Abstract
Telomerase is an RNA-directed DNA polymerase, composed of RNA and protein subunits, that replicates the telomere ends of linear eukaryotic chromosomes. Using a genetic strategy described here, we identify the product of the EST2 gene, Est2p, as a subunit of telomerase in the yeast Saccharomyces cerevisiae. Est2p is required for enzyme catalysis, as mutations in EST2 were found to result in the absence of telomerase activity. Immunochemical experiments show that Est2p is an integral subunit of the telomerase enzyme. Critical catalytic residues present in RNA-directed DNA polymerases are conserved in Est2p; mutation of one such residue abolishes telomerase activity, suggesting a direct catalytic role for Est2p.
Keywords: telomeres, reverse transcriptase
Telomeres are DNA–protein complexes that cap the ends of eukaryotic chromosomes and maintain chromosome stability. Tandem arrays of simple guanine-rich repeat sequences constitute the telomeric DNA (1, 2). In most eukaryotes, these telomeric repeats are synthesized by telomerase, an enzyme composed of one or more protein subunits and an RNA subunit that provides the template for telomeric DNA synthesis (3–5). Thus, telomerase is an RNA-directed DNA polymerase, or reverse transcriptase, that includes the RNA template as a subunit.
Interest in telomeres and telomerase has heightened in recent years with the discovery that telomerase is present at low or undetectable levels in most human somatic tissues while being readily detectable in germ-line cells and in the vast majority of tumor cell samples (6, 7). Somatic cell lineages that lack telomerase lose telomere segments progressively as they pass through successive replication cycles (8, 9), ultimately limiting their lifespan. Conversely, cell populations that resurrect telomerase expression ensure maintenance of telomere length, thereby removing a barrier to their further unlimited replication and resulting in the process termed immortalization (10, 11). These observations have led to a model which proposes that the process of telomere shortening limits the replicative potential of most human somatic cell lineages and that cancer cells overcome this limitation by activating telomerase expression during the course of tumor progression (10, 11).
Validation of this model has been held back by the difficulties associated with identifying the mechanism by which telomerase activity is regulated. Studies of telomerase regulation have focused on the identification of the enzyme subunits. The telomerase RNA subunit gene has been cloned from many species (4), including humans (12), but the expression of the mammalian telomerase RNAs does not correlate well with telomerase activity (12–14). Two proteins, p95 and p80, have also been identified from highly purified fractions of Tetrahymena telomerase and have been found to associate with either enzyme activity or the RNA component (15). A mammalian homolog of p80, TP1/TLP1, has now been cloned and shown to be physically associated with telomerase activity; however, the expression of TP1/TLP1 in different tissues does not correlate with activation of the enzyme (16, 17). Additionally, it is unknown whether any of these proteins are essential for enzyme function.
Surprisingly, there are no obvious sequences homologous to p80 or p95 encoded within the one eukaryotic genome that has been entirely sequenced, that of the yeast Saccharomyces cerevisiae (18). Because the telomerase enzyme of yeast requires both RNA and protein components (19, 20), either yeast possess telomerase protein subunit(s) that are unrelated to, or have diverged from, p95 and p80, or the core components of telomerase remain to be identified in Tetrahymena.
Although the RNA subunit of yeast telomerase is known to be encoded by the gene TLC1 (20) and is essential for telomerase catalysis (19), no yeast protein has heretofore been found to be essential for telomerase enzyme activity. TLC1 mutants (20) exhibit two phenotypes that are shared by mutants of the genes EST1 (21) and CDC13 (22–24): a gradual decrease in telomere length and a consequent decline in cell viability. However, EST1 is not essential for telomerase activity (19), and mutational analysis of CDC13 suggest that the product of this gene may function as a telomere capping protein (22). Therefore, despite the similar mutant phenotype of EST1 and CDC13, these genes do not appear to encode essential components of the telomerase enzyme. A third ORF that exhibits a similar mutant phenotype, EST2 (ever shorter telomeres gene 2), was recently identified (25). We and others (26) now show that this gene encodes the catalytic subunit of telomerase.
MATERIALS AND METHODS
Yeast Strains.
