Summary
Green Fluorescent Protein (GFP) and homologous proteins possess a unique pathway of chromophore formation based on autocatalytic modification of their own amino acid residues. Green-to-red photoconvertible fluorescent protein Kaede carries His-Tyr-Gly chromophore-forming triad. Here we describe synthesis of Kaede red chromophore (2-[(1E)-2-(5-imidazolyl)ethenyl]-4-(p-hydroxybenzylidene)-5-imidazolone) and its analogs that can be potentially formed by natural amino acid residues. Chromophores corresponding to the following tripeptides were obtained: His-Tyr-Gly, Trp-Tyr-Gly, Phe-Trp-Gly, Tyr-Trp-Gly, Asn-Tyr-Gly, Phe-Tyr-Gly, and Tyr-Tyr-Gly. In basic conditions they fluoresced red with relatively high quantum yield (up to 0.017 for Trp-derived compounds). The most red-shifted absorption peak at 595 nm was found for the chromophore Trp-Tyr-Gly in basic DMSO. Surprisingly, in basic DMF non-aromatic Asn-derived chromophore Asn-Tyr-Gly demonstrated the most red-shifted emission maximum at 642 nm. Thus, Asn residue may be a promising substituent, which can potentially diversify posttranslational chemistry in GFP-like proteins.
Keywords: green fluorescent protein, fluorophore, bathochromic shift, fluorescent probes, protein engineering
1. Introduction
In contrast to all other known natural protein-based pigments, proteins of Green Fluorescent Protein (GFP) family form their chromophores by autocatalytic modification of internal amino acid residues [1]. This property makes GFP-like proteins a useful fluorescent marker for live cell labeling [2]. Mechanisms of chromophore formation within GFP-like proteins and properties of the chromophores are of undoubted basic interest.
Chromophore in GFP is formed by cyclization and further dehydration and oxidation of amino acids at positions 65–67 (Ser-Tyr-Gly). These reactions result in 4-(p-hydroxybenzylidene)-5-imidazolone, which includes a newly formed 5-membered heterocycle and Tyr66-derived phenolic ring connected by a methylene bridge (Scheme 1). In all known natural GFP-like proteins positions corresponding to the chromophore-forming Tyr66 and Gly67 are invariant, while the first chromophore-forming residue 65 can vary (here and further the numbering is in accordance to GFP). Studies of last several years clearly demonstrated that 4-(p-hydroxybenzylidene)-5-imidazolone chromophore core is common for all natural GFP-like proteins. At the same time, red-shifted proteins can contain a modification of the protein backbone at position 65 [1].
Scheme 1.
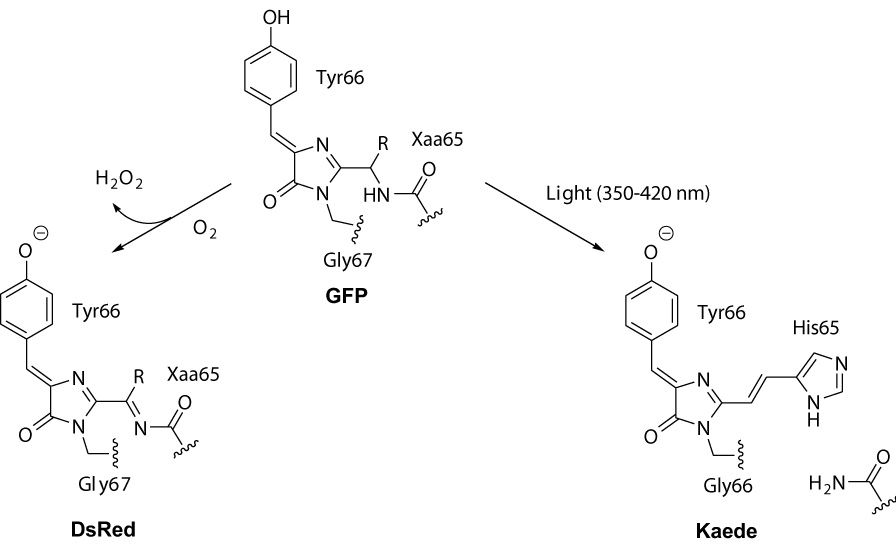
Origination of DsRed-type and Keade-type chromophores from GFP-type precursor in the course of posttranslational modifications of fluorescent proteins.
Two main types of red chromophores within GFP-like proteins are known. One type was first described for DsRed [3]. DsRed-type chromophore consists of GFP chromophore core extended with an acylimine group (Scheme 1) which is formed as a result of dehydrogenation of Cα-N bond of the first chromophore-forming residue with molecular oxygen [3, 4]. This oxidation is catalyzed by nearby residues and occurs in the dark, although it can be greatly facilitated by irradiation with light at about 400 nm [5]. In some proteins, DsRed-like chromophore undergoes further chemical modifications of the acylimine group [6–9].
Second red chromophore type is characteristic for green-to-red photoconvertible fluorescent proteins carrying His-Tyr-Gly chromophore-forming residues. The first protein of this type named Kaede was cloned from stony coral Trachyphyllia geoffroyi in 2002 [10]. In the dark, Kaede is a stable green fluorescent protein with a GFP-like chromophore. However, irradiation with UV or violet light (at approximately 350–420 nm) efficiently converts Kaede into red fluorescent state. Kaede red chromophore is formed by protein backbone break between N and Cα of His65 [11–13]. As a result, GFP-like chromophore core is brought into conjugation with His65 heterocycle (Scheme 1). This reaction is non-oxidative and requires no molecular oxygen. A number of Kaede-like proteins were characterized and shown to be a useful tool to study movements of cells, organelles and proteins [10,14–19], to perform ultra-high resolution fluorescence imaging [20], and to visualize protein degradation in living cells [21].
Considering the role of His65 aromatic ring in Kaede-type chromophore red shift, it is tempting to test other aromatic amino acids at this position. However, up to date all attempts to create functional mutants of Kaede-like proteins carrying Phe, Tyr or Trp as well as any other residues instead of chromophore-forming histidine failed [11,14]. Such mutants are either non-fluorescent or have no ability of green-to-red conversion. This failure might be a result of improper orientation of the bigger (compared to His) aromatic residues. It is known from crystal structure of Kaede-like proteins that position and orientation of the chromophore-forming His is almost unchanged during green-to-red photoconversion [12,13]. Thus, possibly the chromophore environment catalyses formation of the red chromophore by stabilizing the product-like transition state. If so, success in the histidine substitution might be a matter of efforts to create a proper chromophore environment by mutating adjacent amino acid positions. Another explanation is a direct participation of His65 imidazole ring in protonation-deprotonation events during photoconversion [11,14]. In this case, Phe, Tyr or Trp residues can not act as His. Anyhow, absence of corresponding mutants of Kaede-like proteins precludes studying spectral properties of Kaede-like chromophore variants.
