7.5 mM ammonium acetate was purged through the system starting at the flow rate of 55 μl/min for 6 s, followed by 30 μl/min for 60 s, and then increased to 250 μl/min for another 12 s. Data were acquired for 1.3 min. Quantification of phosphatidylcholine (PC), sphingomyelin (SM), lysophosphatidylcholine, phosphatidylethanolamine (PE), PE-plasmalogen, and ceramide (Cer) relied upon lipid class-specific fragments (Table 1). Free cholesterol and cholesteryl esters (CEs) were analyzed by selected reaction monitoring (25). Data analysis, including deisotoping and quantification, was performed as described (23, 26).
TABLE 1.
Lipid class-specific mass spectrometric identification
| Lipid Class | Fragment Ion or Neutral Loss |
|---|---|
| neutral loss, amu | |
| Phosphatidylethanolamine | 141.02 |
| Phosphatidylserine | 185.01 |
| Phosphatidylglycerol | 189.04 |
| fragment ion(s), m/z | |
| Phosphatidylcholine | 184.07 |
| Lysophosphatidylcholine | 184.07 |
| Sphingomyelin | 184.07 |
| Hexosylceramide | 264.25 |
| Ceramide | 264.25 |
| PE-plasmalogen | Boolean scansa |
| PE-O-16:0p/… | 364.25 |
| PE-O-18:1p/… | 390.25 |
| PE-O-18:0p/… | 392.25 |
TLC analysis
TLC analysis of lipid extracts was performed on Merck Silica Gel 60 HPTLC plates (10 × 10 cm). Plates were developed with an ethanol/CHCl3/triethylamine/water (40:35:35:9, v/v/v/v) solvent system, followed by n-hexane-ethylacetate (5:1, v/v). Lipid bands were visualized by spraying the plates with 20% H2SO4 in ethanol and charring at 200°C.
Lipid extraction recipes
Extraction with MTBE
Methanol (1.5 ml) was added to a 200 μl sample aliquot, which was placed into a glass tube with a Teflon-lined cap, and the tube was vortexed. Then, 5 ml of MTBE was added and the mixture was incubated for 1 h at room temperature in a shaker. Phase separation was induced by adding 1.25 ml of MS-grade water. Upon 10 min of incubation at room temperature, the sample was centrifuged at 1,000 g for 10 min. The upper (organic) phase was collected, and the lower phase was reextracted with 2 ml of the solvent mixture, whose composition was equivalent to the expected composition of the upper phase [obtained by mixing MTBE/methanol/water (10:3:2.5, v/v/v) and collecting the upper phase]. Combined organic phases were dried in a vacuum centrifuge. To speed up sample drying, 200 μl of MS-grade methanol was added to the organic phase after 25 min of centrifugation. Extracted lipids were dissolved in 200 μl of CHCl3/methanol/water (60:30:4.5, v/v/v) for storage.
Extraction according to Folch
Methanol (1.5 ml) was added to 200 μl of the sample aliquot and vortexed. Then, 3 ml of CHCl3 was added, the mixture was incubated for 1 h at room temperature in a shaker, and then phase separation was induced by adding 1.25 ml of water. The extract was left for 10 min at room temperature and then centrifuged at 1,000 g for 10 min. The lower (chloroform) phase was collected, and the upper phase was washed with 2 ml of the solvent mixture, whose composition was equivalent to the assumed composition of the lower phase (CHCl3/methanol/water, 86:14:1, v/v/v). Combined organic phases were dried in a vacuum centrifuge and dissolved in 200 μl of CHCl3/methanol/water (60:30:4.5, v/v/v) for storage.
Recovery of lipid standards
Aliquots of 20 μl of 20 μM lipid standard solutions in CHCl3/methanol/water (60:30:4.5, v/v/v) were pipetted into 2 ml Eppendorf tubes and dried in a vacuum centrifuge. A total of 20 μl of MS-grade water was added to each tube, and lipid extraction was performed according to the Folch or MTBE protocol as described above. Collected organic phases were dried and redissolved in 1 ml of MS-mix containing 400 nM of another lipid of the same class that served as an internal standard. As internal standards, PC 18:0/18:0, PE 17:0/17:0, phosphatidylinositol (PI) 17:0/17:0, C24:1 β-d-galactosylceramide, and diacylglycerol 16:0/18 were used. Control samples were prepared by pipetting the same volumes of lipid stock solutions into Eppendorf tubes, drying, and dissolving in MS-mix containing the same amount of the internal standard. Each extraction experiment was performed in triplicate, and each extract was independently analyzed three times. Survey time-of-flight (TOF) MS spectra were acquired for 5 min with an accumulation time of 1 s. The lipid standard recovery was estimated as the ratio of the intensities of monoisotopic peaks of the extracted compound and the internal standard of the same lipid class.