Strain Y0025, in which RAD52 gene expression could be regulated, was constructed by replacing the RAD52 gene of strain L3853 (a gift of G. Fink of the Whitehead Institute; MATa, ura3–52, lys2-Δ201, leu2–3,112, trp1-Δ1, his3-Δ200) with HIS3 by homologous recombination and introducing the plasmid pYCPR+, which encodes the RAD52 gene under its own promoter and the selectable marker URA3 (27). The tlc1Δ/rad52Δ strain expressing Rad52p from a plasmid mentioned in Fig. 1A was derived by replacing the TLC1 gene with LEU2 in a Y0025 background. Similarly, the tlc1Δ strain mentioned in Fig. 1C was generated by replacing the TLC1 locus with a URA3 marker in the L3853 strain. The EST2-HA strain described in the legend of Fig. 2 was constructed by introducing an HA-URA3-HA cassette in-frame into the 3′ end of the EST2 gene by homologous recombination, and then counter-selecting on 5-fluoroorotic acid (FOA) to excise the URA3 gene, generating three copies of the influenza hemagglutinin (HA) tag in frame with EST2 (28). The construction of this epitope-tagged version deleted the 10 carboxyl-terminal amino acids of the protein product of the EST2 gene (Est2p); these codons contain a TA repeat sequence that is present in multiple copies in the yeast genome. A derivation of this strain, EST2-HA tlc1Δ (Fig. 2B) was generated by replacing the TLC1 gene with a URA3 marker.
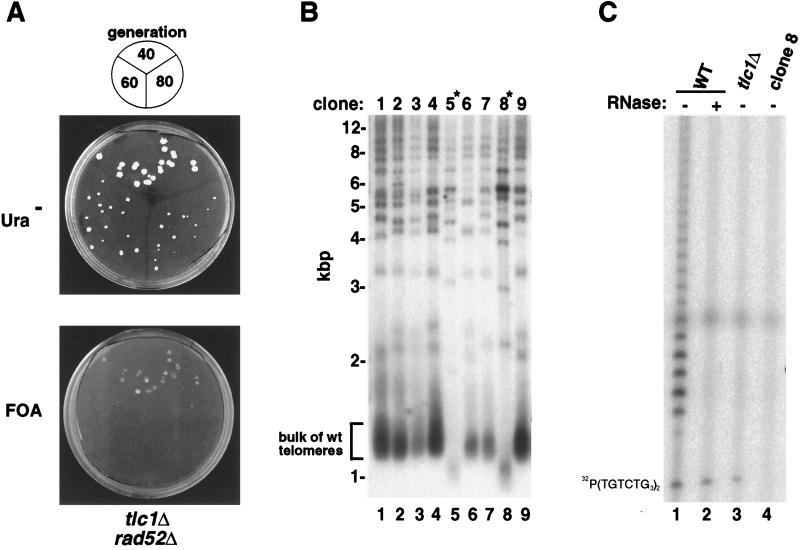
Figure 1.
A genetic screen identifies genes required for telomerase activity. (A) Demonstration of the feasibility of the rad52 synthetic lethal screen. tlc1Δ rad52Δ haploid yeast harboring a plasmid expressing Rad52 protein were aged for 40, 60, or 80 generations and replica-plated on medium lacking uracil (Ura−) to retain the RAD52 plasmid (Upper) or on FOA to evict the RAD52 plasmid (Lower). (B) Identification of mutant clones with short telomeres (∗). Telomere hybridization analysis of XhoI-digested genomic DNA isolated from a subset of mutant clones that depend upon the RAD52-expressing plasmid for viability. The bulk of the wild-type (wt) telomeres appear as heterogeneous size fragments at ≈1.3 kbp. (C) Identification of a mutant clone lacking telomerase activity. Extracts were isolated from wild-type (WT) parental strain Y0025, from a tlc1Δ strain, and from mutant clone 8 and were incubated with (lane 2) or without (lanes 1, 3, and 4) RNase prior to assaying telomerase activity. Position of the γ-32P-labeled primer is shown on the left.