Chemical synthesis of natural compounds often provides useful information on their structure and properties. Synthetic imidazolones related to GFP chromophore have been a subject of extensive research [22–26]. However, up to date, a few compounds corresponding to natural chromophores of red-shifted GFP-like proteins were synthesized. Model compounds that are similar but not identical to DsRed chromophore were described [27]. We earlier synthesized a chromophore of a non-fluorescent purple chromoprotein asFP595 [28]. Here we describe a general synthetic approach to the chromophores of Kaede type. Synthesis of Kaede-like chromophores with different groups corresponding to natural amino acids at positions 65 and 66 made it possible to evaluate their influence on spectral properties of the chromophore and to find structures with the most red-shifted absorption and emission.
2. Materials and methods
2.1. General
NMR spectra were recorded on a Bruker DRX-500 instrument; chemical shifts are expressed in δ scale downfield from tetramethylsilane and are referenced to residual protium in the NMR solvent (CHCl3, δ 7.26; CHD2OD, 3.30; CD3S(O)CD2H, δ 2.50). Analytical and preparative TLC was performed on precoated Silufol UV 254 silica or alumina plates impregnated with a fluorescent indicator (254 nm). TLC plates were visualized by exposure to UV light or treatment with saturated permanganate solution followed by washing with excess water, or with phosphomolybdic acid with subsequent heating, or by specific color change of acidic compounds in ammonia vapors. Column chromatography was performed on Merck Kieselgel 60 (70–230 mesh) silica or Aldrich CAMAG-A-1, (150 mesh, basic, Brockman activity I) alumina. ESI mass-spectra were obtained on Bruker Esquire6 Plus instrument.
2.2. Phosphonium salts
Phosphonium salts were prepared from corresponding bromides and triphenylphosphine by refluxing in toluene until the bromide was consumed, except for (4-imidazolyl)methyltriphenylphosphonium chloride (3a) [29]. N-Boc-3-(bromomethyl)indole (3e) was prepared as described [30].
2.3. Aldehyde 2a
A suspension of imidazolone 1a (2.98 g, 13.9 mmol) and selenium dioxide (1.85 g, 16.7 mmol, 1.2 equiv) in anhydrous dioxane (140 mL) was stirred at reflux for 1 h and carefully decanted from the red solid while hot. The solution was concentrated in vacuo, the crude product was dissolved in dichloromethane (50 mL), treated sequentially with diisopropylethylamine (3.59 g, 27.8 mmol, 2.0 equiv) and tert-butyldimethylsilyl chloride (4.19 g, 27.8 mmol, 2.0 equiv). After stirring for 2 h at room temperature, the volatiles were removed in vacuo and the residue was purified by column chromatography (silica, 2% EtOH:CHCl3) to afford the aldehyde 2a as an orange-red solid (2.73 g, 57%). Rf 0.46, silica, 2% EtOH:CHCl3; 1H NMR (500 MHz, CDCl3) δ 9.68 (s, 1H), 8.28 (d, 2H, J = 8.4 Hz), 7.48 (s, 1H), 7.00 (d, 2H, J = 8.4 Hz), 3.32 (s, 3H), 0.96 (s, 9H), 0.27 (s, 6H).
2.4. Aldehyde 2b
A suspension of imidazolone 1b (707 mg, 2.95 mmol) and selenium dioxide (393 mg, 3.54 mmol, 1.2 equiv) in anhydrous dioxane (50 mL) was stirred at reflux for 1 h and carefully decanted from the red solid while hot. The solvent was removed in vacuo and the residual red oil was purified by column chromatography (basic alumina, 10% EtOH in CHCl3) to afford the aldehyde 3b as a fine red crystalline solid (549 mg, 73%). Rf 0.31, basic alumina, 10% EtOH:CHCl3; 1H NMR (500 MHz, DMSO-d6) δ 12.51 (br s, 1H), 9.69 (s, 1H), 8.53 (s, 1H), 8.07 (s, 1H), 7.53 (d, J = 7.8 Hz), 7.32-7.24 (m, 3H), 3.35 (s, 3H).
2.5. Chromophore HYG (Kaede chromophore)
To a solution of 480 mg (1.39 mmol, 2.1 equiv) of aldehyde 2a in dichloromethane (10 mL) 2M aqueous NaOH (10 mL) was added, and then solid phosphonium salt 3a (250 mg, 0.66 mmol) was added with vigorous stirring. The stirring was continued for 30 min and the organic phase was separated. Dichloromethane was evaporated, the mixture was redissolved in 10 mL THF and a solution of tetrabutylammonium fluoride trihydrate (438 mg, 1.39 mmol) in 5 mL THF was added. The mixture was stirred for 1 min and then neutralized with glacial acetic acid (0.12 mL, 2 mmol). The volatiles were removed in vacuo and the residue was redissolved in ethanol (5 mL). The ethanolic solution (1 mL) was purified by preparative TLC (15% EtOH:CHCl3). The combined target fractions from 5 runs were chromatographed again in the same manner, resulting in 10 mg (5%) of pure product as a red-orange solid. Rf 0.30, silica, 15% EtOH:CHCl3; 1H NMR (500 MHz, CD3OD) 8.10 (d, J = 8.6 Hz, 2H), 7.95 (d, J = 15.6 Hz, 1H), 7.81 (s, 1H), 7.50 (s, 1H), 7,01 (s, 1H), 6.99 (d, J = 15.6 Hz, 1H), 6.84 (d, J = 8.6 Hz, 2H), 3.58 (s, 3H). MS (ESI-MS) calcd for C16H15N4O2+ [M+H+] 295.12, found 295.27.
2.6. Chromophore YYG
To a suspension of aldehyde 2a (500 mg, 1.45 mmol) and (4-(tert-butyldimethylsiloxy)benzyl)triphenylphosponium bromide (3b) (817 mg, 1.45 mmol, 1.0 equiv) in dichloromethane (60 mL) was added 2M NaOH (40 mL). The orange organic layer turned instantly dark-red. The resulting mixture was vigorously stirred for 30 min, and the layers were separated. The aqueous layer was extracted with dichloromethane (2×30 mL). The combined organic extracts were treated with tetrabutylammonium fluoride trihydrate (960 mg, 3.04 mmol, 2.1 equiv) to produce a violet suspension which was neutralized with 30% aqueous CH3COOH. Solvents were removed in vacuo and the crude product was purified by column chromatography (silica, 10% EtOH:CHCl3) and recrystallized from methanol to give the pure product as red-brown crystals (108 mg, 23%). Rf 0.19, silica, 5% EtOH:CHCl3; 1H NMR (500 MHz, DMSO-d6) δ 10.13, (br s, 2H), 8.14 (d, J = 8.7 Hz, 2H), 7.95 (d, J = 15.7 Hz, 1H), 7.71 (d, J = 8.7 Hz, 2H), 6.99 (d, J = 15.7 Hz, 1H), 6.93 (s, 1H), 6.87 (d, J = 8.7 Hz, 2H), 6.85 (d, J = 8.7 Hz, 2H), 3.26 (s, 3H); MS (ESI-MS) calcd for C19H17N2O3+ [M+H+] 321.12, found 321.18.
