PARAMUTATION is the fascinating ability of specific DNA sequences to communicate in trans to establish meiotically heritable expression states. Intriguingly, newly silenced sequences continue to issue instructions to naïve alleles in subsequent generations. The term “paramutation” was first coined in the 1950s by Alexander Brink to describe this puzzling phenomenon at the r1 locus in maize (Brink 1956); an interaction between specific alleles in heterozygotes led to heritable decreases in gene expression of one allele. Not only was the reduced expression state stable through meiosis, but also the low-expressing allele could induce silencing of another high-expressing allele in subsequent generations. The frequency of the change was 100% and the stability of the change was lower than typical mutations; hence the term “paramutation.” A few years later, Ed Coe, Jr., described another maize example in which interaction between alleles at the b1 locus also led to heritable silencing (Coe 1959) and Rudolf Hagemann described interactions at the sulfurea locus in tomato (Hagemann 1969). Since that time other examples of paramutation have been identified in maize and in other species (reviewed in Chandler and Stam 2004; Stam and Mittelsten Scheid 2005; Chandler 2007), yet the two maize loci where paramutation was initially described, r1 and b1, remain the most extensively characterized and best understood. The r1 and b1 loci encode closely related, functionally equivalent transcription factors that activate the anthocyanin pigment biosynthetic pathway (Goff et al. 1990; Ludwig et al. 1990). They are likely related to each other through a duplication resulting from an ancient allotetraploidization event during maize evolution (Gaut and Doebley 1997). The two loci have multiple alleles with distinct expression patterns, which regulate the distribution of anthocyanin pigments during development (Styles et al. 1973; Coe 1979). Recent work demonstrates a key role for RNA in mediating both r1 and b1 paramutation, as the mop1 gene that encodes an RNA-dependent RNA polymerase (RDR; Alleman et al. 2006) is absolutely required for paramutation at both loci (Dorweiler et al. 2000). Yet, there are striking differences in the properties of r1 and b1 paramutation, which hint at distinct mechanisms. In this article, the most striking differences between r1 and b1 paramutation are described and potential mechanisms are discussed relative to our current understanding of the role of RNA interference (RNAi) in mediating transcriptional silencing.
OVERVIEW OF R1 AND B1 PARAMUTATION
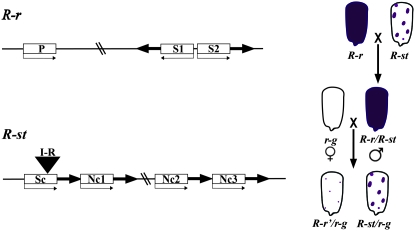
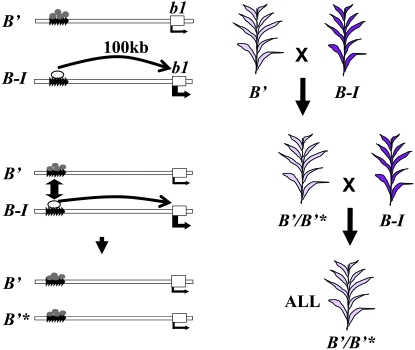
In spite of the close relationship, r1 and b1 have quite distinct gene organizations and there is no evidence of paramutant interactions between the loci (Brink et al. 1960). Specific alleles at each locus participate in paramutation. At r1, all known alleles induce paramutation, respond to paramutation, or cause reversal of paramutation. The alleles that cause paramutation are generally structurally distinct from the alleles that become silenced (reviewed in Chandler et al. 2000; diagrammed in Figure 1). The r1 haplotypes that participate in paramutation are expressed in the aleurone layer of the seed and the paramutant phenotype is strongest in this tissue (reviewed in Brink et al. 1968; Brink 1973). All r1 “alleles” that undergo paramutation contain multiple copies of the coding region (Figure 1) and as such are referred to as haplotypes. In contrast, the b1 alleles that participate in paramutation have a single coding region and identical DNA sequences and are thus epialleles (Stam et al. 2002a,b). These b1 epialleles are expressed in the epidermal layer of the plant body, but not in the seed (Figure 2).