In a separate series of experiments, the PI lipid standard was processed as described above, but instead of water 20 μl of E. coli cell suspension containing 1.8 E07 cells was added to the tube. The recovery was estimated by multiple reaction monitoring in negative ion mode as the ratio of the peak areas of the phosphoinositol fragment with m/z 241.02 obtained from the extracted compound and the internal standard.
Comparison of MTBE and Folch extracts from E. coli
Cells from overnight bacteria culture grown on DMEM were collected by centrifugation, washed with 0.1% ammonium acetate in water, and resuspended in 0.1% ammonium acetate. Six aliquots, each containing 5.4 E10 cells in 200 μl, were placed into glass tubes; three aliquots were extracted according to Folch and the other three aliquots were extracted with MTBE as described above. Lipid extracts were diluted 1:100 with MS-mix and independently analyzed two times.
Lipid profiles were obtained by the interpretation of DDA data sets by LipidInspector software, which emulated specific neutral loss scans, precursor ion scans, and Boolean scans (25). For E. coli extracts, profiles of PE and phosphatidylglycerol (PG) lipid classes were obtained by emulating neutral loss scans of head group fragments with m/z 141.02 and m/z 189.04, respectively (Table 1). The PI profile was determined in negative ion mode by emulating precursor ion scans for the head group fragment with m/z 241.02. Peak areas of individual lipid species were normalized to the sum of peak areas of all detected lipid species of the lipid class.
Comparison of MTBE and Folch extracts from mouse brain
Brain tissue from adult mouse NMRI strain was rinsed in 0.1% ammonium acetate in water, cut into small pieces, and homogenized in 0.1% ammonium acetate in water in a Potter homogenizer on ice. Aliquots (200 μl) of homogenate containing 40 mg of proteins were taken in triplicate and extracted according to the Folch and MTBE protocols. The lipid extracts were diluted 1:100 with MS-mix and analyzed twice. Profiles for PE, PE-plasmalogen, phosphatidylserine (PS), PC, and hexosylceramides were obtained as described above (Table 1).
Comparison of MTBE and Folch lipid extracts of C. elegans
Six aliquots of the suspension in MS water of C. elegans eggs (each of 200 μl or ∼900 eggs) were subjected to three rounds of freezing-thawing, and then three aliquots were extracted according to the Folch protocol and three were extracted according to the MTBE protocol. To obtain lipid profiles, each extract was diluted 1:10 with MS-mix and analyzed in triplicate. PC, PE, PE-plasmalogen, and PS lipid profiles were determined as described above (Table 1).
Comparison of MTBE with the Bligh and Dyer extraction of human blood plasma
Lipid extracts were prepared from 20 μl of human EDTA plasma transferred into glass centrifuge tubes. Before lipid extraction, internal standards [combined solution containing PC 28:0, PC 44:0, PE 28:0, PE 40:0, lysophosphatidylcholine (LPC) 13:0, LPC 19:0, Cer 14:0, Cer 17:0, D7-cholesterol, CE 17:0, and CE 22:0] dissolved in chloroform were placed into the same tubes and evaporated. Additionally, calibration samples were prepared by the addition of known quantities of naturally occurring lipid species.
For Bligh and Dyer extraction, 20 μl of plasma was diluted up to the volume of 800 μl with water. Three milliliters of the methanol-chloroform (2:1, v/v) mixture was added and left for 1 h at room temperature (14). Phase separation was achieved by adding 1 ml of CHCl3 and 1 ml of water. For MTBE extraction, 80 μl of water was added to the plasma samples and extracted as described above, although repetitive extraction of the lower (aqueous) phase was omitted.
Two milliliters of organic phase collected according to both protocols was further processed by a pipetting robot (Tecan Genesis, Männedorf, Switzerland) equipped with four fixed Teflon-coated needles. To avoid cross-contamination, the needle was washed with methanol-chloroform (1:1, v/v) solvent. For both lipid extraction procedures, the organic phase was recovered at a fixed z-position from the lower chloroform and upper MTBE phases, respectively. Both positions were chosen to avoid extract contamination by the aqueous phase. Z-movement of the needles was performed with reduced speed for the chloroform phase to prevent sample dripping. Organic phases were transferred to standard 1.5 ml glass autosampler vials.