2.7. Chromophore FYG
Procedure is essentially the same as for the chromophore YWG; benzyltriphenylphosphonium bromide (3c) was used. Yield 53%. Rf 0.35, silica, 5% EtOH:CHCl3; 1H NMR (500 MHz, DMSO-d6) δ 10.20, (br s, 1H), 8.20 (d, J = 8.7 Hz, 2H), 7.99 (d, J = 15.7 Hz, 1H), 7.84 (d, J = 7.5 Hz, 2H), 7.47-7.40 (m, 3H), 7.22 (d, J = 15.7 Hz, 1H), 6.99 (s, 1H), 6.89 (d, J = 8.7 Hz, 2H), 3.29 (s, 3H); MS (ESI-MS) calcd for C19H17N2O2+ [M+H+] 305.13, found 305.25.
2.8. Chromophore NYG
Procedure is essentially the same as for the chromophore YWG; (carbamoylmethyl)benzyltriphenylphosphonium bromide (3d) was used. Yield 49%. Rf 0.26, silica, 10% EtOH:CHCl3; 1H NMR (500 MHz, DMSO-d6) δ 10.29, (br s, 1H), 8.15 (d, J = 7.8 Hz, 2H), 7.93 (s, 1H), 7. 45 (s, 1H), 7.28 (d, J = 12.8 Hz, 1H), 7.22 (d, J = 12.8 Hz, 1H), 7.08 (s, 1H), 6.85 (d, J = 7.8 Hz, 2H), 3.20 (s, 3H); MS (ESI-MS) calcd for C14H14N3O3+ [M+H+] 272.10, found 272.36.
2.9. Chromophore WYG
To a stirred suspension of aldehyde 2a (224 mg, 0.65 mmol) and ((N-Boc-3-indolyl)methyl)triphenylphoshonium bromide (3e) (372 mg, 0.65 mmol, 1.0 equiv) in t-butyl alcohol (9 mL) was added a solution of potassium t-butoxide (73 mg, 0.65 mmol, 1.0 equiv) in t-butyl alcohol (3 mL). After 1 h of stirring, more potassium t-butoxide (37 mg, 0.33 mmol, 0.5 equiv) was added and stirring was continued for 30 min. Solvent was removed in vacuo, the residue was dissolved in dichloromethane (5 mL) and treated with tetrabutylammonium fluoride trihydrate (215 mg, 0.68 mmol, 1.05 equiv) and stirred for 5 min. To the resulting deep purple solution were added sequentially p-cresol (70 mg, 1.0 equiv) and trifluoroacetic acid (1 mL) and the mixture was refluxed for 30 min, cooled down and neutralized with conc. ammonia (1.5 mL). The resulting precipitate was filtered and recrystallized from methanol to yield the pure product as a red crystalline solid (49 mg, 22%). Rf 0.21, silica, 5% EtOH:CHCl3; 1H NMR (500 MHz, DMSO-d6) δ 11.96, (br s, 1H), 10.16, (br s, 1H), 8.53 (s, 1H), 8.01 (d, J = 15.6 Hz, 1H), 7.99 (d, J = 8.6 Hz, 2H), 7.97 (d, J = 7.8 Hz, 1H), 7.47 (d, J = 7.8 Hz, 1H), 7.18 (t, J = 7.8 Hz, 1H), 7.16 (t, J = 7.8 Hz, 1H), 6.75 (d, J = 15.6 Hz, 1H), 6.70 (s, 1H), 6.24 (d, J = 8.6 Hz, 2H), 3.25 (s, 3H). MS (ESI-MS) calcd for C21H18N3O2+ [M+H+] 344.14, found 344.17.
2.10. Chromophore YWG
To a stirred suspension of aldehyde 2b (76 mg, 0.30 mmol) and (4-(tert-butyldimethylsiloxy)benzyl)triphenylphosphonium bromide (3b) (169 mg, 0.30 mmol) in tert-butyl alcohol (4 mL) was added a solution of potassium t-butoxide (33 mg, 0.20 mmol) in tert-butyl alcohol (2 mL). The reaction mixture was stirred for 30 min, the solvent was removed in vacuo and the residue was purified by column chromatography on alumina (CHCl3) to afford the TBS-protected chromophore. Rf 0.32, silica, 5% EtOH–CHCl3. TBS-protected chromophore was dissolved in CH2Cl2 (5 mL) and treated with tetrabutylammonium fluoride trihydrate (100 mg, 0.32 mmol, 1.05 equiv). The resulting purple solution was neutralized with acetic acid (25 µL, 0.44 mmol) and the reaction mixture was concentrated in vacuo. The product crystallized on treatment with 80% aqueous methanol (2 mL) to afford the title compound as a red-purple crystalline solid (43 mg, 42%). Rf 0.23, silica, 5% EtOH:CHCl3; 1H NMR (500 MHz, DMSO-d6) δ 11.93 (br s, 1H), 9.90 (br s, 1H), 8.54 (s, 1H), 8.22 (d, J = 7.9 Hz, 1H), 7.91 (d, J = 15.6 Hz, 1H), 7.68 (d, J =8.2 Hz, 2H), 7.49 (d, J = 7.9 Hz, 1H), 7.34 (s, 1H), 7.22 (t, J = 7.9 Hz, 1H), 7.20 (t, J = 7.9 Hz, 1H), 6.99 (d, J = 15.6 Hz, 1H), 6.86 (d, J = 7.5 Hz, 2H), 3.18 (s, 3H); MS (ESI-MS) calcd for C21H18N3O2+ [M+H+] 344.14, found 344.16.
2.11. Chromophore FWG
To a stirred suspension of aldehyde 2b (50 mg, 0.20 mmol) and benzyltriphenylphosphonium bromide (3c) (85 mg, 0.20 mmol, 1.0 equiv) in tert-butyl alcohol (3 mL) was added a solution of potassium tert-butoxide (33 mg, 0.30 mmol, 1.5 equiv) in tert-butyl alcohol (1 mL). Instant color change from orange to dark-red was observed. The reaction mixture was stirred for 30 min, the solvent was removed in vacuo and the residue was purified by column chromatography on silica (5% EtOH:CHCl3) to afford the product as fine red-brown crystalline solid (28 mg, 43%). Rf 0.38, silica, 5% EtOH–CHCl3; 1H NMR (500 MHz) δ 12.06 (br s, 1H), 8.26 (s, 1H), 8.07 (d, J = 7.9 Hz, 1H), 7.94 (d, J = 7.8 Hz, 2H), 7.85 (d, J = 7.9 Hz, 1H), 7.64-7.36 (m, 3H), 7.24-7.15 (m, 2H), 7.12 (d, J = 13.4 Hz, 1H), 7.08 (t, J = 7.9 Hz, 1H), 6.48 (d, J = 13.4 Hz, 1H), 3.18 (s, 3H); MS (ESI-MS) calcd for C21H18N3O+ [M+H+] 328.14, found 328.17.