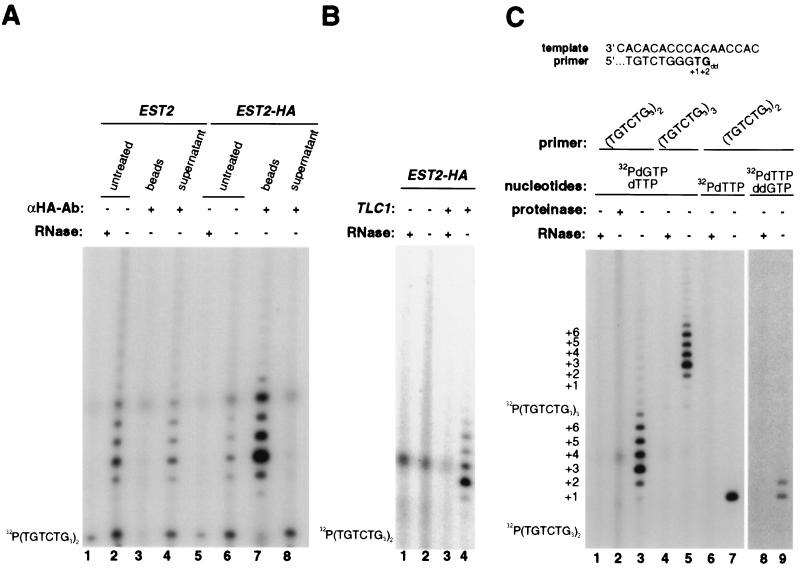
Figure 2.
Est2p is a component of telomerase. (A) Immunoprecipitation and immunodepletion of telomerase activity by anti-HA antibody from EST2-HA yeast. Telomerase activity was assayed from lysates isolated from yeast strains expressing wild-type (EST2, lanes 1–4) or HA-tagged (EST2-HA, lanes 5–8) Est2p. The extracts were left untreated (lanes 2 and 6), treated with RNase (lanes 1 and 5), or incubated with protein A-agarose beads preloaded with anti-HA antibody and then centrifuged to separate the beads (lanes 3 and 7) from the supernatant (lanes 4 and 8). Position of the γ-32P-labeled primer is shown on the left. (B) Telomerase activity of Est2p-HA immunoprecipitates requires functional Tlc1. Telomerase activity was assayed on anti-HA immunoprecipitates from extracts of EST2-HA yeast with either a mutant tlc1Δ allele (lanes 1 and 2) or a wild-type TLC1 allele (lanes 3 and 4). RNase was added to duplicate reactions as a negative control (lanes 1 and 3). Position of the γ-32P-labeled primer is shown on the left. (C) The telomerase activity of Est2p-HA immunoprecipitates uses Tlc1 as a template. (Upper) Predicted alignment of the primer and the telomerase template, showing expected elongation products generated in the presence of [α-32P]dTTP alone or in conjunction with ddGTP. (Lower) Anti-HA immunoprecipitates from extracts of yeast expressing Est2p-HA were preincubated with either proteinase K (lane 1) or RNase (lanes 2, 4, 6, and 8) or were left untreated (lanes 3, 5, 7, and 9) prior to assaying telomerase. Telomerase activity was assayed with primer (TGTCTG3)2 (lanes 1–3 and 6–9) or (TGTCTG3)3 (lanes 4 and 5) in the presence of the deoxynucleoside triphosphate(s) [α-32P]dGTP and dTTP (lanes 1–5), or [α-32P]dTTP alone (lanes 6 and 7), or [α-32P]dTTP and ddGTP (lanes 8 and 9). A longer exposure is shown for lanes 8 and 9. The positions of the γ-32P-labeled primers and the elongation products from +1 to +6 are shown on the left.
Yeast Mutagenesis.
Approximately 230 μg of the appropriately prepared yeast mini-Tn3::lacZ::LEU2 genomic library (29) was introduced by homologous recombination into strain Y0025. Approximately 1 × 106 uracil-independent leucine-independent clones representing successful transposon insertions were pooled, and replated to a total number of ≈1 × 106 clones to “age” the yeast. Then 1 × 105 of such yeast were reseeded for the rad52Δ synthetic lethal screen, as described in the text.
Sequence Analysis.
Genomic DNA flanking the transposon insertion sites was rescued as described (29), sequenced, and compared with the Saccharomyces cerevisiae Genome Database (http://genome-www.stanford.edu/Saccharomyces/) with the blast sequence comparison algorithm. Est2p sequences were compared using blast to multiple genome databases, including the Arabidopsis thaliana Database (http://genome-www.stanford.edu/Arabidopsis/).
Southern Hybridization.
Genomic DNA was isolated and digested with the restriction enzyme XhoI, which liberates the majority of telomeres as small (≈1.3-kbp) heterogeneous fragments. Samples (2 μg) of each DNA were resolved on 0.8% agarose gels for 570 V⋅h and hybridized as described (10), using a 32P kinase-labeled yeast telomeric oligonucleotide CACCACACCCACACACCACA.