In total, 1,600 μl of the organic phase was recovered from each sample, of which 1,000 μl was used for phospholipid analysis and the remaining 600 μl was used for cholesterol analysis. The solvent was removed by vacuum centrifugation, and lipids were dissolved in 1,000 and 600 μl of methanol-chloroform (3:1, v/v) containing 7.5 mM ammonium acetate for phospholipid and cholesterol analysis, respectively.
To determine the recoveries of PCs, SMs, PEs, LPCs, Cers, cholesterol, and CEs, three plasma samples with low, medium, and high lipid content (ranked by a separately determined total cholesterol index as follows: low, <83 mg/dl; medium, 207–282 mg/dl; high, >282 mg/dl) were extracted in triplicate.
Absolute quantities of lipid species were determined by extracting 10 aliquots of a plasma sample 10 times as described above.
RESULTS
Lipid extraction with MTBE
Previous observations indicated that MTBE could replace chloroform in popular solvent systems used for lipid extraction. The established extraction routine is, in general, similar to the Folch or Bligh and Dyer protocols. In the first step, samples were extracted within a one-phase system allowing optimal contact between extraction solvent and biological material.
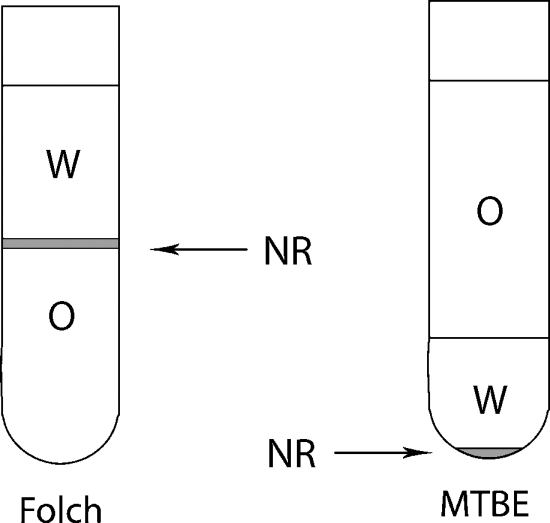
Lipids were recovered into the MTBE phase, which, because of its lower density, was the upper phase of the two-phase solvent system. In contrast to the Folch recipe, nonextractable matrix resided in the aqueous phase at the bottom of the extraction vial, whereas the organic phase enriched with lipids was easily accessible by the micropipette from the top (Fig. 1). This feature was especially useful for extracting lipids from C. elegans, which left behind voluminous nonextractable debris that heavily contaminated the chloroform fraction collected with the Folch recipe; therefore, an additional microfiltering step was required to clear it up before ESI-MS analysis.
Fig. 1.
Methyl-tert-butyl ether (MTBE) lipid extraction method. Phase distribution in the MTBE and Folch methods. NR, insoluble (protein) residue; O, organic phase; W, water phase.
Validation of the MTBE extraction protocol
The MTBE extraction procedure was validated in three ways. First, we determined the absolute recoveries of several synthetic lipid standards of various classes under fully controlled experimental settings and compared them with the recoveries achieved by the Folch method. Once we established that the recovery of both methods was almost quantitative, we used the Folch recipe as a reference to determine whether lipid yields were influenced by various biological matrices and whether they depend on the structural properties of lipid species. Finally, MTBE extraction was automated and applied for high-throughput screening of human blood plasma lipidomes.
Absolute recoveries of lipid standards by the MTBE and Folch methods
Using synthetic standards, we first determined what absolute lipid recoveries were achievable using the Folch and MTBE recipes. To this end, 20 pmol of lipid standards of five lipid classes was extracted, and their recoveries were determined by mass spectrometry using internal standards of the same class and similar mass and whose concentrations were exactly known (Table 2). Depending on the lipid class, the recovery achieved by both methods was very similar, varying within 90–98%. The only notable exception was the PI standard, which was more extractable by MTBE. The same tendency was observed in extracting the PI standard spiked into E. coli total lipid extract: 67.3 ± 4.7% was recovered by Folch and 81.3 ± 8.1% by MTBE. Therefore, we concluded that, in further experiments, we could benchmark MTBE extraction efficiency relative to the Folch method, relying upon the relative abundance of peaks of individual species.
TABLE 2.
