Abstract
Entamoeba histolytica is a single cell eukaryote that is the etiologic agent of amoebic colitis. Core promoter elements of E. histolytica protein encoding genes include a TATA-like sequence (GTATTTAAAG/C) at −30, a novel element designated GAAC (GAACT) that has a variable location between TATA and the site of transcription initiation, and a putative initiator (Inr) element (AAAAATTCA) overlying the site of transcription initiation. The presence of three separate conserved sequences in a eukaryotic core promoter is unprecedented and prompted examination of their roles in regulating transcription initiation. Alterations of all three regions in the hgl5 gene decreased reporter gene activity with the greatest effect seen by mutation of the GAAC element. Positional analysis of the TATA box demonstrated that transcription initiated consistently 30–31 bases downstream of the TATA region. Mutation of either the TATA or GAAC elements resulted in the appearance of new transcription start sites upstream of +1 in the promoter of the hgl5 gene. Mutation of the Inr element resulted in no change in the site of transcription initiation; however, in the presence of a mutated TATA and GAAC regions, the Inr element controlled the site of transcription initiation. We conclude that all three elements play a role in determining the site of transcription initiation. The variable position of the GAAC element relative to the site of transcription initiation, and the multiple transcription initiations that resulted from its mutation, indicate that the GAAC element has an important and apparently novel role in transcriptional control in E. histolytica.
Entamoeba histolytica is a protozoan parasite that is the etiologic agent of amoebic colitis and liver abscess. It is second only to malaria as a parasitic protozoan cause of death (1); however, fundamental research to characterize this organism at a molecular level lags far behind that of other parasites. Understanding the mechanisms of gene expression may provide insight into the ability of the organism to survive changes in its environment, transform from a cyst to a trophozoite, and cause asymptomatic versus invasive disease.
E. histolytica is an early diverging member of the eukaryotic tree and has many unusual characteristics with regard to gene organization and transcriptional control. It has a small genome (1.5 × 107 bp) (2), which is AT rich (67% within coding regions and 78% overall) (3, 4) and an RNA polymerase II that is resistant to α-amanitin (5). Thus far only three small introns have been identified in protein-encoding genes (6–8), and there appears to be no trans-splicing or polycistronic transcription (9). The mRNA is also unique, compared with that of multicellular eukaryotes, with a very small (5–21 bp) 5′ untranslated region and a short 3′ untranslated region with an average length of 33 bases (9). We previously have shown that amoebic promoter sequences do not function in a mammalian system and that the cytomegalovirus, human immunodeficiency virus long terminal repeat, actin promoter of Dictyostelium (R.R. Vines and W.A.P., unpublished results), and the simian virus 40 promoter are nonfunctional in amoebic trophozoites (10). In addition the E. histolytica putative TATA binding protein (GenBank accession no. Z48307) has significant sequence divergence from the TATA binding protein of Drosophila melanogaster, Caenorhabditis elegans, and Plasmodium falciparum (11). The identification of unusual or novel core promoter regions therefore would not be completely unexpected in this organism.
The core promoter region in metazoans is the target of a variety of regulatory proteins that work in concert to direct the complex mechanisms of transcriptional control. Two well defined regions in this area are the TATA box located 25–30 bp upstream of the site of transcription initiation, and the initiator (Inr) directly overlapping the transcription start site. Consensus higher eukaryotic TATA (TATAa/tAa/t) and Inr sequences (PyPyA+1NT/APyPy) have been identified. The most highly conserved nucleotides in the Inr element of >500 eukaryotic genes appear to be −1(C), +1(A), and +3(T) (12). Transcription initiation relies on the assembly of RNA polymerase II and a variety of other transcription factors (TFIID, TFIIA, TFIIB, TFIIE, TFIIF, and TFIIH) into a preinitiation complex. The assembly of the preinitiation complex may occur in a sequential manner on the DNA sequence of the core promoter, or a “holoenzyme” complex may form, which then binds specifically to the core promoter region (13). Further fine control over transcription regulation is provided by the numerous TATA binding protein-associated factors, which play an important role in up-regulation of transcription by their interaction with specific activator and coactivator proteins. Both the TATA box and Inr region therefore appear to direct the formation of the preinitiation complex, control the site of transcription initiation, and regulate activation by upstream activator proteins (14).
Analysis of the core promoters of 37 protein encoding genes of E. histolytica revealed three conserved regions in the core promoter (15). These sequences are (i) GTATTTAAA(G/C), the putative TATA element at −30, (ii) GAACT, named GAAC with a variable location in the core promoter, and (iii) AAAAATTCA, the putative Inr overlying the site of transcription initiation. The putative TATA and Inr regions of E. histolytica have little sequence similarity with the consensus TATA box and Inr of higher eukaryotes. E. histolytica has significant core promoter sequence divergence even compared with other protozoa. The core promoter of the slime mold Dictyostelium contains a consensus TATA element, lacks an Inr element, and has stretches of poly(dT) at or immediately upstream from the start of transcription initiation (16, 17). In contrast, the core promoter of Acanthamoeba contains a consensus TATA element and a conventional higher eukaryotic Inr element (18). Genetic analysis of Trichomonas has revealed a core promoter that lacks a TATA motif but has an Inr with sequence homology to the Inr of higher eukaryotes and functions as an Inr in a heterologous in vitro transcription system (19). The three core promoter elements identified in E. histolytica thus appear to be unique. Characterization of these elements was undertaken to define their role in regulating the site of transcription initiation.