Figure 1.—
Paramutation at r1. The structures of the paramutable (R-r) and paramutagenic (R-st) alleles are diagrammed as well as the crosses and phenotypes used to monitor paramutation. The open boxes represent the r1 genes with the small arrows indicating the direction of transcription: P stands for plant and is a gene expressed in vegetative plant tissues; S1 and S2 are two genes expressed in the seed that are organized in an inverted repeat; Sc stands for self-color and is the most highly expressed gene in the R-st complex; Nc1–Nc3 stand for near colorless, three r1 genes that are expressed at low levels in R-st. I-R indicates the transposable element inserted into Sc, which is responsible for the spotted phenotype. The thick arrows in R-st represent the large repeats spanning the r1 genes. Only the repeats associated with the coding regions are shown. There are additional small, related sequences related to transposable elements that are not diagrammed, but are described in detail with primary references cited in Chandler et al. (2000). A comprehensive list of r1 haplotypes with their tissue-specific expression patterns and paramutation properties can be found in Neuffer et al. (1997).
Figure 2.—
Paramutation at b1. The paramutable (B-I) and paramutagenic (B′) epialleles have identical DNA sequences, but distinct chromatin structures represented by different proteins (circles) associated with the seven tandem repeats (arrowheads) that mediate paramutation and are located 100 kbp upstream of the coding region (open box). The arrows below the gene boxes indicate the direction of transcription and the thickness of the arrows indicate the relative transcription levels observed with the two epialleles. The crosses and phenotypes used to monitor paramutation are diagrammed with B′*, indicating a B′ allele that was B-I in the previous generation. B′ and B′* are indistinguishable in their ability to paramutate B-I.
When the R-r allele, which confers dark purple seeds, is crossed to R-stippled (R-st), which confers purple spotted seeds (because of a transposon insertion that is not involved in paramutation), R-r is heritably changed such that it confers lightly pigmented seeds upon subsequent outcrosses (designated R-r′), while the R-stippled allele segregates unchanged (Figure 1). Intriguingly, when R-r′ is crossed to R-r, R-r can be changed to R-r′; this is referred to as secondary paramutation to distinguish it from the primary paramutational interaction between R-st and R-r. At the b1 locus, the low-expressing B′ allele (which can also derive spontaneously from the high-expressing B-I allele) changes B-I into B′ (new B′ alleles are designated B′*) with the silencing observed in the F1 (Figure 2). The newly altered B′* allele is indistinguishable from B′ in its ability to paramutate naïve B-I alleles in subsequent generations. Thus, at b1, secondary paramutation is indistinguishable from primary paramutation. This contrasts with r1 paramutation; the strength of R-r′ secondary paramutation (the ability of R-r′ to paramutate a naïve R-r allele) is fully penetrant only when R-r′ has been heterozygous with R-st for multiple generations (Brown and Brink 1960).
Two assays are routinely used to monitor paramutation: (1) the ability of a paramutagenic allele to cause a heritable change in the expression of a paramutable allele (as measured at r1 and b1 by a reduction in pigment); and (2) the heritable alteration of the paramutable allele into a paramutagenic allele. At b1, these two phenotypes always occur simultaneously and completely. In contrast, at r1, the extent of paramutagenicity measured as the level of R-r pigmentation depends on the haplotype and the circumstances of the crosses (discussed in the next section and reviewed in Chandler et al. 2000). Specific nomenclature is used to describe the various steps in the paramutation process. Establishment describes the trans-interactions between alleles that produce the distinct expression states. In the case of both r1 and b1, the new states show reduced expression (designated R-r′ and B′), and the ability to maintain that silencing phenotype is referred to as “maintenance of silencing.” The extent to which the silencing and ability to cause paramutation is maintained in subsequent generations is referred to as heritability. The reason for distinguishing among establishment, maintenance, and heritability is that certain situations or genetic backgrounds can influence these processes differentially. The frequent association between paramutation and genes involved in pigmentation is likely to reflect ascertainment bias, because of the ease of scoring visible pigment phenotypes and the dispensable nature of anthocyanin for plant development.