Recovery of lipid standards by MTBE and Folch extraction protocols
| Recoveries
|
||
|---|---|---|
| Lipid | Folch Extraction | MTBE Extraction |
| Phosphatidylethanolamine 18:0/22:6 | 90.9 ± 2.1 | 92.6 ± 3.5 |
| Phosphatidylcholine 16:0/16:0 | 91.6 ± 1.6 | 87.1 ± 3.3 |
| Phosphatidylinositol 16:0/16:0 | 68.6 ± 5.0 | 91.7 ± 2.9 |
| Glu(β) C12 Ceramide | 96.2 ± 3.9 | 98.9 ± 2.2 |
| Diacylglycerol 16:0/16:0 | 96.3 ± 2.1 | 92.6 ± 3.5 |
Values shown are % (means ± SD). Recoveries of 20 pmol of lipid standards after Folch or MTBE extraction were determined by comparison with the intensities of monoisotopic peaks of the corresponding internal standards.
The yield of extracted lipids is independent of the biological matrix
The MTBE and Folch protocols were applied to recover lipids from several popular model organisms, such as E. coli, mouse brain, and C. elegans, representing an example of cell culture, intact animal, and animal tissue, respectively. Exactly the same samples were extracted in parallel by both methods, recovered lipids were analyzed by TLC, and individual lipid species were quantified by shotgun DDA profiling on a quadrupole time-of-flight mass spectrometer (22).
The E. coli lipidome consists mostly of PE (∼75%) and PG (∼20%) (27). Almost identical (with respect to detected species and absolute signal intensities) survey TOF mass spectra were acquired, suggesting that the extraction yields were similar for both methods and were independent of lipid class and the individual fatty acid composition of the species. This notion was independently confirmed by TLC analysis (see supplementary Fig. IA). Furthermore, the m/z profiles of background peaks were also similar.
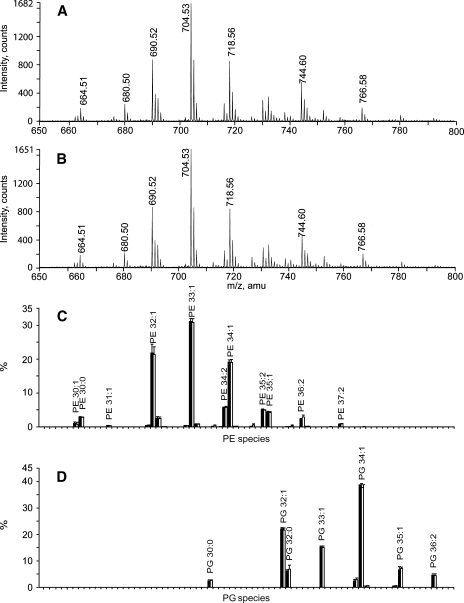
DDA-driven lipid profiling in positive ion mode revealed the same total number and mol% content of PE and PG species (Fig. 2C, D). In total, we quantified 27 species of PEs and 11 species of PGs (see supplementary Fig. IIA), which again agreed with recently published evidence (28).
Fig. 2.
Glycerophospholipids from E. coli recovered by Folch and MTBE extraction. A: Time-of-flight (TOF)-MS spectrum of the Folch lipid extract. B: TOF MS spectrum of the MTBE lipid extract. C: Phosphatidylethanolamine (PE) profiles of Folch-extracted (black bars) and MTBE-extracted (white bars) lipids; intensities were normalized to the sum of all PE species (means ± SD; n = 6). D: Phosphatidylglycerol (PG) lipid profiles, with notations as in C.