Our work is based on promoter analysis of the hgl gene family (15). The hgl multigene family encodes the heavy subunit of the Gal-GalNAc-specific lectin. The lectin mediates in vitro adhesion to epithelial and immune cells and is required for contact dependent lysis of a variety of target cells in vitro. The putative TATA region of E. histolytica FeSOD gene has been demonstrated to be recognized in a sequence-specific manner by a nuclear protein (9). Mutation of the TATA and Inr sequences in the hgl gene family decreased reporter gene activity (15, 20). The effect of these mutations on the site of transcription initiation was not determined. The third conserved region (GAAC) in the core promoter has not been subjected to mutational analysis.
MATERIALS AND METHODS
Cultivation of E. histolytica and Transient and Stable Transfection.
E. histolytica strain HM-1:IMSS trophozoites were cultured in TYI-S-33 medium containing penicillin (100 units/ml) (GIBCO/BRL) and streptomycin (100 μg/ml) (GIBCO/BRL) as previously described (21). Trophozoites in logarithmic phase of growth were washed once in incomplete cytomix (10) and resuspended to 2.7 × 106/ml in incomplete cytomix containing glycerol (0.375%) and DEAE-Dextran (3.1 μg/ml). Plasmid DNA (40 μg) for transfection was purified via an anion exchange column (Qiagen) and its concentration estimated by optical density at 260 nm. For the transient transfection experiments, multiple DNA preparations (two independent preparations of each plasmid) were used, and each transfection was done a minimum of four times. Amoebae were electroporated at 500 V/cm and 500 μF before being added to 12 ml of culture medium containing 8.0 μM E-64c [(2S, 3S)-trans-epoxysuccinyl-l-leucylamido-3-methylbutane) (Sigma). Trophozoites were harvested at 10 hr postelectroporation and lysed in luciferase assay lysis buffer (Promega) with the addition of leupeptin (0.375 μg/ml) (Sigma) and E-64 [trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane] (37.5 μM) (Sigma). Lysates were frozen overnight at −20°C. After thawing on ice for 10 min, cellular debris was pelleted, and the samples were allowed to warm to room temperature. Luciferase activity was measured according to the manufacturer’s instructions (Promega) using a Turner Luminometer (model TD-20E).
Stable transfection was achieved using the two-promoter vector pTP-Luc plasmid (5′ hgl5-luciferase-hgl 3′ and 5′actin-neor-actin 3′) (22). Trophozoites were electroporated as described above except that two consecutive pulses were used. The amoebae were then diluted into 50 ml of TYI-S-33 media in 25 cm2 flasks and incubated overnight at 37°C. After 24 hr, G418 was added to the media at an initial concentration of 6 μg/ml with subsequent stepwise increases (23). Primer extension analysis was performed on RNA isolated from stably transfected parasites maintained at 24 μg/ml G418.
Plasmid Construction.
Mutational and positional analyses were undertaken on the 5′ upstream region of hgl5 fused to a luciferase reporter gene (plasmid BΔ1R8.D3′-272) (15). The mutations in the Inr, GAAC, and TATA elements were made using two complete rounds of PCR using primers with mutations in the desired region as described previously (15). One unique primer was synthesized per mutation to replace the desired region with a mutated sequence. The primer used for the TATA mutation was (5′-AAGGCAATTGAAACAAAACAAGACAAT-3′). The first round of PCR used the above primer with the primer 5′-CTTTCTTTATGTTTTTGGCG-3′, which hybridized with the coding region of luciferase (bases 1,727 to 1,746 of pGEM-luc, Promega). This resulted in a 100-bp fragment, which then was used as a primer for the second round of PCR using the primer 5′-CTACTGAAGCTTAGTAAAGAATAGTATTGA-3′ (containing a HindIII restriction site shown underlined for cloning), which hybridized at the 5′-end of the 272-bp promoter of the BΔ1R8.D3′-272. The constructs where Inr and GAAC were mutated were made in a similar manner using a unique primer for the first round of PCR, which incorporated the desired point mutations. Each primer had at least 14 bases of sequence homology upstream and downstream of the mutations to allow for hybridization with the backbone. These colonies were screened by restriction enzyme analysis where appropriate, or by sequence analysis. All constructs used in our experiments were sequenced in their entirety to rule out PCR-induced mutations. The plasmids in which the TATA element was moved upstream were made using the construct BΔ1R8.D3′-272 in which bases −21 to −13 were replaced with a SalI restriction site. Introduction of this restriction site in the plasmid did not affect reporter gene activity (15). Using this SalI restriction site between the TATA and GAAC elements, oligonucleotides of varying lengths with SalI compatible ends were inserted, which increased the distance between the TATA and Inr elements from 14 bases (wild type) to 29 and 54 bases. The oligonucleotide (5′-TCGAGAAGATCTTCT-3′) increased the spacing by 15 bases, and the (5′-CAAGCTTAGATATCAGTCGACAAGCTTAGATATCAGTCGA-3′) oligonucleotide increased the spacing by 40 bases. These insertions maintained an approximately 60% AT-rich sequence in the core promoter and also contained unique restriction sites for screening purposes.