Telomerase Assays.
Six liters of yeast cultures was harvested at an OD of 0.4–0.6 at 600 nm, resuspended in TMG buffer (19), disrupted with a bead beater, and centrifuged for 90 min at 100,000 × g at 4°C as described (19). The supernatant was decanted, flash frozen, and stored at −70°C. Extracts typically had a protein concentration of 16–20 mg/ml. Telomerase activity was assayed essentially as outlined by Blackburn and co-workers (19, 30) except that 4 μg of extract was assayed, and the reaction products were purified and resolved on 15% acrylamide/7 M urea sequencing gels as described elsewhere (6). In the case of extracts pretreated with RNase or proteinase K, 4 μg of extract was incubated with 3.5 μg/ml RNase A or 0.5 mg/ml proteinase K for 5 min at 30°C.
Immunoprecipitation and Immunodepletion.
A 540-μg portion of yeast extract was diluted in 270 μl of TMG buffer supplemented with 150 mM NaCl (buffer TMGN) and incubated at 4°C with 20 μl of packed protein A-agarose beads preloaded with anti-HA antibody (monoclonal antibody 12CA5). After 2 h, the mixture was briefly centrifuged to separate the agarose beads from the supernatant extract. The beads were then washed four times with buffer TMGN and resuspended in 50 μl of TMGN. As a control, a duplicate extract was diluted and left untreated on ice for 2 h. Four micrograms of supernatant or untreated extracts and 3–10 μl of beads were assayed for telomerase activity.
Site-Directed Mutagenesis.
A genomic copy of EST2 was PCR amplified with primers ACGGGATCCCGTAACCATGTCAACTATGAAAATCTTATTCGAG and CGGGATCCTTATCCTGCATAGTCCGGGACGTCATAGGGATAGCCCGCATAGTCAGGAACATCGTATGGGTATGCTTGCAAGTGTTGAATTTCC, generating an HA-tagged EST2 ORF flanked by BamHI sites. The PCR product was cut with BamHI and inserted into the BamHI site of pBluescript SK+ (Stratagene). This plasmid was amplified by inverse long-range PCR with primers GAATTTCATGAAATAAAGCTCTGGTAAGA and GAAGTCAAATCTTGCTATGATTCCATACC to generate the D530E mutation and a diagnostic BstBI site. The resulting product was then digested with DpnI, which cuts only methylated DNA, before ligation and bacterial transformation. The BamHI EST2-HA fragment containing the point mutation was subcloned into the plasmid p426-GAL (31) and the insert was sequenced. Clones containing only the desired mismatch were used to transform either est2::Tn3 rad52Δ or est2::Tn3 yeast strains.
RESULTS AND DISCUSSION
A Genetic Screen Identifies Genes Required for Telomerase Activity.
Because neither genetic nor sequence homology approaches had defined a protein subunit of yeast telomerase, we initiated a screen for yeast genes that are essential for telomerase catalytic function. We reasoned that mutations in such genes would lead to three phenotypes: (i) absence of telomerase enzymatic activity, (ii) a consequent decrease in telomere length, and (iii) dependence on the RAD52 gene product for prolonged viability. This third prediction is based on the observation that the loss of telomeric DNA observed in yeast lacking a functional telomerase RNA component can be compensated by the DNA recombination machinery, of which RAD52 is a central component. Accordingly, the simultaneous inactivation of RAD52 and the telomerase RNA component results in cell lethality once telomeres become critically short in both S. cerevisiae (25) and Kluyveromyces lactis (32).
To determine if mutants in telomerase function could be revealed by a genetic screen for yeast that require RAD52 function, we generated a yeast strain that lacked a functional TLC1 telomerase RNA subunit gene but also carried an inactive chromosomal rad52Δ allele, which was complemented by a plasmid-borne wild-type RAD52 gene linked to a URA3 gene. As expected, this strain survived with each successive passage because the RAD52-dependent mechanism for telomere maintenance was intact, although, in accord with the observations of others (25), the viability of the yeast did decline at later generations (Fig. 1A Upper). However, when RAD52 function was inactivated through eviction of the RAD52-encoding plasmid (by counterselecting against the URA3 marker with FOA) there was a dramatic decrease in viability at early passage and complete inviability by later generations (Fig. 1A Lower). We concluded that, as anticipated, telomerase-deficient cells perished upon the loss of RAD52 function, and this phenotype could therefore be used to screen for mutants carrying lesions in a variety of genes affecting telomerase function.