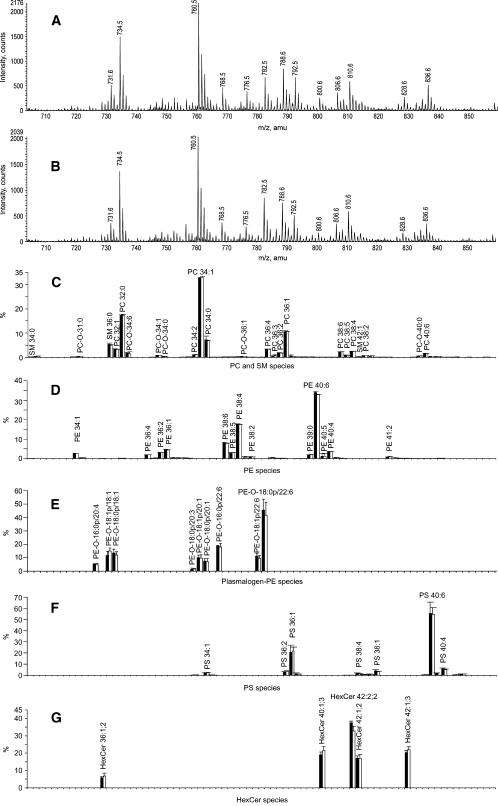
Next, the Folch and MTBE protocols were applied to extract lipids from adult mouse brain tissue. TOF MS analysis of the extracts produced almost identical spectra (Fig. 3A, B) and was further corroborated by TLC analysis (see supplementary Fig. IIB). The DDA-driven profiling revealed 158 lipid species from eight lipid classes (see supplementary Fig. IIB). A total of 110 of 158 species represented lipids from the most abundant PC, SM, and PE classes (Fig. 3C, D), whereas the rest were assigned to relatively minor species from hexosylceramide and PS classes. Importantly, the relative abundance of major and minor species recovered by both methods was almost identical, suggesting that corresponding organic phases (MTBE and CHCl3) were not saturated with lipids.
Fig. 3.
Comparison of the lipid composition of mouse brain tissue obtained by Folch and MTBE lipid extraction protocols. A, B: Positive ion mode survey scans of the Folch and MTBE lipid extracts. C: Normalized profiles of phosphatidylcholine (PC) and sphingomyelin (SM) species. D: Normalized profiles of PE species in Folch and MTBE extracts. E: Normalized profiles of PE-plasmalogen species. F: Normalized profile of phosphatidylserine (PS) species. G: Profiles of hexosylceramides (HexCer). Folch extract is designated with black bars, and MTBE extract is designated with white bars (means ± SD; n = 6).
MTBE and Folch extraction of intact C. elegans also produced identical patterns of lipid peaks, detecting, in total, 116 lipid species from five major lipid classes (Fig. 4A, B; see supplementary Figs. IIC, III). Importantly, profiles of glycerophospholipids identified in positive ion mode, including species with moieties of polyunsaturated fatty acids, were identical in both extracts (Fig. 4C–F).
Fig. 4.
Glycerophospholipids of C. elegans embryos recovered by Folch and MTBE protocols. A, B: Positive ion mode survey scans of the Folch and MTBE lipid extracts. C: Normalized profiles of PC lipid species. D: Profiles of PE lipid species. E: Normalized profiles of PE-plasmalogen lipid species. F: Normalized profiles of PS lipid species. Folch extract is designated with black bars, and MTBE extract is designated with white bars (means ± SD; n = 6).
Automated MTBE extraction of lipids from human plasma
The MTBE protocol was further adapted for automated extraction of human blood plasma and used in clinical lipidomics screens (23, 24). In parallel, the same plasma samples were extracted by the Bligh and Dyer method using the same robotic setup.
The absolute extraction recovery was tested by processing three plasma samples with low, medium, and high lipid content. Both Bligh and Dyer and MTBE methods recovered the same amount of lipids of major classes (Table 3), with very similar coefficient of variation, which was <6% (see supplementary Fig. IV). MTBE extraction recovered slightly more ceramides from high-lipid-content plasma samples compared with the Bligh and Dyer recipe.
TABLE 3.
Recovery of lipids after extraction of plasma samples according to the MTBE or Bligh and Dyer protocol
| Ratio MTBE/BDb
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Lipid Contenta | Total | Phosphatidylcholine | Sphingomyelin | Phosphatidylethanolamine | Lysophosphatidylcholine | Ceramide | Cholesteryl Ester | Free Cholesterol |
| Low | 101 | 97 | 96 | 106 | 104 | 108 | 102 | 101 |
| Medium | 102 | 97 | 103 | 97 | 113 | 125 | 99 | 102 |
| High | 100 | 99 | 98 | 98 | 94 | 144 | 101 | 98 |
MTBE, Methyl-tert-butyl ether.
Lipid content refers to the total cholesterol content of the plasma samples (low, 83 mg/dl; medium, 207 mg/dl; high, 282 mg/dl) determined by enzymatic assays.
Ratios (%) of lipid concentrations recovered after automated MTBE or Bligh and Dyer (BD) extraction. The values are means of two independent experiments each performed in triplicate.