To make the construct in which both the TATA and GAAC elements were moved upstream, an oligonucleotide with XbaI and BglII restriction sites was introduced between the wild-type GAAC and Inr elements using two rounds of PCR as described above. The first round of PCR included the common primer in luciferase and a primer 5′-AAACAAGACAATGAACTAGATCTAGATAGAAAGACAAAGATATGAAATTATTATT-3′ and introduced BglII and XbaI restriction sites as shown in bold above. The location of the GAAC and Inr elements are underlined at the 5′ and 3′ end of the insert, respectively. The insert was 67% AT rich and was introduced through using two rounds of PCR as described in the reference. This construct then had the TATA and GAAC elements mutated using the two rounds of PCR technique and similar primers as mentioned above.
To achieve stable transfection, all mutated core promoters of interest were incorporated into the pTP-Luc vector (22). This was done through the use of the BamHI (5′ end of luciferase) and HindIII (5′ end of hgl5) restriction sites, which allowed replacement of the 1-kb fragment of the hgl5 promoter region with the mutated or altered promoter of 272 bp. Colonies were screened by identification of the introduced restriction site in the plasmid and by noting a change in the size of the 5′ noncoding region of the hgl5 gene. Each construct had its entire promoter region of approximately 280 bp (including the BamHI and HindIII cloning restriction sites) sequenced to rule out extraneous mutations.
Nuclear Run-On Analysis.
Nuclei were harvested from 5 × 107 logarithmically growing trophozoites (stably transfected with the wild-type plasmid maintained at 24 μg/ml G418) and stored at −70°C (9). These were thawed on ice, and the nuclear run-on was done as described (24). RNA extraction was performed using the guanidinium isothiocyanate method (RNagen Kit, Promega). Approximately 1.2 pmol of DNA probes (genomic DNA from E. histolytica strain HM-1:IMSS, luciferase, neomycin, and 272-bp promoter of the hgl5 gene) were purified, denatured, and dot-blotted onto Zeta-probe GT Genomic blotting membrane (Bio-Rad). The membrane was incubated with the prehybridization solution at 65°C for 20 min; denatured RNA probe was added, and the mixture was incubated overnight at 65°C. The membrane was washed at 65°C according to the manufacturer’s instructions and exposed on a PhosphorImager (Molecular Dynamics).
Northern Blot Analysis.
Polyadenylated mRNA from stably transfected amoebae was isolated using the PolyATract System 1000 (Promega). One microgram of poly(A)+ RNA was electrophoresed through a formaldehyde gel and transferred to a Zeta-probe GT Genomic blotting membrane (Bio-Rad). The membrane was incubated in the prehybridization at 56°C for 20 min. The radiolabeled denatured DNA probes were added to the hybridization mixture and incubated overnight at 56°C. The DNA probes consisted of the coding regions of luciferase and neomycin, which were extracted by digestion of the pTP-Luc plasmid (22), and pTCV1 plasmid (23) by BamHI and SalI. These probes were labeled with random primers, the Klenow fragment of DNA polymerase I, and [α-32P]dCTP. The membranes were washed at 56°C according to the manufacturer’s instructions and exposed on a PhosphorImager (Molecular Dynamics). Message levels were analyzed by densitometric analysis.
Primer Extension Analysis.
Polyadenylated mRNA from stably transfected amoebae was isolated using the PolyATract System 1000 (Promega). Primer extension was performed using the Superscript II RNase H− Reverse Transcriptase System (GIBCO/BRL): poly(A)+ RNA was resuspended in water, heat-denatured to 90°C for 2 min and 65°C for 10 min, and cooled slowly to 45°C. The primer 5′-AGGATAGAATGGCGCCGG-3′, which is complementary to the luciferase mRNA 45 bp downstream from the luciferase ATG start codon, was labeled with T4 kinase and [γ-32P]ATP according to the manufacturer’s instructions (Boehringer Mannheim). It then was annealed with the cooled, denatured poly(A)+ RNA at 55°C for 30 min. The entire reaction was then cooled to room temperature for 10 min, and the reaction was extended at 42°C for 50 min according to the manufacturer’s instructions. The sample was ethanol-precipitated overnight, resuspended in water and loading buffer, heat-denatured at 90°C for 10 min, and run next to the appropriate sequencing ladder on a 6% polyacrylamide gel. Sequencing was performed using the Circumvent Thermal Cycle Sequencing System (New England Biolabs) using the [α-35S]dATP incorporation method. To rule out contaminating or nonspecific extension products, control samples with tRNA were used and certain mRNA samples (including mutated TATA and mutated GAAC) were treated in DNase buffer (50 mM Tris⋅HCl, pH 7.4/1 mM EDTA, pH 8.0/10 mM MgCl2/1 mM 1,4-DTT) with 10 units of RNase-free DNase at 37°C for 60 min followed by overnight ethanol precipitation before primer extension experiments. In certain experiments, the mRNA sample was treated with RNase enzyme and then used as a control sample to show specificity of the primer extension reactions.