STABILITY DIFFERENCES BETWEEN R1 AND B1 PARAMUTATION
The r1 and b1 systems differ in several properties, which hint at basic mechanistic differences. One dramatic example is the extreme stability of B′ as compared with the instability of the R-r′ silenced state. B-I is truly converted to a B′ epiallele whereas R-r′ states exist as a continuum, depending on the counter-allele and the genetic history. In wild-type genetic backgrounds, B-I is always changed to B′* (Coe 1966). B′ and B′* are indistinguishable, and both are extremely stable; they have never been observed to change back to B-I in wild-type genetic backgrounds and this is true whether they are maintained as homozygous or heterozygous with an allele that does not participate in paramutation (Coe 1966). Spontaneous changes of B-I to B′ occur at high frequencies, often 1–10% (Coe 1966), and spontaneous B′ alleles are as fully paramutagenic as B′* alleles segregating from B′/B-I plants (Figure 2). Thus, the low-expression state associated with B′ is invariably associated with strong paramutagenicity (Coe 1966).
In contrast, R-r′ is unstable with its pigment level and reversion frequency dependent on the strength of the R-st derivative to which it was crossed, the number of generations of heterozygosity with R-st, or when R-r′ is homozygous. When R-r′ is exposed to repeated generations of crosses with paramutagenic haplotypes, its seed pigmentation level continues to decrease to almost colorless and the haplotype becomes more paramutagenic (Mikula 1961; McWhirter and Brink 1962). Furthermore, expression of R-r is more reduced following passage through trisomics containing two doses of the paramutagenic allele relative to passage through disomic heterozygotes with one dose of the paramutagenic allele (Kermicle et al. 1995). Paramutation can also be reversed gradually if R-r′ is maintained as hemizygous or heterozygous with alleles that contain a single copy of the r1 locus, such that seed color becomes even darker than nonparamutant R-r (Styles and Brink 1966, 1969). This set of experiments, together, suggests that there is communication between the various r1 haplotypes and that this communication influences expression levels.
SEQUENCES REQUIRED FOR PARAMUTATION
One common feature of r1 and b1 paramutation is that tandem repeats are involved in paramutation at each locus, yet the actual sequences and properties of these repeats are quite distinct between the two loci. Within the r1 paramutagenic haplotypes, the repeats are of unknown length but each is likely quite large, spanning the r1-coding regions (Figure 1; Eggleston et al. 1995). The regions associated with paramutagenicity have been mapped for two r1 haplotypes using unequal crossing over between the multiple r1 genes in paramutagenic and nonparamutagenic haplotypes to generate a series of alleles with differing numbers of tandem repeats (Kermicle et al. 1995; Kermicle 1996; Panavas et al. 1999). Intriguingly, these results suggest that no specific region is required for r1 paramutation, but that only repeat number matters: paramutagenicity is reduced as repeat number is reduced and paramutagenicity is increased as repeat number is increased. The region within R-r required to respond to paramutation has also been mapped and lies within the inverted repeat structure of the S component (Brown and Brink 1960; Robbins et al. 1991; Kermicle 1996). Again, the r1 genes themselves lie within the sequences associated with paramutation. The results are quite different at b1 (Figure 2), in which specific sequences, an array of seven tandem repeats of 853 bp each, are required for paramutation; these are located >100 kb upstream of the single b1-coding region and share no sequence identity with the coding region (Stam et al. 2002a). Analyses of recombinant alleles differing only in the number of the upstream b1 tandem repeats demonstrated that tandem repeats are required for paramutation and the high expression of B-I (Stam et al. 2002a). Alleles that do not participate in paramutation have a single copy of this sequence, which is unique in the maize genome, and thus is not shared with other genes that also undergo paramutation, such as r1. The observation that tandem repeats are associated with paramutation might suggest a mechanism of repeat expansion and contraction. However, as discussed below, the numbers of repeats do not change with paramutation.