2.12. Optical spectral measurement
Absorption and excitation–emission spectra were recorded at room temperature using Beckman DU520 UV/VIS spectrophotometer and Varian Cary Eclipse fluorescence spectrophotometer, respectively. Emission spectra were corrected for photomultiplier response. Fluorescence quantum yields of chromophores in basic DMF were determined by using purified DsRed2 as a reference standard (quantum yield 0.55) [31].
3. Results and discussion
3.1. Synthesis
Kaede-like chromophores corresponding to the following tripeptides were obtained: His-Tyr-Gly, Trp-Tyr-Gly, Phe-Trp-Gly, Tyr-Trp-Gly, Asn-Tyr-Gly, Phe-Tyr-Gly, and Tyr-Tyr-Gly. They were designated according to the one-letter amino acid code as HYG, WYG, FWG, YWG, NYG, FYG, and YYG, respectively (Fig. 1).
Figure 1.
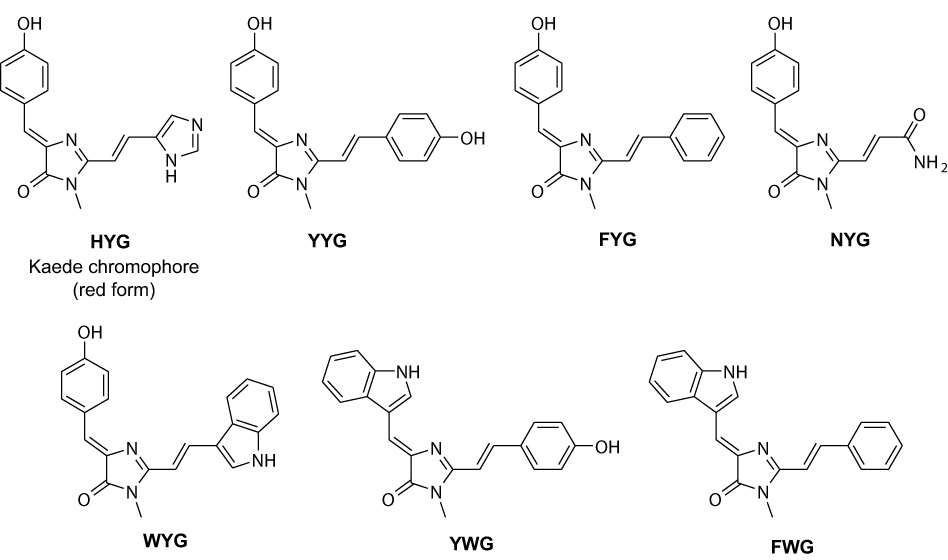
Structures of Kaede and Kaede-like chromophores synthesized.
Oxidation of readily available 2-methylimidazolones 1 [31] with selenium dioxide in refluxing dioxane provided aldehydes 2 (Scheme 2). In case of imidazolone 1a, the desired aldehyde underwent decomposition to unidentified products on attempt of chromatographic purification. The TBS-protected aldehyde 2a, synthesized by treatment of the crude product with TBSCl and diisopropylethylamine, was stable to chromatography and was isolated in 57% combined yield. However, it decomposes significantly in 1 month even if stored at −20°C. A number of other methods to bring about such oxidation failed. O-protected substrates also did not produce the desired products with SeO2 or other oxidants. Indolic imidazolone 1b gave the corresponding aldehyde 2b in 73% yield. Target chromophores were obtained via Wittig reaction of aldehydes 2a and 2b with appropriate phosphonium salts 3 (Table 1). In all cases, the reaction proceeded stereoselectively to give trans-product (E:Z > 15:1), with characteristic coupling constant of about 15 Hz. Low yield of the Kaede chromophore HYG is probably due to a competing deprotonation of imidazole nitrogen in 3e and expulsion of PPh3 molecule, producing the electrophilic diazafulvene intermediate 4 [29] rather than desired nucleophilic phosphorous ylide 5 (Scheme 3). If sterically hindered potassium tert-butoxide is used as base, this pathway seems to become major and no chromophore is formed. In this case, biphasic mixture of aqueous NaOH and CH2Cl2 was employed. For other chromophores, the results obtained using either of these two bases were comparable.
Scheme 2.
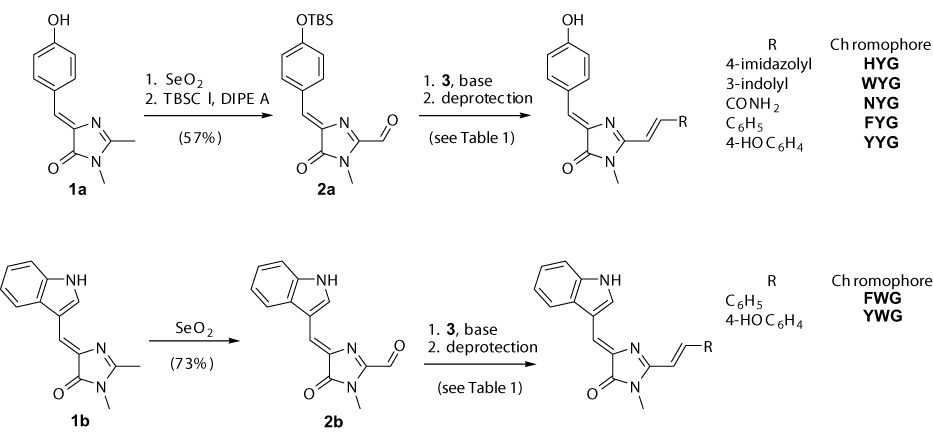
Synthesis of Kaede-type chromophores.
Table 1.
Yields and conditions of Wittig reaction.
| Aldehyde | Phosphonium salt | Product (yield, %) |
|---|---|---|
| 2a |
 (3a) (3a) |
HYG (5)[a] |
| 2a | 4-(TBSO)C6H4CH2PPh3Br (3b) | YYG (23)[a] |
| 2a | BnPPh3Br (3c) | FYG (53)[b] |
| 2a | H2NCOCH2PPh3Br (3d) | NYG (49)[b] |
| 2a |
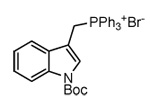 (3e) (3e) |
WYG (18)[b,c] |
| 2b | 4-(TBSO)C6H4CH2PPh3Br (3b) | YWG (42)[b] |
| 2b | BnPPh3Br (3c) | FWG (44)[b] |
2M aqueous NaOH / CH2Cl2.
t-BuOK / t-BuOH.