RESULTS
Nuclear Run-On Analysis of the hgl5 Gene Transcript.
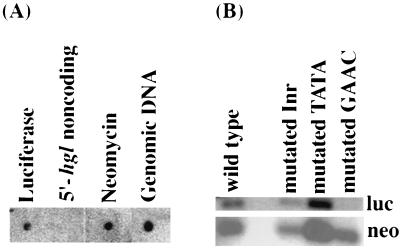
Nuclear run-on analysis can be used to detect nascent mRNA. This technique was used to determine if the gene product of the hgl5 gene of E. histolytica was transcribed monocistronically. Radiolabeled RNA transcripts that hybridized with DNA probes to the ORFs of luciferase, neomycin, and total E. histolytica genomic DNA were detected as expected, whereas no RNA hybridization was detected to the 5′ 272-bp promoter region of the hgl5 gene (Fig. 1A). These data indicated that the hgl5 gene is monocistronically transcribed as previously reported for the hgl2 gene (9). Thus, primer extension can be expected to map the transcription start sites of nascent transcripts and not sites of final processing of mature transcripts.
Figure 1.
(A) Nuclear run-on analysis of trophozoites stably transfected with the wild-type plasmid. Approximately 1.2 pmol of total genomic E. histolytica DNA, luciferase, and neomycin cDNA fragments, and the noncoding promoter region of hgl5 gene DNA were dot-blotted and hybridized with radiolabeled nascent RNA. (B) Northern blot analysis of reporter gene (luciferase) and control (neomycin) mRNA transcribed from the E. histolytica hgl5 promoter constructs containing wild-type, mutated Inr, TATA, and GAAC elements. Poly(A)+ mRNA (1 μg) was hybridized with oligonucleotides containing the neomycin (neo) (0.8 kb) and luciferase (luc) (1.6 kb) coding regions.
Effects of Mutations of Core Promoter Elements on Luciferase Expression and mRNA Levels.
Reporter gene expression and mRNA levels from cells transfected with the wild-type and mutated core promoter constructs were compared. Mutation of the Inr and TATA resulted in modestly decreased luciferase levels, whereas mutation of the GAAC region resulted in a marked decrease in luciferase levels (Fig. 1B). In constructs with mutated Inr and TATA regions, there was a 8.3% and 26% reduction in reporter gene mRNA and a 27% and 52% reduction in luciferase expression, respectively. In the construct with a mutated GAAC region, the luciferase mRNA was reduced by 84.6%, and reporter gene expression was decreased by 82%–91% compared with wild type. Thus, mutation of the GAAC region had the most marked effect on reporter gene mRNA and protein levels.
Effect of Mutations of Core Promoter Elements on the Site of Transcription Initiation.
Alignment of the 5′-flanking regions of E. histolytica protein-encoding genes revealed two conserved core promoter elements with positional similarity to classical Inr and TATA sequences and a third element (GAAC) (15). Mutation of all three core promoter elements (Inr, TATA, and GAAC) affected reporter activity and mRNA levels. The decreased reporter activity could be due to changes in the site of transcription initiation, mRNA stability, or translational efficiency. Because all three conserved elements are within the core promoter, they could be involved in controlling the site of transcription initiation. The technique of primer extension therefore was used to determine the effect of mutations in the core promoter on the choice of transcription start sites. Mapping the 5′ end of mature mRNA with primer extension should accurately reflect the transcription start site because E. histolytica mRNAs are monocistronically transcribed and lack trans-splicing (9).
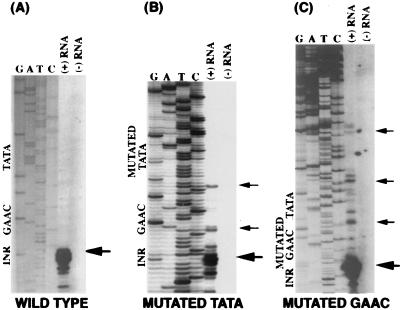
A major extension product for the wild-type (hgl5 promoter-luciferase) mRNA was observed at nucleotides A or C, 30–31 bases downstream of the putative TATA element (Fig. 2A). This is the same location of transcription initiation as the endogenous hgl5 gene (15). No extension product was seen when RNA was excluded from the reaction or when the RNA sample was treated with RNase. In the construct where the putative TATA element was mutated (Fig. 2B), primer extension products were observed upstream, as well as at the wild-type transcription start site. These results were consistently reproduced and not abolished by DNase treatment of the samples before primer extension.