POTENTIAL COMMUNICATION MECHANISMS
What might be the basis for the allele and haplotype communication? Two possibilities are the RNA-based model or the DNA pairing model, but these are not mutually exclusive (reviewed in Chandler and Stam 2004). Both Brink and Coe explored various mechanisms for the cause of r1 and b1 paramutation using the genetic approaches available to them at the time. Both were on the right track in understanding that paramutation must involve direct or indirect contact between paramutagenic and paramutable alleles. Brink understood that during paramutation R-st and R-r must communicate early in development in new heterozygotes. By attempting to block paramutation through the use of translocated chromosomes that would disrupt pairing, he tested whether chromosome pairing might mediate the communication. The experiment did not lead to conclusive results because the prior genetic history of the alleles used confounded the interpretation (J. Kermicle, personal communication). Brink also tested for the existence of a “cytoplasmic particle,” loosely interpreted as a “trans-acting” or “communication molecule” that would be produced by R-st and transmit paramutation to R-r, but found no evidence for such (Brink et al. 1964). On the basis of studies at the b1 locus, Coe favored a model in which the paramutagenic allelic transferred a physical entity, which he hypothesized could be DNA or RNA (Coe 1968). We now know that there are no genetic changes among b1 alleles, but there is good evidence that RNA signals are involved in allele interactions (see below).
REPEAT COUNTING MECHANISMS
At the b1 locus, the full penetrance and stability of paramutation is conferred by the seven tandem repeats, as paramutation with b1 alleles with three tandem repeats was less penetrant and less stable (Stam et al. 2002a). This result suggests that there is a mechanism for counting the numbers of repeats. Potential counting mechanisms could involve chromatin marks such as DNA and histone modifications, number of specific proteins bound, or RNA signals, whose level depends on the number of repeats.
Brink et al. (1968) suggested that the r1 haplotypes exist in a wide continuum of states and that the ability to move along the continuum is an inherent property of the haplotype itself with the extent and direction of movement along the continuum influenced by the nature of the other allele present. Brink proposed that the r1 locus had two components: (1) the gene complex encoding the protein involved in anthocyanin synthesis and (2) a heterochromatic segment assumed to consist of varying numbers of a repeating unit called a metamere, which functioned to repress r1. He further proposed that the degree of repression was proportional to the number of metameres, which could change through misreplication during somatic mitosis (Brink et al. 1968). This hypothesis sprang from Brink's being influenced by the description of position-effect variegation (PEV) in Drosophila (reviewed in Lewis 1950). The influence likely resulted from the mosaic pattern of PEV in Drosophila eyes being strikingly similar to the mottled phenotype of paramutant R-r′ kernels and the idea that both PEV and paramutation were caused by aberrations in gene expression systems.
Brink clearly sought a hypothesis that incorporated the essence of heterochromatin and repeated sequences without the requirement for large centric blocks that were known to be part of the PEV phenomenon. He attempted to avoid these inconsistencies with an alteration of terminology, for example, “ortho” and “parachromatin,” in which parachromatin is the part of the genome responsible for chromatin condensation and gene silencing (Brink et al. 1960). He called parachromatin the “nexus between chromosomal heredity and development” and provided that it could be “altered in accordance with the cellular substrate and could react to extra-chromosomal stimuli that define developmental progression.” This description sounds provocatively similar to RISC-complex-mediated gene silencing (Zaratiegui et al. 2007).
The large tandem repeats associated with paramutagenic alleles are consistent with the metamere hypothesis, but molecular studies have demonstrated that the number of repeats does not change during plant development (M. Alleman, unpublished data); thus differences in r1 expression are likely to be mediated by epigenetic mechanisms (such as changes in DNA methylation and histone modifications). Potentially, the larger number of tandem repeats in paramutagenic alleles facilitates the communication with the inverted repeat at the sensitive locus—either directly through pairing or through some type of RNA-signaling molecule. At r1, a larger number of repeats could result in an increase in the amount of communication signal sent by paramutagenic haplotypes or an increased frequency of direct pairing. The inverted repeat nature of the paramutable haplotypes may make them particularly receptive to this signal.