After Boc cleavage (see Experimental Section).
Scheme 3.
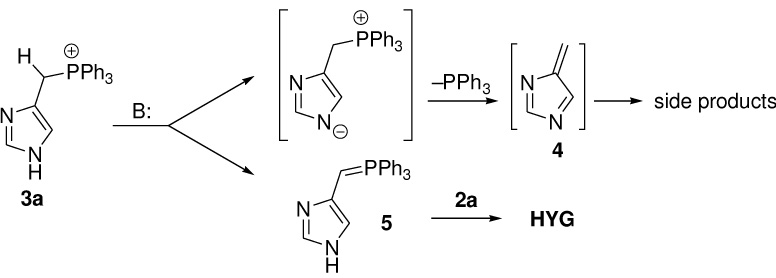
Reaction pathways for phosphonium salt 3a.
3.2. Spectral properties
As expected, wild type Kaede chromophore HYG demonstrated clear pH dependence of absorption spectrum. In water solutions, acidic form absorbed at 430 nm (Table 2). With an increase of pH, this peak transformed into a peak at 490 nm with isosbestic point at 447 nm and pKa 7.7 (Fig. 2A). We attributed this transition to deprotonation of phenolic oxygen that is a common process for chromophores within other GFP-like proteins. No other spectral transitions were observed in interval pH 3–14. Nature of solvent was found to have a minor impact on absorption maximum and extinction coefficient of the acidic form of HYG (Fig. 2B). In contrast, in basic conditions HYG chromophore possessed clearly different absorption spectra in different solvents. Strong absorption red shift was observed in the series water-ethanol-isopropanol-DMF-DMSO (Fig. 2C, Table 2). Compared to HBMPI and HBMPDI [27], absorption maxima are higher due to extension of conjugation to another aromatic ring or electron-withdrawing substituent.
Table 2.
Spectral properties of Kaede-like chromophores.
| Chrommophore | Absorption max, nm (extinction coefficient, mM−1cm−1) in neutral solutions[a] |
pKa (water) | Absorption max, nm (extinction coefficient, mM−1cm−1) in basic solutions[b] |
Emission max, nm | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water | EtOH | iPrOH | DMF | DMSO | Water | EtOH | iPrOH | DMF | DMSO | (QY)[c] | ||
| HYG | 430 | 443 | 444 | 446 | 448 | 7.7 | 490 | 502 | 514 | 557 | 565 | 582 |
| (41) | (40) | (41) | (40) | (37) | (53) | (79) | (80) | (77) | (80) | (0.005) | ||
| WYG | 460 | 462 | 460 | 460 | 465 | 8.1 | 494 | 509 | 532 | 580 | 595 | 615 |
| (7) | (15) | (16) | (14) | (14) | (21) | (24) | (27) | (27) | (27) | (0.017) | ||
| FWG | 455 | 456 | 460 | 456 | 462 | 11.6 | 498[d] | 508 | 526 | 556 | 563 | 625 |
| (11) | (23) | (25) | (20) | (19) | (13) | (23) | (26) | (22) | (24) | (0.016) | ||
| YWG | 462 | 466 | 468 | 464 | 468 | 8.3 | 496 | 509 | 533 | 567 | 577 | 610 |
| (8) | (30) | (26) | (23) | (22) | (27) | (34) | (40) | (38) | (44) | (0.017) | ||
| NYG | 424 | 433 | 433 | 431 | 435 | 7.6 | 510 | 531 | 548 | 564 | 543 | 642 |
| (27) | (19) | (18) | (21) | (18) | (35) | (36) | (30) | (34) | (25) | (0.005) | ||
| FYG | 425 | 437 | 451 | 437 | 438 | 7.8 | 492 | 513 | 526 | 553 | 572 | 635 |
| (28) | (29) | (26) | (27) | (28) | (43) | (45) | (44) | (49) | (50) | (0.009) | ||
| YYG | 436 | 459 | 483 | 445 | 447 | 7.8 | 506 | 522 | 531 | 556 | 588 | 625 |
| (30) | (35) | (42) | (22) | (29) | (49) | (61) | (60) | (59) | (59) | (0.016) | ||
In water at pH 5 or in pure solvents.
In water at pH 10 or in solvents containg 50 mM NaOH.
In basic (50 mM NaOH) DMF.
At pH 13.
Figure 2.
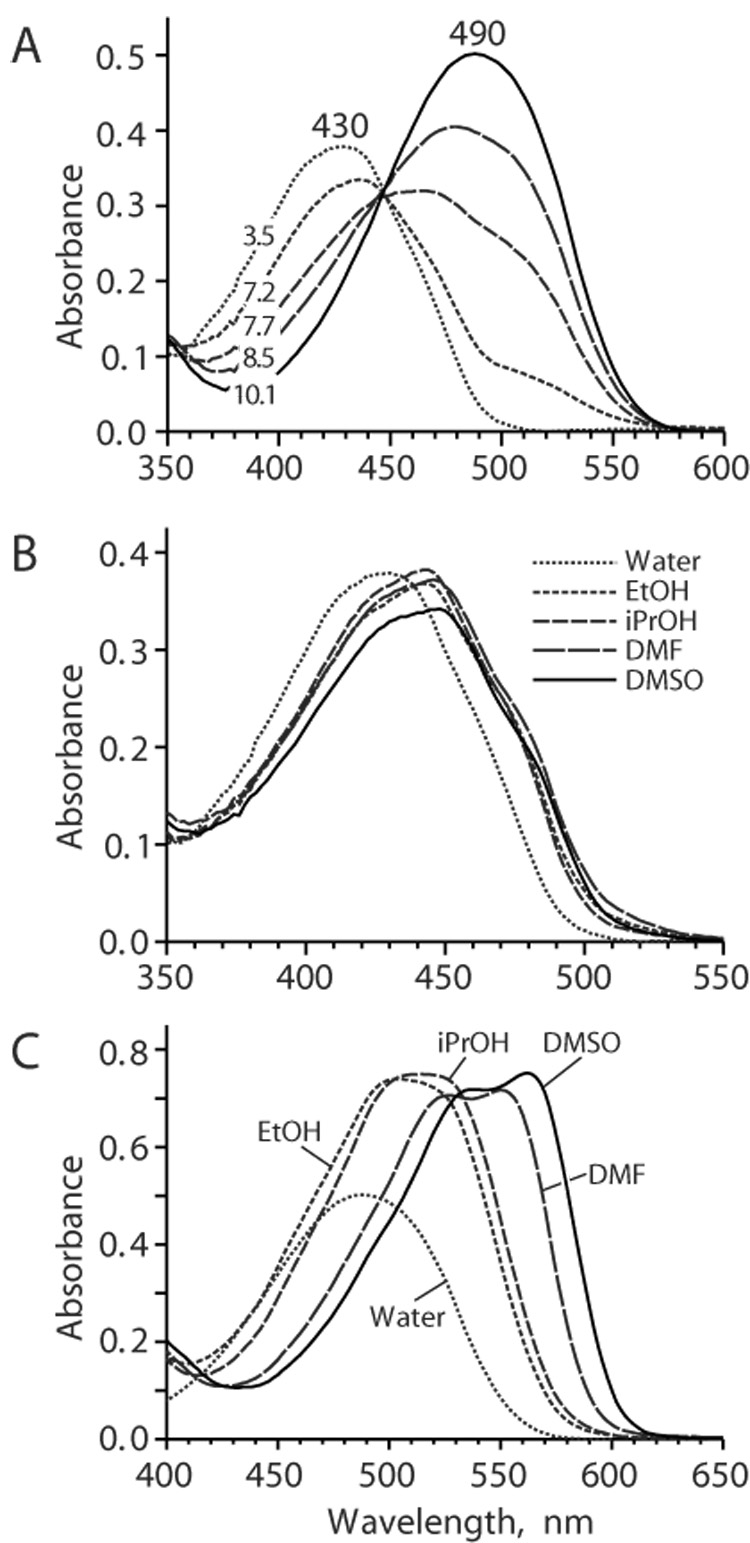
Absorption spectra for HYG chromophore. (A) Absorption spectra for buffered water solutions of HYG chromophore at different pH as designated on the graphs. (B and C) Absorption spectra for HYG chromophore in different solvents at pH 3.5 (B) or pH 10.1 (C).