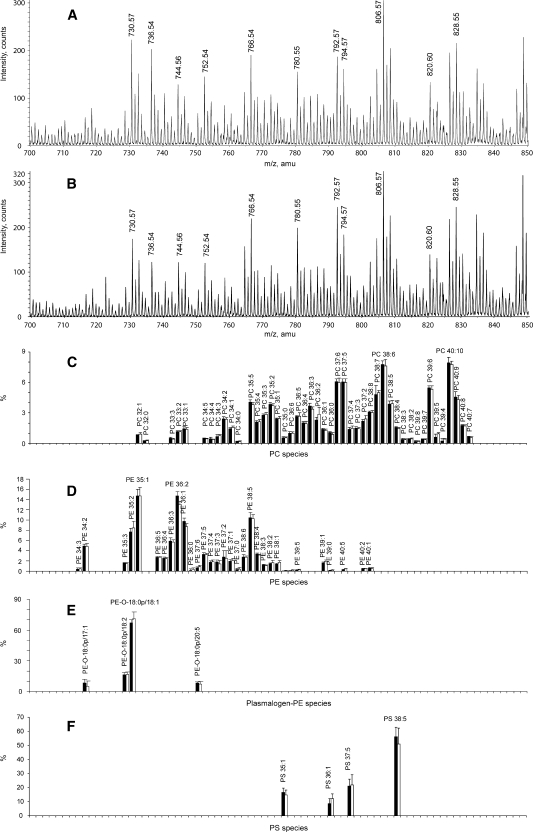
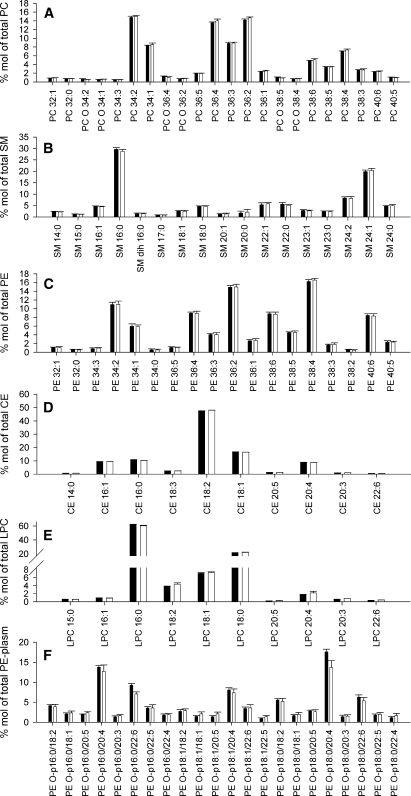
To estimate the accuracy of automated lipid profiling, 10 equal aliquots of the same plasma sample were extracted according to either the MTBE or the Bligh and Dyer method. Mass spectrometric analysis of the extracts revealed >130 lipid species of six major lipid classes (PC, SM, PE, CE, Cer, and LPC; see supplementary Fig. IID), which were detected above the limit of quantification (coefficient of variation ⩽ 20%). Species with relative abundance > 0.5 mol% of the total content of the corresponding lipid class are presented in Fig. 5. No differences in the lipid profiles of the most abundant lipid classes, PC, SM, CE, and LPC, or for the low-abundance PE and PE-plasmalogens, were observed (Fig. 5).
Fig. 5.
Comparison of major lipid components of human plasma obtained by Bligh and Dyer or MTBE automated lipid extraction protocols. A: PC lipid species profile normalized to total PC content. B: SM lipid species profile normalized to total SM content. C: PE lipid species profile normalized to total PE content. D: Cholesteryl ester (CE) lipid species profile normalized to total CE content. E: Lysophosphatidylcholine (LPC) lipid species profile normalized to total LPC content. F: PE-plasmalogen profile normalized to total PE-plasmalogen content. Profiles are displayed in mol% for lipid extracts obtained by the Bligh and Dyer or MTBE protocol. All species at >0.5 mol% are displayed. Bligh and Dyer extract is designated with black bars, and MTBE extract is designated with white bars (means ± SD; n = 10).
Therefore, we conclude that the MTBE recipe was well suited for automated lipid extraction of biological fluids and produced the same or slightly better recoveries as the established Bligh and Dyer method.