TRANS-ACTING FACTORS REQUIRED FOR PARAMUTATION
Characterization of trans-acting factors required for paramutation has begun to provide mechanistic clues. Recent results strongly suggest that the communication signal is likely to be RNA molecules produced and interpreted by genes involved in the RNAi transcriptional silencing pathway. Several trans-acting factors that affect paramutation have been identified (Dorweiler et al. 2000; Hollick and Chandler 2001; Hollick et al. 2005). Two of these genes have been cloned: mop1, which encodes an RDR (Alleman et al. 2006), and rmr1, which encodes a putative chromatin-remodeling protein (Hale et al. 2007). Both of these proteins are closely related to proteins involved in RNAi-mediated transcriptional gene silencing in Arabidopsis thaliana, suggesting that the RNAi pathway mediating heritable chromatin structures is required for paramutation. The tandem repeats mediating b1 paramutation are transcribed on both strands (Alleman et al. 2006) and MOP1 is required for an increased quantity of 24-nucleotide silent RNAs (siRNAs) coming from these repeats (M. Arteaga-Vazquez and V. Chandler, unpublished data). In addition to affecting paramutation at multiple loci (Dorweiler et al. 2000), the MOP1 protein also influences the epigenetic regulation of Mutator transposons (Lisch et al. 2002; Slotkin et al. 2005; Woodhouse et al. 2006a,b) and certain transcriptionally silenced transgenes (McGinnis et al. 2006), processes associated with RNAi mechanisms in multiple species.
Both establishment of paramutation and maintenance of the transcriptional silencing at B′ absolutely depend on mop1 (Dorweiler et al. 2000), suggesting a role for RNA in both processes. However, mop1 has a more subtle role in the heritability of the reduced expression state. When mop1 mutations are outcrossed to wild type to reintroduce the wild-type MOP1 protein, most of the progeny have B′ expression levels (Dorweiler et al. 2000). This result suggests that the MOP1 protein can efficiently resilence the “active” epiallele transmitted from the mop1 mutant plant. Another possibility is that the “active” allele remembers that it was B′ in the previous generation in spite of its high expression promoted by the absence of MOP1 and is therefore expressed at a low level in the progeny. The latter hypothesis would require that some type of heritable mark remain at the B′ allele even when it is expressing at a B-I level and would require that mark to be efficiently transmitted through meiosis. Recently we noted, when growing thousands of progeny in the absence of the MOP1 protein, that at a low frequency the B′ epiallele is changed to a B-I state that is heritable and immune to resilencing in the presence of the MOP1 protein. This contrasts with wild-type backgrounds in which B′ has never been observed to change to B-I. Similarly, in the absence of the MOP1 protein, a transcriptionally silent transgene can be reactivated such that it stays active for multiple generations even in the presence of the MOP1 protein introduced by outcrosses (McGinnis et al. 2006). This result is consistent with the establishment of chromatin states that are relatively immune to silencing.
MOP1 is also absolutely required to establish r1 paramutation (Dorweiler et al. 2000) as R-r is not changed to R-r′ by R-st in homozygous mop1 mutant plants. Intriguingly, the MOP1 protein is not required to maintain the reduced expression associated with R-r′ (J. Kermicle, personal communication). There are several potential explanations. First, an RNA mechanism may not be involved in maintaining the silencing of R-r. Second, at r1, the silencing might be stabilized through the more extensive DNA methylation that occurs. A third possibility could be that the R-r′ allele does not require the RDR to maintain an RNA signal; potentially, the inverted repeat structure seen at R-r (Figure 1) could generate hairpin RNA in sufficient quantities. This latter hypothesis is similar to what is seen with the MuK locus of maize, which generates a double-stranded RNA hairpin homologous to the fully functional autonomous Mutator element, MuDR, silencing it (Slotkin et al. 2005); this silencing does not require MOP1 (Woodhouse et al. 2006a,b). Further experiments to test the role of other RNAi pathway mutants should reveal whether an RNAi pathway is involved in maintaining R-r′ silencing. Another complication in interpreting the r1 result is that the phenotype is observed only in paternally transmitted alleles, which are subject to genomic imprinting (Alleman and Doctor 2000). DNA methylation within the inverted repeats in R-r does correlate with its silencing (Walker 1998) and this methylation may contribute to the maintenance of silencing in the absence of the MOP1 pathway. Another possibility is that the silencing mediated by imprinting is epistatic to the silencing mediated by MOP1 and thus MOP1's absence is inconsequential.