Figure 2.
Primer extension analysis of mRNA transcribed from hgl5 promoter constructs containing wild-type (A), mutated TATA (B), and mutated GAAC (C) sequences. The extension products are located to the right of the sequencing ladders in the (+) RNA lane. Control reactions without RNA are in the lanes labeled (−) RNA. The location of each element (Inr, GAAC, and TATA) is labeled on the DNA sequence. The main primer extension product at GAAAGAC+1 AA (Inr element) is marked with a large arrow. Minor extension products from the mutated TATA and mutated GAAC constructs are labeled with small arrows. Each primer extension experiment was performed at least twice. Five micrograms of poly(A)+ mRNA was used in A and B, and 20 μg of poly(A)+ mRNA was used in C. Samples that were DNase-treated before primer extension showed identical transcription initiation sites (not shown).
Analysis of the promoter in which the GAAC element was mutated (Fig. 2C) revealed that the majority of the transcripts originated at the nucleotides A and C within the Inr. Once again, however, and to a greater extent than seen previously in the TATA mutant, new primer extension bands appeared upstream in the promoter region (see Fig. 2C). In comparing the extension products from mutated TATA and mutated GAAC constructs, the majority of the upstream extension products appeared consistently and reproducibly at adenine residues.
Mutation of the Inr element, in the presence of wild-type TATA and GAAC elements, did not alter the site of transcription initiation (data not shown). These results were consistent with involvement of both the TATA and GAAC elements in controlling the site of transcription initiation, as mutation of these elements resulted in new upstream transcription start sites.
Positional Analysis of the TATA and Inr Elements.
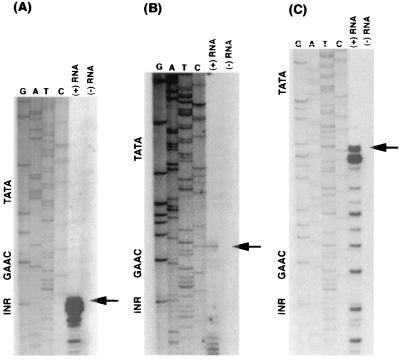
To determine the effects of positional manipulation of the TATA element on the site of transcription initiation, we performed primer extension analysis on constructs in which the TATA element was moved upstream by 15 and 40 nucleotides. As seen in Fig. 3A, primer extension revealed that in the wild-type construct transcription initiated 30–31 bases downstream of the TATA element. When the wild-type TATA element was moved upstream by 15 nucleotides (Fig. 3B), the site of transcription initiation also moved upstream by 15 nucleotides. Similarly when the wild-type TATA element was moved upstream by 40 nucleotides (Fig. 3C) the transcription initiation site moved upstream by 40 nucleotides. The 15-bp insertion between the TATA and Inr regions resulted in a greater decrement in transcription as compared with insertion of 40 nucleotides, as seen by the decreased intensity of the primer extension product in Fig. 3 B versus C. This result may be due to rotational positioning (1.5 versus 4 helical turns between TATA and Inr) of the core promoter elements relative to each other.
Figure 3.
Primer extension analysis of mRNA transcribed from hgl5 promoter constructs where the TATA element has been moved upstream. The position of the Inr, GAAC, and TATA elements are labeled on the left side of the sequencing ladder. The extension product of each reaction is located to the right of the appropriate sequencing ladder in the (+) RNA lane and control reactions are shown in the (−) RNA lane. The main primer extension product is labeled with a large arrow. Each reaction was done using 5 μg of poly(A)+ mRNA.
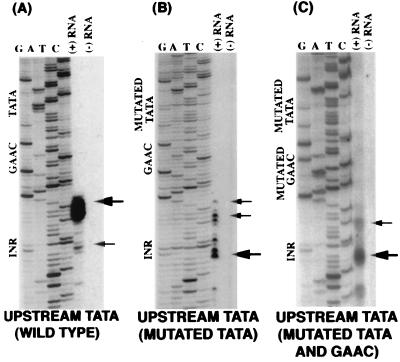
In the above-mentioned experiments, the TATA element was moved upstream independently of the GAAC element. Positional analysis of the TATA element also was done in conjunction with the GAAC element. In Fig. 4A primer extension analysis is shown on a construct where the TATA and GAAC elements both were moved upstream by 10 nucleotides. Transcription start site was mapped and consistent with previous results, the majority of transcription initiated 30–31 bp downstream of the TATA element with a minor band at the Inr element.
Figure 4.