Other Kaede-like chromophores demonstrated similar dependences on pH and solvent with some exceptions (Table 2 and Fig. 3). As expected, chromophore FWG showed no spectral changes at interval pH 5–10 since this chromophore does not have acidic phenolic hydroxyl. Instead, a spectral transition to a red-shifted form was observed with pKa 11.6 probably associated with deprotonation of indole. At low pH Trp-containing chromophores (WYG, FWG and YWG) demonstrated transition with pKa ~2.5–3.5 into spectral forms characterized with considerably higher extinction coefficient and red-shifted absorption maxima compared to corresponding absorption peaks at pH 4–7 (Table 3 and Fig. 3C). Most probably, this transition can be attributed to appearance of positively charged chromophore state due to protonation of N-3 of the imidazolone ring, which is stabilized by indole. Other Kaede-like chromophores also possessed similar spectral transition but at considerably lower pH (pKa ~1.5) as reported for GFP chromophore [23] and for HBMPI and HBMPDI [27] chromophores. NYG chromophore had the most red-shifted absorption in DMF, not in DMSO as others.
Figure 3.
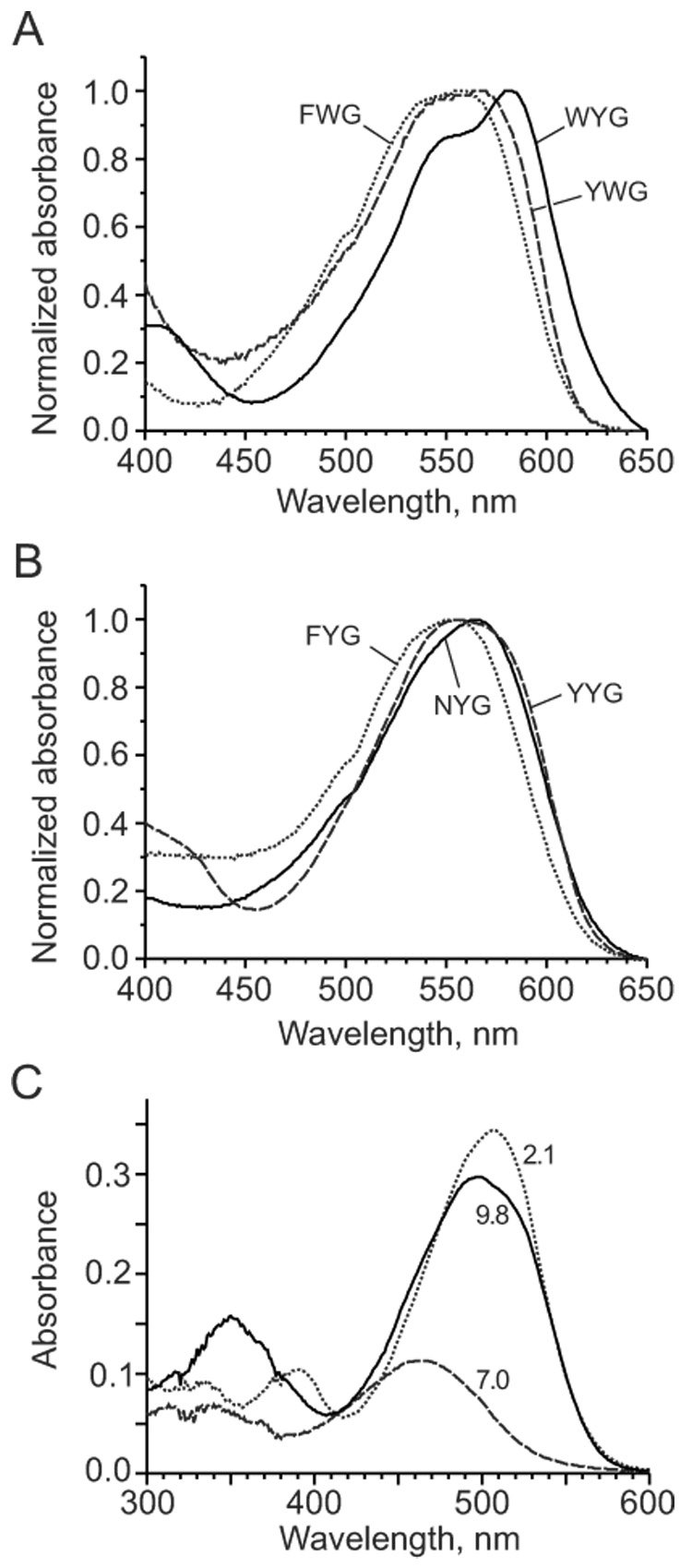
Absorption spectra for Kaede-like chromophores. (A) Absorption spectra for Trp-containing chromophores (WYG, FWG and YWG) in basic DMF. (B) Absorption spectra for chromophores FYG, NYG and YYG in basic DMF. (C) Absorption spectra for buffered water solutions of WYG chromophore (15 µM) at pH 2.1 (dotted line), 7.0 (dashed line), and 9.8 (solid line).
Table 3.