OPEN QUESTIONS
Given the dramatic differences between r1 and b1 paramutation, it is striking that both involve repeated sequences of some type and that an RNA trans-effect links the two processes. Transgene silencing and heterochromatin silencing in multiple species also involve repeated sequences, and RNA-directed chromatin changes are mediated by components of the RNAi pathway (Henderson and Jacobsen 2007; Zaratiegui et al. 2007). While similar proteins are involved in maize paramutation and RNA-directed DNA methylation in A. thaliana, it is interesting that paramutation has yet to be described in A. thaliana. The FWA locus in A. thaliana is the system most similar to b1 paramutation in that methylated tandem repeats producing siRNAs from a silenced endogenous locus can communicate with an incoming transgene to silence it (Henderson and Jacobsen 2007). Yet a silenced endogenous FWA locus does not silence an active endogenous allele. Why is the active FWA locus immune to silencing while the FWA transgene is silenced?
Many additional intriguing questions remain. What process enables the extremely efficient and highly heritable trans-communication associated with paramutation in maize? What is the nature of the RNA that triggers paramutation? Why are tandem repeats required? What are the heritable molecules or marks? Why does paramutation exist and is it really rare? Thoughts on some of these questions are discussed in Chandler (2007). Current approaches directed toward identifying additional key genes required for paramutation and understanding the relationships between tandem repeat RNA and chromatin structure offer hope for eventually understanding this intriguing process.
Acknowledgments
Paramutation studies in the Chandler laboratory were supported by grants to V.C. from the National Science Foundation (NSF) and the National Institutes of Health. M.A. received a sabbatical supplement from the NSF. We thank Jerry Kermicle for helpful discussions.
References
- Alleman, M., and J. Doctor, 2000. Genomic imprinting in plants: observations and evolutionary implications. Plant Mol. Biol. 43 147–161. [DOI] [PubMed] [Google Scholar]
- Alleman, M., L. Sidorenko, K. McGinnis, V. Seshadri, J. E. Dorweiler et al., 2006. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442 295–298. [DOI] [PubMed] [Google Scholar]
- Brink, R. A., 1956. A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41 872–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R. A., 1973. Paramutation. Annu. Rev. Genet. 7 129–152. [DOI] [PubMed] [Google Scholar]
- Brink, R. A., D. F. Brown, J. Kermicle and W. H. Weyers, 1960. Locus dependence of the paramutant R-phenotype in maize. Genetics 45 1297–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R. A., J. L. Kermicle and D. F. Brown, 1964. Tests for a gene-dependent cytoplasmic particle associated with R paramutation in maize. Proc. Natl. Acad. Sci. USA 51 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R. A., E. D. Styles and J. D. Axtell, 1968. Paramutation: directed genetic change. Paramutation occurs in somatic cells and heritably alters the functional state of a locus. Science 159 161–170. [DOI] [PubMed] [Google Scholar]
- Brown, D., and R. Brink, 1960. Paramutagenic action of paramutant R-r and R-g alleles in maize. Genetics 45 1313–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V. L., 2007. Paramutation: from maize to mice. Cell 128 641–645. [DOI] [PubMed] [Google Scholar]
- Chandler, V. L., and M. Stam, 2004. Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. Genet. 5 532–544. [DOI] [PubMed] [Google Scholar]
- Chandler, V. L., W. B. Eggleston and J. E. Dorweiler, 2000. Paramutation in maize. Plant Mol. Biol. 43 121–145. [DOI] [PubMed] [Google Scholar]
- Coe, E. H., Jr., 1959. A regular and continuing conversion-type phenomenon at the B locus in maize. Proc. Natl. Acad. Sci. USA 45 828–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E. H., Jr., 1966. The properties, origin and mechanism of conversion-type inheritance at the B locus in maize. Genetics 53 1035–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E. H., Jr., 1968. Heritable repression due to paramutation in maize. Science 162 925. [DOI] [PubMed] [Google Scholar]
- Coe, E. H., Jr., 1979. Specification of the anthocyanin biosynthetic function by b and r in maize. Maydica 24 49–58. [Google Scholar]
- Dorweiler, J. E., C. C. Carey, K. M. Kubo, J. B. Hollick, J. L. Kermicle et al., 2000. Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12 2101–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston, W. B., M. Alleman and J. L. Kermicle, 1995. Molecular organization and germinal instability of R-stippled maize. Genetics 141 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and J. F. Doebley, 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S. A., T. M. Klein, B. A. Roth, M. E. Fromm, K. C. Cone et al., 1990. Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 9 2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann, R., 1969. Somatic conversion (paramutation) at the sulfurea locus of lycopersicon esculentum Mill. III. Studies with trisomics. Can. J. Genet. Cytol. 11 346–358. [Google Scholar]