DISCUSSION
Recent developments in mass spectrometry have enabled comprehensive quantitative profiling of eukaryotic lipidomes. Although lipid extraction from cells, fluids, or tissues is a serious bottleneck in the automated lipidomics pipelines, it received little attention, and the methods palette is typically confined to the Folch or Bligh and Dyer recipes. The MTBE extraction procedure reported here allowed faster and cleaner recovery of most of the major lipid classes and was well suited for shotgun profiling, in which total extracts were infused directly into a mass spectrometer with no prior cleanup. The advantage of MTBE extraction over conventional two-phase chloroform-containing solvent systems came from the low density of the lipid-containing organic phase that forms the upper layer during phase separation. This greatly simplified its collection and minimized dripping losses. Furthermore, compared with chloroform, MTBE is nontoxic and noncarcinogenic (29, 30), which reduces the environmental burden as well as the health risks for exposed personnel. It is also noncorrosive and chemically stable, forms no peroxides during storage, and hence presents no danger of degrading labile lipids (31).
Rigorous testing that encompassed in four diverse biological matrices >400 species from 12 major lipid classes convincingly demonstrated that the MTBE protocol delivered similar or better recoveries, compared with the gold-standard Folch or Bligh and Dyer recipes, and revealed no specific limitations of the method. It enabled efficient processing of cells, biological fluids, and tissues and was easy to automate using a conventional micropipetting robot, hence paving the way to shotgun high-throughput profiling of complex lipidomes in a fully automated manner (9, 12, 22).
Supplementary Material
Acknowledgments
The authors are grateful to Ronny Herzog (Max Planck Institute of Molecular Cell Biology and Genetics) and Jeff Oegema (Scionics Computer Innovations), who developed the software for interpreting mass spectra, and to members of the Kurzchalia and Shevchenko laboratories for useful discussion and expert support. The authors also thank Julio Sampaio (Max Planck Institute of Molecular Cell Biology and Genetics) for critical reading of the manuscript.
Published, JLR Papers in Press, February 16, 2008.
Footnotes
This work was funded, in part, by Grant SFB TR13 from the Deutsche Forschungsgemeinschaft to T.V.K. and A.S. (projects B2 and D1, respectively) and by a Human Frontier Science Program grant to T.V.K.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures.
References
- 1.Han X., and R. W. Gross. 2005. Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes. Expert Rev. Proteomics. 2 253–264. [DOI] [PubMed] [Google Scholar]
- 2.Wenk M. R. 2005. The emerging field of lipidomics. Nat. Rev. Drug Discov. 4 594–610. [DOI] [PubMed] [Google Scholar]
- 3.Piomelli D., G. Astarita, and R. Rapaka. 2007. A neuroscientist's guide to lipidomics. Nat. Rev. Neurosci. 8 743–754. [DOI] [PubMed] [Google Scholar]
- 4.van Meer G. 2005. Cellular lipidomics. EMBO J. 24 3159–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yetukuri L., M. Katajamaa, G. Medina-Gomez, T. Seppanen-Laakso, A. Vidal-Puig, and M. Oresic. 2007. Bioinformatics strategies for lipidomics analysis: characterization of obesity related hepatic steatosis. BMC Syst Biol. 1 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sommer U., H. Herscovitz, F. K. Welty, and C. E. Costello. 2006. LC-MS-based method for the qualitative and quantitative analysis of complex lipid mixtures. J. Lipid Res. 47 804–814. [DOI] [PubMed] [Google Scholar]
- 7.Hermansson M., A. Uphoff, R. Kakela, and P. Somerharju. 2005. Automated quantitative analysis of complex lipidomes by liquid chromatography/mass spectrometry. Anal. Chem. 77 2166–2175. [DOI] [PubMed] [Google Scholar]
- 8.Haimi P., A. Uphoff, M. Hermansson, and P. Somerharju. 2006. Software tools for analysis of mass spectrometric lipidome data. Anal. Chem. 78 8324–8331. [DOI] [PubMed] [Google Scholar]
- 9.Schwudke D., J. T. Hannich, V. Surendranath, V. Grimard, T. Moehring, L. Burton, T. Kurzchalia, and A. Shevchenko. 2007. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal. Chem. 79 4083–4093. [DOI] [PubMed] [Google Scholar]