Primer extension analysis of mRNA transcribed from hgl5 promoter constructs with shifted wild-type, mutated TATA, and mutated TATA and GAAC elements. The locations of the Inr, GAAC, and TATA elements are labeled on the left of the sequencing ladder. The extension product of each reaction and appropriate controls are to the right of the appropriate sequencing ladder. Major and minor extension products are indicated by large and small arrows, respectively. Primer extension was performed using 5 μg of poly(A)+ mRNA in A, 10 μg of poly(A)+ mRNA in B, and 80 μg of poly(A)+ mRNA in C. Longer exposures of experiments in which the TATA element was mutated revealed new upstream transcription start sites that were similar in location to those seen in Fig. 1B.
We concluded from these positional analyses that the TATA element was dominant in controlling the site of transcription initiation. The role of the Inr element in regulating transcription initiation therefore was studied in the presence of a mutated TATA element. As shown in Fig. 4A, in the construct where a wild-type TATA and GAAC were moved upstream by 10 nucleotides, two sites of transcription initiation were identified. In the construct where a mutated TATA and wild-type GAAC were moved upstream by 10 nucleotides (Fig. 4B) a significant portion of the primer extension product was now present at the A and C nucleotides in the Inr element (large arrow) with other extension products, which occurred 30–31 bases downstream of the TATA element (small arrow). To determine whether the GAAC or Inr controlled the endogenous transcription start site, the GAAC element was mutated in the construct with the upstream-mutated TATA and wild-type GAAC regions. In Fig. 4C, the primer extension results indicated that the transcription initiated at the endogenous site (large arrow) when the TATA and GAAC elements were mutated. These results indicated that when the TATA and GAAC elements were mutated, the wild-type Inr controlled the selection of transcription start sites.
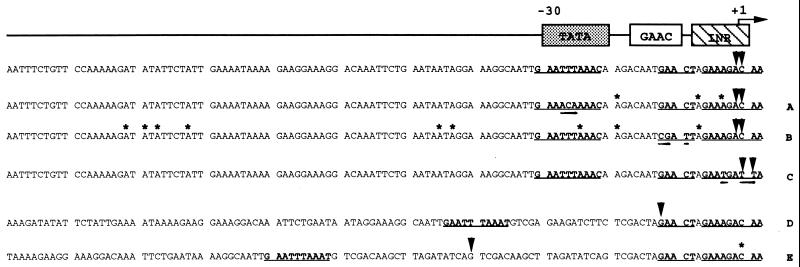
In Fig. 5, a schematic of the core promoter of E. histolytica and a summary of the primer extension analyses are shown. The sequences of the wild-type and mutated TATA, GAAC, and Inr constructs are outlined, and transcription initiation sites are indicated. In each of the above mentioned constructs the major site of transcription initiation was at nucleotides A and C within the Inr element (+1). When the TATA and GAAC elements were mutated new transcription start sites became apparent. These new sites were upstream of the wild-type start site (+1), were consistent between the two constructs, and occurred mostly at adenine residues. Mutation of the GAAC element resulted in transcription initiating farther upstream (−90) than mutation of the TATA region (−19). Positional analysis of the TATA element revealed that transcription initiated precisely 30–31 bases downstream of this region.
Figure 5.
Summary of results of primer extension experiments. Major (arrowheads) and minor (asterisks) primer extension products for each promoter construct are shown. The first sequence is the wild-type hgl5 promoter from −110 to +1. The three conserved elements TATA (−30), GAAC (−15), and Inr (+1) are shown in bold and underlined. Primer extension products of constructs with specific mutations in TATA, GAAC, and Inr (double underlined) are shown in A, B, and C, respectively. In lines D and E, the primer extension results are indicated for the constructs in which the TATA element has been moved upstream by 14 and 40 nucleotides, respectively. In these lines the TATA, GAAC, and Inr elements are in bold.
DISCUSSION
The major conclusion from this study is that three conserved regions in the core promoter of the E. histolytica hgl5 gene regulate the site of transcription initiation. This is in contrast to previously described eukaryotic promoters where only two elements (TATA and Inr) have been demonstrated to control the site of transcription initiation. The TATA and Inr elements in E. histolytica have little sequence homology with apparently functionally equivalent elements in other eukaryotes, and the third element (GAAC) and its sequence are unprecedented in eukaryotes. The GAAC region is present in 31 of 37 sequenced core promoters and has variable positioning in the core promoter (15). Transcription initiation did not occur in the wild-type manner when GAAC was mutated; instead transcription initiated at multiple sites occurring as far upstream as −90. Because the TATA and Inr elements were intact in this construct and theoretically should be able to direct formation of a stable preinitiation complex, the marked decrease in gene activity, and appearance of new transcription start sites by mutation of the GAAC region points to a central and critical role of this element. Disruption of the GAAC element may allow other sites that are normally disfavored to become functional transcription start sites. Possible roles for this region include stabilization of the preinitiation complex via interactions with TFIID, TFIIB, other general transcription factors, or RNA polymerase II. The variable positioning of this element in various core promoters suggests that this region acts in a position-independent manner. One hypothesis is that the GAAC element functions in “tethering” the TFIID/TATA binding protein to the wild-type TATA element, facilitating organized, regulated gene expression. Disruption of GAAC function could be postulated to result in inefficient transcription occurring from TFIID binding randomly to AT-rich sequences in the promoter of the hgl5 gene.