Absorption characteristics of indolic Kaede-like chromophores in acidic solutions (in water at pH 2 or in solvents containg 50 mM HCl).
| Chromophore | pKa (water) | Absorption max, nm (extinction coefficient, mM−1cm−1) |
||||
|---|---|---|---|---|---|---|
| Water | EtOH | iPrOH | DMF | DMSO | ||
| WYG | 3.5 | 507 | 512 | 521 | 509 | 500 |
| (23) | (27) | (30) | (25) | (23) | ||
| FWG | 2.6 | 491 | 486 | 509 | 480 | 477 |
| (24) | (21) | (27) | (25) | (21) | ||
| YWG | 2.8 | 508 | 510 | 523 | 511 | 496 |
| (32) | (32) | (40) | (26) | (22) | ||
All Kaede-like chromophores in basic DMF and DMSO demonstrated a well-detectable red fluorescence with a quantum yield well above that of HBMPI, HBMPDI [27], and GFP and asFP595 chromophores [22,28] (Table 2 and Fig. 4). The highest fluorescence quantum yield (about 0.017) was measured for Trp-derived chromophores WYG and YWG (for comparison, we measured quantum yield of GFP chromophore in basic DMF to be 0.00005). Probably, bulky Trp side chain prevents to some extent fast isomerization of the chromophore molecules and thus decreases non-radiative decay of the excited state.
Figure 4.
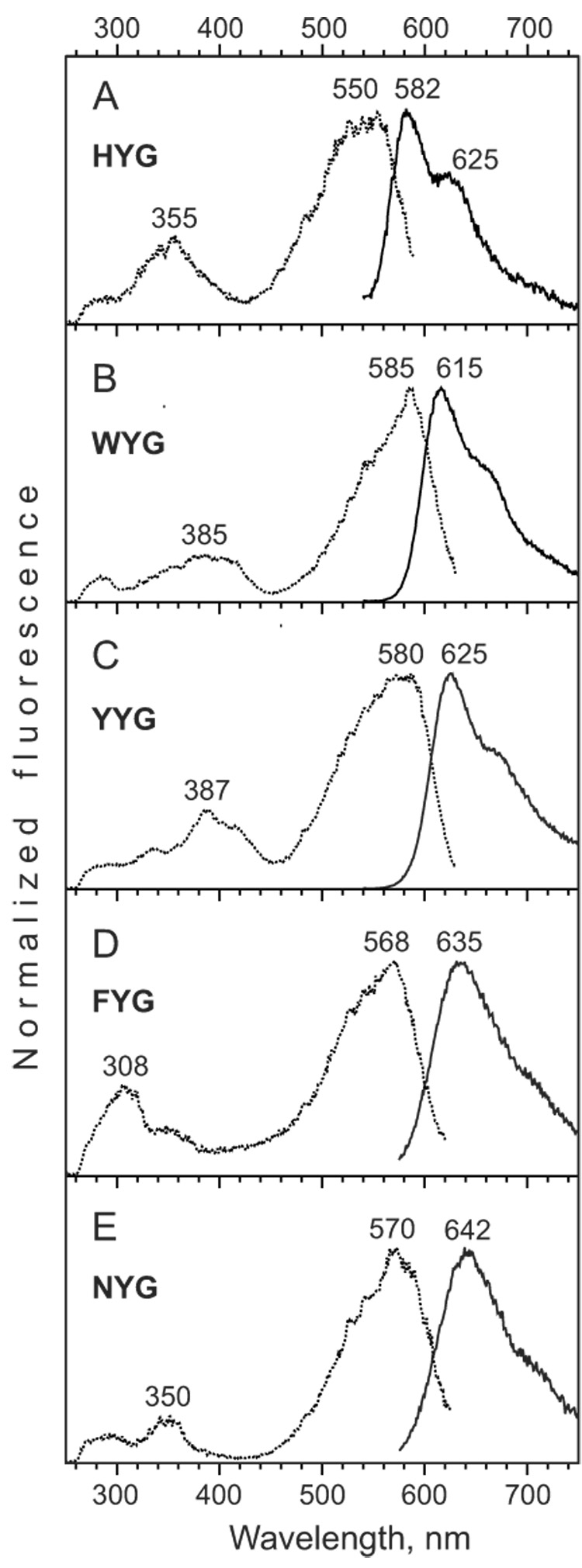
Fluorescence spectra for Kaede-like chromophores. (A–E) Excitation (dotted lines) and emission (solid lines) spectra for HYG, WYG, YYG, FYG, and NYG solutions in basic DMF.
Emission spectra of HYG chromophore in DMF and DMSO had maxima at 582 and 598 nm, respectively. Thus, DMF appears to be close to the chromophore environment within Kaede protein, which has emission maximum at 582 nm [10]. Earlier, the same similarity between spectra of synthetic chromophore in basic DMF and native protein was found for asFP595 [28]. In contrast to other Kaede-like chromophores as well as GFP and asFP595 chromophores, HYG emission spectrum contained a shoulder at 620–640 nm which is separated from the main peak very clearly (Fig. 4A). This well-defined long wavelength shoulder or even minor peak is characteristic for all Kaede-like proteins [10,14,17,33]. Thus, His residue appears to determine this unusual shape of the emission curve.
All “non-natural” Kaede-like chromophores demonstrated considerably red-shifted emission spectra compared to HYG chromophore (Table 2 and Fig. 4). Surprisingly, the most red-shifted emission was detected for NYG chromophore. In basic DMF NYG possessed excitation and emission maxima at 570 and 642 nm, respectively, demonstrating a large Stokes shift (Fig. 4E). In case of successful substitution H65N in a Kaede-like protein such mutant could represent a new group of green-to-far-red photoconvertible fluorescent proteins.
The panel of Kaede-like chromophores allows for direct comparison of some substituents (Table 2 and Table 3). “Symmetrical” compounds WYG and YWG indeed possess almost identical absorption maxima in all solvents in positively charged and neutral states and in almost all solvents in negatively charged states. Only basic DMF and DMSO provide red-shifted absorption for WYG. As expected, p-hydroxyphenyl group gives red-shifted absorption in comparison with phenyl group (compare YYG and FYG). FWG and FYG chromophores demonstrate that neutral indole ensures stronger absorption red shift compared to neutral p-hydroxyphenyl; however, in the negatively charged states their spectral behavior is very similar.
Fluorescent proteins are chemically and optically complex molecules. The availability of model compounds has in the past proven useful for advancing our understanding of their behavior [22–24,34,35]. In particular, the chromophores described here might provide a useful reference for prediction of the optical properties of GFP-related chromophores and proteins using quantum chemical approaches.
To the best of our knowledge, the present study describes deprotonated indole-containing GFP-like chromophore for the first time. We measured pKa 11.6 for deprotonation of the indole group in WYG that is much lower than of indole itself (pKa 17 in water). We found that model compound corresponding to chromophore within CFP (GFP-Y66W, structure 1b, Scheme 2) also shows about 80-nm absorption red shift in a strong base (not shown). Apparently, this state corresponds to deprotonated indole moiety. Potentially, introducing Arg and Lys residues in the vicinity of chromophore-forming Trp66 into ECFP (or similar mutants) could result in novel (and potentially useful) fluorescent protein variant where Trp-containing chromophore exists in anionic state.