We therefore proceeded to mutagenize the rad52Δ mutant cells carrying the complementing RAD52 plasmid. The mutagenesis was accomplished by transforming the yeast with a library of yeast genomic fragments, each of which carried one or more copies of an inserted mini-Tn3::lacZ::LEU2 transposon. These chimeric fragments inactivated chromosomal genes through homologous recombination (29). When the approach to regulate RAD52 gene expression described above was used, 245 clones from an initial population of 105 transposon-mutagenized cells were found to require the RAD52-expressing plasmid for continued viability. We tested these RAD52-dependent cell clones for changes in telomere structure by Southern hybridization analysis of their telomeres. An example of such a Southern blot is shown in Fig. 1B. In clones 5 and 8, the bulk of the telomeres, which normally appear as a smear at ≈1.3 kbp (33, 34), were much shorter than those observed in other clones (Fig. 1B) or in the parental strain (not shown). [The higher molecular weight bands are combinations of single telomeres and interstitial telomeric repeats (33, 34).] A total of 16 such clones were identified.
Mutants exhibiting telomere shortening were then screened for the presence of telomerase activity. Using the protocol described by Cohn and Blackburn (19), we assayed telomerase activity by incubating yeast extracts with the single-stranded telomere primer (TGTCTGGG)2, in the presence of [α-32P]dGTP and dTTP, the only nucleotides required for telomerase function. As previously shown (19), extracts from wild-type yeast elongated the primer by 4–16 nucleotides (Fig. 1C, lane 1); this activity was sensitive to low amounts (3.5 μg/ml) of RNase A (Fig. 1C, lane 2) and depended upon a functional TLC1 gene product (Fig. 1C, lane 3).
When telomerase activity was assayed in mutants exhibiting telomere shortening, a number of clones were identified that lacked detectable telomerase activity. Two such clones, clone 5 and clone 8, are described here (Fig. 1C, lane 4, and not shown). By comparison with dilutions of wild-type extract (not shown), we estimated that extracts from telomerase negative extracts contained at most 5% of wild-type telomerase activity. While the actual levels of telomerase in the mutant yeast are likely to be much lower, such lower levels could not be quantified with this assay.
Thus, this genetic screen had identified transposon-mutated genes that exhibited the three phenotypes predicted for telomerase mutants: dependence on RAD52, gradual telomere shortening, and the absence of telomerase activity. Analysis of the DNA sequences flanking the transposon insertion sites showed that clone 5, which lacked telomerase activity, contained a transposon insertion site within the TLC1 telomerase RNA subunit gene. Five additional clones exhibiting telomere shortening also contained transposon insertions at distinct sites within TLC1. This result provided strong and direct validation of the utility of our strategy to screen for genes that directly affect telomerase function.
The transposon insertion abolishing telomerase activity in clone 8 was found to reside within a novel ORF on chromosome XII, which, during the course of this work, was identified as the EST2 gene, previously reported and named by others because of its association with telomere shortening (25). Transposon insertions in seven additional clones contained mutations at six distinct sites within EST2. The absence of detectable telomerase activity in the est2::Tn3 mutants was found to cosegregate with the mutation upon tetrad analysis (not shown).
Est2p Is a Component of Telomerase.
In light of the fact that no protein had previously been found to be essential for telomerase activity in any organism, we wished to determine whether the product of EST2 was actually a component of telomerase or simply an upstream regulator required for telomerase activity. To distinguish between these possibilities, we replaced the endogenous EST2 gene with a variant encoding a protein with three HA epitope tags at its carboxyl terminus (EST2-HA). This modified EST2 allele, and thus its product, was fully functional. The EST2-HA yeast strain showed no overt phenotype, its telomeres were of wild-type length (not shown), and extracts from these cells exhibited normal levels of telomerase activity (Fig. 2A, lane 6).