- Hale, C. J., J. L. Stonaker, S. M. Gross and J. B. Hollick, 2007. A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 5 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I. R., and S. E. Jacobsen, 2007. Epigenetic inheritance in plants. Nature 447 418–424. [DOI] [PubMed] [Google Scholar]
- Hollick, J. B., and V. L. Chandler, 2001. Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics 157 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick, J. B., J. L. Kermicle and S. E. Parkinson, 2005. Rmr6 maintains meiotic inheritance of paramutant states in Zea mays. Genetics 171 725–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J. L., 1996. Epigenetic silencing and activation of a maize r gene, pp. 267–287 in Epigenetic Mechanisms of Gene Expression, edited by V. E. A. Russo, R. A. Martienssen and A. D. Riggs. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kermicle, J. L., W. B. Eggleston and M. Alleman, 1995. Organization of paramutagenicity in R-stippled maize. Genetics 141 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, E. B., 1950. The phenomenon of position effect. Adv. Genet. 3 73–115. [DOI] [PubMed] [Google Scholar]
- Lisch, D., C. C. Carey, J. E. Dorweiler and V. L. Chandler, 2002. A mutation that prevents paramutation in maize also reverses Mutator transposon methylation and silencing. Proc. Natl. Acad. Sci. USA 99 6130–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, S. R., B. Bowen, L. Beach and S. R. Wessler, 1990. A regulatory gene as a novel visible marker for maize transformation. Science 247 449–450. [DOI] [PubMed] [Google Scholar]
- McGinnis, K. M., C. Springer, Y. Lin, C. C. Carey and V. Chandler, 2006. Transcriptionally silenced transgenes in maize are activated by three mutations defective in paramutation. Genetics 173 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter, K., and R. Brink, 1962. Continuous variation in level of paramutation at the R locus in maize. Genetics 47 1053–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikula, B. C., 1961. Progressive conversion of R-locus expression in maize. Proc. Natl. Acad. Sci. USA 47 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer, M. G., E. H. Coe and S. R. Wessler, 1997. Mutants of Maize. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Panavas, T., J. Weir and E. L. Walker, 1999. The structure and paramutagenicity of the R-marbled haplotype of Zea mays. Genetics 153 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, T. P., E. L. Walker, J. L. Kermicle, M. Alleman and S. L. Dellaporta, 1991. Meiotic instability of the R-r complex arising from displaced intragenic exchange and intrachromosomal rearrangement. Genetics 129 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin, R. K., M. Freeling and D. Lisch, 2005. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat. Genet. 37 641–644. [DOI] [PubMed] [Google Scholar]
- Stam, M., and O. Mittelsten Scheid, 2005. Paramutation: an encounter leaving a lasting impression. Trends Plant Sci. 10 283–290. [DOI] [PubMed] [Google Scholar]
- Stam, M., C. Belele, J. E. Dorweiler and V. L. Chandler, 2002. a Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16 1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., C. Belele, W. Ramakrishna, J. E. Dorweiler, J. L. Bennetzen et al., 2002. b The regulatory regions required for B′ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics 162 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles, E. D., and R. A. Brink, 1966. The metastable nature of paramutable R alleles in maize. I. Heritable enhancement in level of standard Rr action. Genetics 54 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles, E. D., and R. A. Brink, 1969. The metastable nature of paramutable r alleles in maize. IV. Parallel enhancement of R action in heterozygotes with r and in hemizygotes. Genetics 61 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles, E. D., O. Ceska and K. T. Seah, 1973. Developmental differences in action of r and b alleles in maize. Can. J. Genet. Cytol. 15 59–72. [Google Scholar]
- Walker, E. L., 1998. Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse, M. R., M. Freeling and D. Lisch, 2006. a Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 4 e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse, M. R., M. Freeling and D. Lisch, 2006. b The mop1 (mediator of paramutation1) mutant progressively reactivates one of the two genes encoded by the MuDR transposon in maize. Genetics 172 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui, M., D. V. Irvine and R. A. Martienssen, 2007. Noncoding RNAs and gene silencing. Cell 128 763–776. [DOI] [PubMed] [Google Scholar]