One of the most interesting results of the present work is that non-aromatic asparagines moiety provides a strong red shift in NYG chromophore. In case of Asn65, Kaede-like chromophore includes additional C=O group in the system of conjugated double bonds. It was shown earlier that C=O group indeed ensures a very strong (about 100 nm) red shift for asFP595-like chromophore compared to GFP chromophore [28]. Considering also a small size of Asn side chain, this residue appeared to be a promising substituent, which can potentially diversify posttranslational chemistry in GFP-like proteins. It is tempting to insert Asn not only at position 65 in a Kaede-like protein, but also at position 66 (instead of chromophore-forming Tyr) in a green FP. In this case one would expect blue fluorescent mutant with a different (compared to all other blue fluorescent variants) chromophore structure and photophysical behavior. These mutagenesis projects are currently underway in our laboratory.
Acknowledgements
We thank Dr. Andrey Formanovsky, Igor Prokhorenko and Vadim Kublitsky for their help. This work was supported by grants from Molecular and Cell Biology Program RAS, Howard Hughes Medical Institute grant HHMI 55005618, National Institutes of Health USA (GM070358), Russian Foundation for Basic Research grants 05-04-49316 and 08-04-01691. K.A.L is supported by Russian Science Support Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Remington SJ. Curr. Op. Struct. Biol. 2006;16:1–8. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Chudakov DM, Lukyanov S, Lukyanov KA. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. Proc. Natl. Acad. Sci. USA. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Proc. Natl. Acad. Sci. USA. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verkhusha VV, Chudakov DM, Gurskaya NG, Lukyanov S, Lukyanov KA. Chem. Biol. 2004;11:845–854. doi: 10.1016/j.chembiol.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Quillin ML, Anstrom DM, Shu X, O'Leary S, Kallio K, Chudakov DM, Remington SJ. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 7.Remington SJ, Wachter RM, Yarbrough DK, Branchaud B, Anderson DC, Kallio K, Lukyanov KA. Biochemistry. 2005;44:202–212. doi: 10.1021/bi048383r. [DOI] [PubMed] [Google Scholar]
- 8.Pletneva N, Pletnev S, Tikhonova T, Popov V, Martynov V, Pletnev V. Acta Crystallogr. D Biol. Crystallogr. 2006;62:527–532. doi: 10.1107/S0907444906007852. [DOI] [PubMed] [Google Scholar]
- 9.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 10.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. Proc. Natl. Acad. Sci. USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno H, Mal TK, Tong KI, Ando R, Furuta T, Ikura M, Miyawaki A. Mol. Cell. 2003;12:1051–1058. doi: 10.1016/s1097-2765(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 12.Nienhaus K, Nienhaus GU, Wiedenmann J, Nar H. Proc. Natl. Acad. Sci. USA. 2005;99:9156–9159. doi: 10.1073/pnas.0501874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi I, Mizuno H, Tong KI, Furuta T, Tanaka F, Yoshimura M, Miyawaki A, Ikura M. J. Mol. Biol. 2007;372:918–926. doi: 10.1016/j.jmb.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 14.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Röcker C, Salih A, Spindler K-D, Nienhaus GU. Proc. Natl. Acad. Sci. USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wacker SA, Oswald F, Wiedenmann J, Knochel WA. Dev. Dyn. 2007;236:473–480. doi: 10.1002/dvdy.20955. [DOI] [PubMed] [Google Scholar]
- 16.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. EMBO Rep. 2005;6:233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Nat. Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 18.Mutoh T, Miyata T, Kashiwagi S, Miyawaki A, Ogawa M. Exp. Neurol. 2006;200:430–437. doi: 10.1016/j.expneurol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 19.Hatta K, Tsujii H, Omura T. Nat. Protoc. 2006;1:960–967. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- 20.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Gurskaya NG, Merzlyak EM, Staroverov DB, Mudrik NN, Samarkina ON, Vinokurov LM, Lukyanov S, Lukyanov KA. Method for real-time monitoring of protein degradation at the single cell level. Biotechniques. 2007;42:446, 448, 450. doi: 10.2144/000112453. [DOI] [PubMed] [Google Scholar]
- 22.Niwa H, Inouye S, Hirano T, Matsuno T, Kojima S, Kubota M, Ohashi M, Tsuji FI. Proc. Natl. Acad. Sci. USA. 1996;93:13617–13622. doi: 10.1073/pnas.93.24.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He X, Bell AF, Tonge PJ. J. Phys. Chem. B. 2002;106:6056–6066. [Google Scholar]
- 24.Follenius-Wund A, Bourotte M, Schmitt M, Iyice F, Lami H, Bourguignon JJ, Haiech J, Pigault C. Biophys. J. 2003;85:1839–1850. doi: 10.1016/S0006-3495(03)74612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hager B, Schwarzinger B, Falk H. Monatsh. Chem. 2006;137:163–168. [Google Scholar]
- 26.Prüger B, Bach T. Synthesis. 2007;2007:1103–1106. [Google Scholar]
- 27.He X, Bell AF, Tonge PJ. Org. Lett. 2002;4:1523–1526. doi: 10.1021/ol0200403. [DOI] [PubMed] [Google Scholar]
- 28.Yampolsky IV, Remington SJ, Martynov VI, Potapov VK, Lukyanov S, Lukyanov KA. Biochemistry. 2005;44:5788–5793. doi: 10.1021/bi0476432. [DOI] [PubMed] [Google Scholar]
- 29.Harusawa S, Kawamura M, Koyabu S, Hosokawa T, Araki L, Sakamoto Y, Hashimoto T, Yamamoto Y, Yamatodani A, Kurihara T. Synthesis. 2003;2003:2844–2850. [Google Scholar]
- 30.Zhang P, Liu R, Cook JM. Tetrahedron Lett. 1995;36:3103–3106. [Google Scholar]
- 31.Kojima S, Ohkawa H, Hirano T, Maki S, Niwa H, Ohashi M, Inouye S, Tsuji FI. Tetrahedron Lett. 1998;39:5239–5242. [Google Scholar]
- 32.Bevis BJ, Glick BS. Nat. Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 33.Labas YA, Gurskaya NG, Yanushevich YG, Fradkov AF, Lukyanov KA, Lukyanov SA, Matz MV. Proc. Natl. Acad. Sci. USA. 2002;99:4256–4261. doi: 10.1073/pnas.062552299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helms V. Curr. Opin. Struct. Biol. 2002;12:169–175. doi: 10.1016/s0959-440x(02)00305-6. [DOI] [PubMed] [Google Scholar]
- 35.Nemukhin AV, Topol IA, Burt Stanley K. J. Chem. Theory Comput. 2006;2:292–299. doi: 10.1021/ct050243n. [DOI] [PubMed] [Google Scholar]