To test whether Est2p, the product of the EST2 gene, is a component of the telomerase enzyme, extracts from EST2-HA and control yeast were incubated with excess anti-HA antibody, after which the immunoprecipitates and remaining supernatants were separated and assayed for telomerase activity. The anti-HA antibody immunoprecipitated telomerase activity from extracts of yeast expressing tagged Est2p-HA (Fig. 2A, lane 7) but not from extracts expressing untagged Est2p (Fig. 2A, lane 3). The immunoprecipitation was specific for the anti-HA antibody, since a control anti-Myc epitope antibody failed to immunoprecipitate activity from either extract (not shown). Essentially all of the telomerase activity remained in the supernatant of the anti-HA immunoprecipitation from wild-type extracts (Fig. 2A, lane 4), whereas incubation with the anti-HA antibody almost completely immunodepleted the telomerase activity from the Est2p-HA extracts (Fig. 2A, lane 8). Therefore, these data indicated that Est2p not only is physically associated with telomerase but also is a constituent of most if not all of the active telomerase complexes in these cells.
The enzymatic activity immunoprecipitated with Est2p was clearly that of a telomerase, because this activity depended upon the presence of the Tlc1 telomerase RNA subunit and because the sequence of the reaction products was complementary to the Tlc1 template (Fig. 2 B and C). As has previously been described for yeast telomerase (2), the telomerase activity of the Est2p immunoprecipitate required the presence of a functional TLC1 gene (Fig. 2B). Also as previously described (19), the telomerase activity was abolished by pretreatment with proteinase K or RNase (Fig. 2C, lanes 1 and 2) and extended primers ending in TGGG by 4–16 nt, irrespective of the length of the primer (Fig. 2C, lanes 3 and 5). By replacing the normal nucleotides used in the assay with [α-32P]dTTP alone or in conjunction with the chain terminator ddGTP (30), we were able to determine that the sequence of shortest reaction products produced by the Est2p-containing complexes were the complement of the RNA template sequence of telomerase. As expected on the basis of the RNA template sequence (Fig. 2 C Upper), the primer (TGTCTGGG)2 was elongated by only one nucleotide (+1) in the presence of [α-32P]dTTP in an RNase-sensitive manner (Fig. 2C, lanes 6 and 7), and by only one (+1) to two nucleotides (+2) if the chain terminator ddGTP was added to the reaction (Fig. 2C, lanes 8 and 9). Identical banding patterns were observed in wild-type extracts (not shown).
Est2p Is the Catalytic Subunit of Yeast Telomerase.
Telomerase is an RNA-directed DNA polymerase that extends telomeric DNA substrates by using an internal RNA template. Sequence homologies between Est2p and RNA-directed DNA polymerases (reverse transcriptases) suggested that Est2p, shown above to be a subunit of telomerase, is likely to act as a catalytic subunit. A search of the nonredundant Arabidopsis protein sequence database using 100-aa blocks of Est2p and the blast search algorithm revealed significant homology (P = 4.5 × 10−3) between Est2p (amino acids 525–564) and Arabidopsis maturase (amino acids 65–104), a group II intron-encoded reverse transcriptase (35). Identification of Est2p Asp-530 as a completely invariant residue in a motif conserved among all reverse transcriptases (36, 37) as well as all other polymerases (38) permitted additional reverse transcriptase motifs to be identified as well. In particular, two motifs found in Est2p are characterized by a total of three aspartic acids (Asp-530, -670, and -671) which are postulated to catalyze phosphoryl transfer in reverse transcriptases (Fig. 3A). These three aspartic acid residues are invariant among RNA-dependent DNA polymerases (36, 37), are found in the catalytic cleft of the HIV-1 reverse transcriptase (39), and are essential for activity, as even conservative mutations to glutamic acid abolish enzymatic activity in HIV-1 reverse transcriptase without affecting other functions of the enzyme (40).
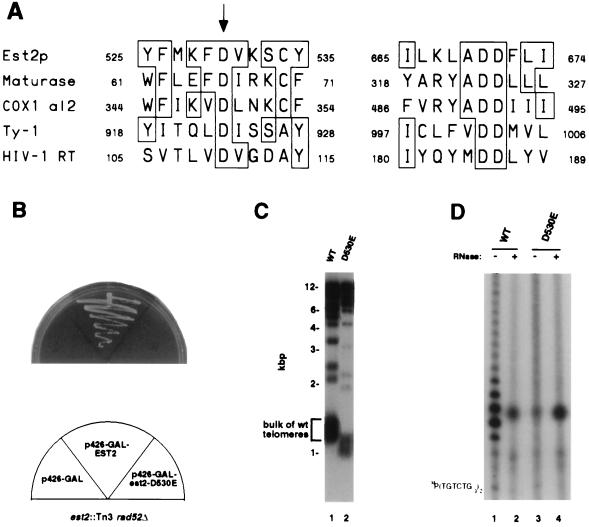
Figure 3.