- 10.Brugger B., G. Erben, R. Sandhoff, F. T. Wieland, and W. D. Lehmann. 1997. Quantitative analysis of biological membrane lipids at the low picomole level by nanoelectrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 94 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han X., K. Yang, J. Yang, K. N. Fikes, H. Cheng, and R. W. Gross. 2006. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J. Am. Soc. Mass Spectrom. 17 264–274. [DOI] [PubMed] [Google Scholar]
- 12.Ejsing C. S., E. Duchoslav, J. Sampaio, K. Simons, R. Bonner, C. Thiele, K. Ekroos, and A. Shevchenko. 2006. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal. Chem. 78 6202–6214. [DOI] [PubMed] [Google Scholar]
- 13.Folch J., M. Lees, and G. H. Sloane Stanley. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226 497–509. [PubMed] [Google Scholar]
- 14.Bligh E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37 911–917. [DOI] [PubMed] [Google Scholar]
- 15.Watson A. D. 2006. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J. Lipid Res. 47 2101–2111. [DOI] [PubMed] [Google Scholar]
- 16.Nagano K., H. Kano, H. Arito, S. Yamamoto, and T. Matsushima. 2006. Enhancement of renal carcinogenicity by combined inhalation and oral exposures to chloroform in male rats. J. Toxicol. Environ. Health A. 69 1827–1842. [DOI] [PubMed] [Google Scholar]
- 17.Schmid P., E. Hunter, and J. Calvert. 1973. Extraction and purification of lipids. III. Serious limitations of chloroform and chloroform-methanol in lipid investigations. Physiol. Chem. Phys. Med. NMR. 5 151–155. [Google Scholar]
- 18.Thomann W. R., and G. B. Hill. 1986. Modified extraction procedure for gas-liquid chromatography applied to the identification of anaerobic bacteria. J. Clin. Microbiol. 23 392–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuyukina M. S., I. B. Ivshina, J. C. Philp, N. Christofi, S. A. Dunbar, and M. I. Ritchkova. 2001. Recovery of Rhodococcus biosurfactants using methyl tertiary-butyl ether extraction. J. Microbiol. Methods. 46 149–156. [DOI] [PubMed] [Google Scholar]
- 20.Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulston, J., and J. Hodgkin. 1988. Methods. In The Nematode Caenorhabditis elegans. W. B. Wood, editor. Cold Spring Harbor Laboratory Press, New York. 587–606.
- 22.Schwudke D., J. Oegema, L. Burton, E. Entchev, J. T. Hannich, C. S. Ejsing, T. Kurzchalia, and A. Shevchenko. 2006. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal. Chem. 78 585–595. [DOI] [PubMed] [Google Scholar]
- 23.Liebisch G., B. Lieser, J. Rathenberg, W. Drobnik, and G. Schmitz. 2004. High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim. Biophys. Acta. 1686 108–117. [DOI] [PubMed] [Google Scholar]
- 24.Liebisch G., M. Binder, R. Schifferer, T. Langmann, B. Schulz, and G. Schmitz. 2006. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim. Biophys. Acta. 1761 121–128. [DOI] [PubMed] [Google Scholar]
- 25.Schwudke D., G. Liebisch, R. Herzog, G. Schmitz, and A. Shevchenko. 2007. Shotgun lipidomics by tandem mass spectrometry under data-dependent acquisition control. Methods Enzymol. 433 175–191. [DOI] [PubMed] [Google Scholar]
- 26.Liebisch G., W. Drobnik, M. Reil, B. Trumbach, R. Arnecke, B. Olgemoller, A. Roscher, and G. Schmitz. 1999. Quantitative measurement of different ceramide species from crude cellular extracts by electrospray ionization tandem mass spectrometry (ESI-MS/MS). J. Lipid Res. 40 1539–1546. [PubMed] [Google Scholar]
- 27.Ingram L. O. 1977. Changes in lipid composition of Escherichia coli resulting from growth with organic solvents and with food additives. Appl. Environ. Microbiol. 33 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oursel D., C. Loutelier-Bourhis, N. Orange, S. Chevalier, V. Norris, and C. M. Lange. 2007. Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization. Rapid Commun. Mass Spectrom. 21 1721–1728. [DOI] [PubMed] [Google Scholar]
- 29.Boorman G. A. 1999. Drinking water disinfection byproducts: review and approach to toxicity evaluation. Environ. Health Perspect. 107 (Suppl. 1): 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greim H., and U. Reuter. 2001. Classification of carcinogenic chemicals in the work area by the German MAK Commission: current examples for the new categories. Toxicology. 166 11–23. [DOI] [PubMed] [Google Scholar]
- 31.Hamid, H., and M. Ali. 2004. Handbook of MTBE and Other Gasoline Oxygenates. CRC Press, New York.
- 32.Zemski Berry K. A., and R. C. Murphy. 2004. Electrospray ionization tandem mass spectrometry of glycerophosphoethanolamine plasmalogen phospholipids. J. Am. Soc. Mass Spectrom. 15 1499–1508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.