The other two elements in the amoebic core promoter, TATA and Inr, have sequences that are divergent from those of other eukaryotes but appeared to function in a relatively classical manner. Our results demonstrated that the TATA element controlled transcription initiation 30–31 bp downstream. This phenomenon occurred consistently even with positional manipulation of this region and was present regardless of whether TATA was moved upstream alone or in conjunction with GAAC (Figs. 3 and 4). Thus the effect of the TATA element in controlling the site of transcription initiation was not due to its proximity to other regulatory regions. The relative dominance of the TATA element is further evidenced by the observation that mutation of the Inr in the presence of a wild-type TATA resulted in relatively minor decrements in gene activity (15, 20) and no change in the site of transcription initiation.
Although the sequence TATAAA is the consensus eukaryotic TATA element, it has been demonstrated that most AT-rich sequences of 6 base pairs or longer can impart TATA activity in the proximity of other control elements (25, 26). Therefore, it would not be completely unexpected in an AT-rich organism such as E. histolytica that mutation of the wild-type TATA element might unveil cryptic TATA elements. This phenomenon may have occurred in the hgl5 gene as shown by mutation of the TATA and GAAC elements and appearance of new upstream transcription start sites (Fig. 2 B and C). Analysis of the upstream region of the hgl5 promoter revealed the sequence GAATAATAGG at −50, which has sequence similarity to the proposed TATA box of the hgl5 gene (GAATTTAAAC), and conceivably could function as a TATA box in situations where the wild-type TATA element is mutated. Interestingly, mutation of the TATA and GAAC elements resulted in new transcription initiation sites, which were the same between the two constructs and in the majority of the cases were adenine residues (Figs. 2 and 5). This result may represent the preference of the transcriptional machinery for this residue, as has been described previously for the metazoan system (27). Adenine also appears to be a preferred start site in E. histolytica as it was present in 10 of 15 genes where the start site has been mapped (15).
The Inr appeared to have a secondary role in the control of transcription initiation in the E. histolytica hgl5 gene. The Inr was demonstrated only to control the site of transcription initiation in situations where the TATA and/or GAAC elements were mutated (Fig. 4 B and C). Interestingly the Inr elements in E. histolytica can be described by five subpopulations: CE1-a, CE1-b, CE1-c, CE1-d, and CE1-e (15). The hgl5 gene Inr belongs to the subfamily CE1-e with the consensus sequence (ATAGACAA). The relative sequence divergence among established Inr regions of higher eukaryotes and the description of multiple Inr families is well known (28). However, the Inr of E. histolytica protein encoding genes has sequence divergence even at the most highly conserved nucleotides −1(C), +1(A), and +3(T), which have been found in the Inr region of >500 eukaryotic genes (12) and in primitive eukaryotes such as Trichomonas vaginalis (19).
Functional Inr elements have been reported in genes that lack a TATA region, and their ability to orchestrate transcription initiation through interaction with transcription factors such as TFIID, TFIIB, TFIIF, TFIII, and RNA polymerase II is described (14). However, the presence of both an Inr and a TATA element in one gene calls into question the role of the weaker region, although there are metazoan and viral examples of core promoters with both regions such as the adenovirus major late promoter (29). It may be that in such cases the role of the Inr is to increase promoter strength via interactions with activator proteins. The Inr also may play a dominant role in as yet undescribed genes of E. histolytica, which lack both a TATA and GAAC element.
In conclusion, E. histolytica has unusual characteristics regarding its regulation of transcription initiation. The elements we describe here are an interesting contrast to the classical core promoter elements seen in higher eukaryotes. The necessity of three regulatory regions in this protozoan parasite is unclear at present. However, each element with its variable effects on transcription initiation may contribute to a finely tuned transcriptional machinery, which we are just beginning to decipher. The presence of a GAAC element and its postulated novel role in transcriptional control provides exciting data regarding the unique mechanisms of transcriptional regulation in this parasite.
Acknowledgments
We thank David Auble and Mitchell Smith for excellent discussions and scientific input. This work was supported by National Institutes of Health Grant AI 37941. U.S. is a National Foundation for Infectious Diseases Fellow, and W.A.P. is a Burroughs Wellcome Scholar in Molecular Parasitology.