Reverse transcriptase homology of Est2p is essential for telomerase function. (A) Homology between reverse transcriptase catalytic residues and Est2p. Alignment of regions of Est2p containing the conserved reverse transcriptase motifs A and C (36), or 3 and 5 (37) with the same regions of Arabidopsis maturase, S. cerevisiae COX1 group II intron 2 and retrotransposon Ty-1, and HIV-1 reverse transcriptase (36, 37). Boxes denote residues conserved with Est2p. The three aspartic acid residues conserved among all the proteins shown are completely invariant among all RNA-dependent DNA polymerases (36, 37). The arrow denotes the Asp-530 residue that was mutagenized. (B) Failure of the D530E mutant to rescue inviability of est2 rad52 mutants. est2 rad52 mutant yeast were transformed with the negative control plasmid p426-GAL (left), with a positive control p426-GAL expressing wild-type EST2 (center), and with a p426-GAL plasmid expressing the D530E mutant (right). Loss of viability in the unrescued yeast is immediate, not delayed, because the est2 rad52 mutants have already been passaged ≈30 generations in culture. (C) Telomere shortening in est2 yeast expressing D530E mutant but not wild-type EST2. Telomere hybridization analysis of XhoI-digested genomic DNA isolated from an EST2 control (lane 1) or from est2 mutants expressing the D530E mutant (lane 2). (D) Lack of telomerase activity in yeast expressing the D530E mutant. Telomerase activity was assayed in extracts from an EST2 wild-type control (lanes 1 and 2) or from yeast expressing the D530E mutant (lanes 3 and 4). Extracts were pretreated with RNase (lanes 1 and 3) or left untreated (lanes 2 and 4). Position of the γ-32P-labeled primer is shown on the left.
Given the sequence homology of Est2p with reverse transcriptases, mutation of an amino acid in Est2p which corresponds to a catalytic residue in reverse transcriptases should abolish telomerase activity. To test this prediction we first generated an Est2p expression construct which could rescue est2 rad52 mutants from cell death. The codon for residue Asp-530, the one amino acid of the motifs described which is invariant among all classes of polymerases (38), was mutated to encode Glu (D530E) in this plasmid copy of EST2. est2 yeast expressing the D530E mutant (p426-GAL-est2-D530E) were inviable in a rad52 mutant background (Fig. 3B). Consistent with this loss of viability, yeast expressing the D530E mutant est2 suffered telomere shortening (Fig. 3C). Finally, telomerase activity was absent in extracts of yeast expressing the D530E mutant (Fig. 3D). Taken together, these data argue that the Est2p aspartic acid residues that are homologous to aspartic acid resides in the catalytic cleft of reverse transcriptases are essential for telomerase enzyme function, strongly suggesting that Est2p acts as the catalytic subunit of yeast telomerase and that its mechanism of catalysis is similar to that of other RNA-directed DNA polymerases.
Conclusions.
The present evidence, taken together, indicates that Est2p is an integral subunit of the S. cerevisiae telomerase and represents, with great likelihood, the catalytic subunit of the enzyme responsible for polymerase activity. We conclude this because Est2p is (i) required for template-dependent telomerase activity, (ii) associated with telomerase activity, and (iii) functionally conserved with reverse transcriptases. In agreement with these observations, Lingner et al. (26) have independently identified Est2p as the catalytic subunit of yeast telomerase. Homologs of Est2p are likely to be the catalytic subunits of telomerase in eukaryotes that depend upon telomerase. Indeed, Est2p homologs have been found in two other evolutionarily distant organisms, the ciliate Euplotes aediculatus (26) and, more recently, in humans (unpublished results).
The identification of Est2p as a protein required for telomerase catalysis will aid in the understanding of this enzyme’s function and in identifying other subunits in yeast and other organisms.