ABBREVIATIONS
- Inr
initiator
- TF
transcription factors
References
- 1.World Health Organization. The World Health Report. Geneva: World Health Organization; 1995. [Google Scholar]
- 2.Dvorak J A, Kobayashi S, Alling D W, Hallahan C W. J Eukaryot Microbiol. 1995;42:610–616. doi: 10.1111/j.1550-7408.1995.tb05915.x. [DOI] [PubMed] [Google Scholar]
- 3.Gelderman A H, Keister D B, Bartgis I L, Diamond L S. J Parasitol. 1971;57:906–911. [PubMed] [Google Scholar]
- 4.Tannich E, Horstmann R D. J Mol Evol. 1992;34:272–273. doi: 10.1007/BF00162976. [DOI] [PubMed] [Google Scholar]
- 5.Lioutas C, Tannich E. Mol Biochem Parasitol. 1995;73:259–261. doi: 10.1016/0166-6851(95)00101-6. [DOI] [PubMed] [Google Scholar]
- 6.Lohia A, Samuelson J. Gene. 1993;127:203–207. doi: 10.1016/0378-1119(93)90720-n. [DOI] [PubMed] [Google Scholar]
- 7.Plaimauer B, Ortner S, Wiedermann G, Scheiner O, Duchene M. Mol Biochem Parasitol. 1994;66:181–185. doi: 10.1016/0166-6851(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 8.Urban B, Blasig C, Forster B, Hamelmann C, Horstmann R D. Mol Biochem Parasitol. 1996;80:171–178. doi: 10.1016/0166-6851(96)02684-9. [DOI] [PubMed] [Google Scholar]
- 9.Bruchhaus I, Leippe M, Lioutas C, Tannich E. DNA Cell Biol. 1993;12:925–933. doi: 10.1089/dna.1993.12.925. [DOI] [PubMed] [Google Scholar]
- 10.Purdy J E, Mann B J, Pho L T, Petri W A., Jr Proc Natl Acad Sci USA. 1994;91:7099–7103. doi: 10.1073/pnas.91.15.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAndrew M B, Read M, Sims P F G, Hyde J E. Gene. 1993;124:165–171. doi: 10.1016/0378-1119(93)90390-o. [DOI] [PubMed] [Google Scholar]
- 12.Bucher P. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 13.Pugh B F. Curr Opin Cell Biol. 1996;8:303–311. doi: 10.1016/s0955-0674(96)80002-0. [DOI] [PubMed] [Google Scholar]
- 14.Nikolov D B, Burley S K. Proc Natl Acad Sci USA. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purdy J E, Pho L T, Mann B J, Petri W A., Jr Mol Biochem Parasitol. 1996;78:91–103. doi: 10.1016/s0166-6851(96)02614-x. [DOI] [PubMed] [Google Scholar]
- 16.Early A E, Williams J G. Nucleic Acids Res. 1989;17:6473–6484. doi: 10.1093/nar/17.16.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sucic J F, Selmin O, Rutherford C L. Dev Genet. 1993;14:313–322. doi: 10.1002/dvg.1020140409. [DOI] [PubMed] [Google Scholar]
- 18.Wong J M, Liu F, Bateman E. Nucleic Acids Res. 1992;20:4817–4824. doi: 10.1093/nar/20.18.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quon D V K, Delgadillo M G, Khachi A, Smale S T, Johnson P J. Proc Natl Acad Sci USA. 1994;91:4579–4583. doi: 10.1073/pnas.91.10.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buβ H, Lioutas C, Dobinsky S, Nickel R, Tannich E. Mol Biochem Parasitol. 1995;72:1–10. doi: 10.1016/0166-6851(95)00060-e. [DOI] [PubMed] [Google Scholar]
- 21.Diamond L S, Harlow D R, Cunnick C C. Trans R Soc Trop Med Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- 22.Ramakrishnan G, Vines R R, Mann B J, Petri W A., Jr Mol Biochem Parasitol. 1997;84:93–100. doi: 10.1016/s0166-6851(96)02784-3. [DOI] [PubMed] [Google Scholar]
- 23.Vines R R, Purdy J E, Ragland B D, Samuelson J, Mann B J, Petri W A., Jr Mol Biochem Parasitol. 1995;71:265–267. doi: 10.1016/0166-6851(95)00057-8. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg M E, Bender T P. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1997. pp. 4.10.1–4.10.11. [Google Scholar]
- 25.Kollmar R, Farnham P J. Proc Soc Exp Biol Med. 1993;203:127–139. doi: 10.3181/00379727-203-43583. [DOI] [PubMed] [Google Scholar]
- 26.Smale S T. In: Transcription: Mechanisms and Regulation. Conaway R C, Conaway J W, editors. New York: Raven; 1994. pp. 63–81. [Google Scholar]
- 27.O’Shea-Greenfield A, Smale S T. J Biol Chem. 1992;267:1391–1402. [PubMed] [Google Scholar]
- 28.Weis L, Reinberg D. FASEB J. 1992;6:3300–3309. doi: 10.1096/fasebj.6.14.1426767. [DOI] [PubMed] [Google Scholar]
- 29.Lee R F, Concino M F, Weinmann R. Virology. 1988;165:51–56. doi: 10.1016/0042-6822(88)90657-5. [DOI] [PubMed] [Google Scholar]