Acknowledgments
We express special thanks to members of the Weinberg and Fink laboratories for help, advice, and encouragement. In particular, we thank H. Madhani, G. T. Milne, R. Perlman, and A. Sherman for their guidance in yeast genetics. G. T. Milne and D. T. Weaver are thanked for the pYCPR+ plasmid. We also thank L. Ziaugra for assistance with DNA sequencing and S. L. Dickinson for technical assistance. Last, we thank R. Young, S. Bacchetti, and T. Cech for critical reading of the manuscript. C.M.C. thanks I. York for helpful discussions. This work was supported by a grant from the National Cancer Institute to R.A.W. C.M.C. is a K. M. Hunter Postdoctoral Fellow of the National Cancer Institute of Canada; M.M. is a Damon Runyon–Walter Winchell Cancer Research Foundation Postdoctoral Fellow; R.A.W. is an American Cancer Society Research Professor.
ABBREVIATIONS
- Est2p
protein product of the EST2 gene
- FOA
5-fluoroorotic acid
- HA
influenza hemagglutinin
References
- 1.Blackburn E H. Annu Rev Biochem. 1984;53:163–194. doi: 10.1146/annurev.bi.53.070184.001115. [DOI] [PubMed] [Google Scholar]
- 2.Zakian V A. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 3.Greider C W, Blackburn E H. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 4.Greider C W. In: Telomerase Biochemistry and Regulation. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 35–68. [Google Scholar]
- 5.Greider C W, Blackburn E H. Nature (London) 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 6.Counter C M, Hirte H W, Bacchetti S, Harley C B. Proc Natl Acad Sci USA. 1994;91:2900–2904. doi: 10.1073/pnas.91.8.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 8.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 9.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Nature (London) 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 10.Counter C M, Avilion A A, Le Feuvre C E, Stewart N G, Greider C W, Harley C B, Bacchetti S. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harley C B, Kim N W, Prowse K R, Weinrich S L, Hirsch K S, West M D, Bacchetti S, Hirte H W, Counter C M, Greider C W, Piatyszek M A, Wright W E, Shay J W. Cold Spring Harbor Symp Quant Biol. 1994;59:307–315. doi: 10.1101/sqb.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- 12.Feng J, Funk W D, Wang S S, Weinrich S L, Avilion A A, Chiu C P, Adams R R, Chang E, Allsopp R C, Yu J, Le S, West M D, Harley C B, Andrews W H, Greider C W, Villeponteau B. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 13.Blasco M A, Rizen M, Greider C W, Hanahan D. Nat Genet. 1996;12:200–204. doi: 10.1038/ng0296-200. [DOI] [PubMed] [Google Scholar]
- 14.Avilion A A, Piatyszek M A, Gupta J, Shay J W, Bacchetti S, Greider C W. Cancer Res. 1996;56:645–650. [PubMed] [Google Scholar]
- 15.Collins K, Kobayashi R, Greider C W. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 16.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Program A E, Bass M B, Arruda I, Robinson M O. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama J-i, Saito M, Nakamura H, Matsuura A, Ishikawa F. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 18.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- 19.Cohn M, Blackburn E H. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 20.Singer M S, Gottschling D E. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 21.Lundblad V, Szostak J W. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 22.Nugent C I, Hughes T R, Lue N F, Lundblad V. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 23.Virta-Pearlman V, Morris D K, Lundblad V. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 24.Lin J J, Zakian V A. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 27.Milne G T, Ho T, Weaver D T. Genetics. 1995;139:1189–1199. doi: 10.1093/genetics/139.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 29.Burns N, Grimwade B, Ross-Macdonald P B, Choi E Y, Finberg K, Roeder G S, Snyder M. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 30.Prescott J, Blackburn E H. Genes Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 31.Mumberg D, Muller R, Funk M. Nucleic Acids Res. 1994;22:5767–8. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McEachern M J, Blackburn E H. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 33.Chan C S, Tye B K. Cell. 1983;33:563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- 34.Walmsley R M, Petes T D. Proc Natl Acad Sci USA. 1985;82:506–510. doi: 10.1073/pnas.82.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennell J C, Moran J V, Perlman P S, Butow R A, Lambowitz A M. Cell. 1993;73:133–146. doi: 10.1016/0092-8674(93)90166-n. [DOI] [PubMed] [Google Scholar]
- 36.Poch O, Sauvaget I, Delarue M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joyce C M, Steitz T A. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 39.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 40.Larder B A, Purifoy D J, Powell K L, Darby G. Nature (London) 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]