Abstract
Single-locus sporophytic self-incompatibility inhibits inbreeding in many members of the mustard family (Brassicaceae). To investigate the genetics of self-incompatibility in the wild mustard Leavenworthia alabamica, diallel crosses were conducted between full siblings. Patterns of incompatibility were consistent with the action of single-locus sporophytic self-incompatibility. DNA sequences related to S-locus receptor kinase (SRK), the gene involved in self-pollen recognition in mustards, were cloned and sequenced. A single sequence with high identity to SRK and several other groups of sequences (Lal1, Lal2, Lal3, Lal8, and Lal14) were isolated from L. alabamica. We propose that either Lal2 sequences are divergent alleles of SRK or Lal2 is in tight linkage with SRK because (1) Lal2 alleles cosegregate with S-alleles inferred from dialleles in all 97 cases tested in five families; (2) Lal2 sequences are highly diverse at both synonymous and nonsynonymous sites and exhibit patterns of selective constraint similar to those observed at SRK in Brassica and Arabidopsis; and (3) transcripts of one Lal2 allele were detected in leaves and the styles of open flowers, but were most abundant in the stigmas of maturing buds. We discuss the utility of the S-linked polymorphism at Lal2 for studying the evolutionary forces acting on self-incompatibility in Leavenworthia.
INBREEDING avoidance is thought to have contributed to the evolution and maintenance of genetically controlled self-incompatibility (SI) systems in flowering plants (Richards 1986). In SI systems, plants that share alleles at the self-incompatibility locus (i.e., S-locus) are incapable of producing seed. SI evolved early during the diversification of angiosperms, and it has been proposed that this system may have contributed to the success of flowering plants by contributing to the maintenance of population–genetic variation (Darlington and Mather 1949; Igic and Kohn 2001, 2006). SI systems are diverse in terms of the molecular mechanisms underlying self-recognition, as well as in associated floral traits that promote pollen transfer between compatible mates (Hiscock and Tabah 2003). A feature of most SI systems is the presence of a single linked complex of genes (or more rarely, several unlinked loci) that code for proteins involved in the recognition and rejection of self-pollen. Since individuals that possess rare S-alleles have a strong fertility advantage, negative frequency-dependent selection is expected to maintain a large amount of genetic diversity at S-loci in natural populations. Indeed, these loci are among the most polymorphic in eukaryotic organisms (Wright 1939; Lawrence 2000).
Single-locus sporophytic SI has been studied in a number of genera in the mustard family (Brassicaceae) (Bateman 1954, 1955; Thompson 1957; Lloyd 1967; Sampson 1967; Schierup et al. 2001). Molecular genetic characterization of SI in Brassica and Arabidopsis species has shown that the S-locus consists of a tightly linked cluster of genes encoding proteins that function together as a receptor-ligand system (Schopfer et al. 1999; Takasaki et al. 2000; for a review see Fobis-Loisy et al. 2004). One of these genes, the S-locus receptor kinase (SRK), codes for a membrane-bound protein expressed in the stigma that binds specifically to the S-locus cysteine-rich ligand (SCR/SP11), which is expressed in the tapetum of anthers (Kachroo et al. 2001; Takayama et al. 2001). The successful binding of SCR by SRK initiates a signaling cascade that prevents pollen tubes from penetrating the stigmatic surface, although the roles of other genes in this pathway are still under investigation (Murase et al. 2004; Liu et al. 2007). It is thought that the linked system of co-evolved SRK and SCR genes has been maintained by natural selection for at least 25–40 million years (Uyenoyama 1995). As would be expected in a system in which selection is strongly negatively frequency dependent, nucleotide polymorphism is pronounced among S-alleles in Brassica campestris and B. oleracea (Hinata et al. 1995), Raphanus sativus (Okamoto et al. 2004), Arabidopsis lyrata and A. halleri (Schierup et al. 2001; Castric and Vekemans 2004; Prigoda et al. 2005), and Capsella grandiflora (Paetsch et al. 2006). Many of these investigations have also revealed dominance interactions among S-alleles in the pollen and/or the stigma, a phenomenon consistent with a sporophytic mechanism of SI.
Sequence-level variation among S-alleles sampled within and between species provides insight into the historical evolution of a locus under strong balancing selection (Richman et al. 1996; Igic and Kohn 2001). Since S-alleles are expected to be favored when rare by negative frequency dependence, and thus be maintained in populations for long periods, coalescence times at these loci might often predate speciation events and could provide insight into the long-term effective sizes of species (Richman and Kohn 2000). In fact, alleles of SRK sampled from A. lyrata and C. grandiflora show a pronounced pattern of trans-specific and transgeneric polymorphism and are exceedingly diverse at synonymous and nonsynonymous sites (Schierup et al. 2001; Paetsch et al. 2006). SRK alleles sampled from species of Brassica and Raphanus also show a pattern of transgeneric polymorphism, although the amount of sequence variation found within each species is much more modest (Hinata et al. 1995; Okamoto et al. 2004). A historical bottleneck in the lineage leading to Brassica and Raphanus may have caused the loss of sequence-level variation within species. There is also a great deal of divergence between SRK sequences sampled from species of Arabidopsis and Capsella compared to those sampled from species of Brassica and Raphanus (Fobis-Loisy et al. 2004). This marked divergence between lineages, coupled with the fact that sequences flanking the S-locus in Arabidopsis and Brassica are derived from alternative chromosomal regions, suggests that a translocation of the entire S-locus region may have occurred (Kusaba et al. 2001).
This article describes sequence-level variation in SRK-like genes in a species of Leavenworthia, a small genus from a portion of the Brassicaceae that has served as a model for evolutionary studies on the loss of SI (Rollins 1963; Lloyd 1965). In terms of molecular characterization of SRK and SRK-like genes, the genus Leavenworthia is unstudied. We expect that a locus orthologous to SRK in L. alabamica should satisfy the following criteria on the basis of patterns observed in both Arabidopsis and Brassica (Charlesworth et al. 2000): (1) sequences of putative S-alleles should exhibit high sequence identity to known alleles of SRK; (2) sequences should cosegregate with S-alleles as inferred from diallele crosses between siblings; (3) the putative SRK ortholog must exhibit high levels of synonymous and especially nonsynonymous diversity and a pattern of selective constraint similar to SRK; and (4) sequences of the putative SRK ortholog should be expressed in the stigmas of maturing buds (although we note that some SRK alleles in A. lyrata are also expressed in leaves as well as in buds (Prigoda et al. 2005). We present evidence for each of these expectations and conclude that one of the studied loci (Lal2) either is orthologous to SRK or is closely linked to SRK and therefore may be a useful tool for typing diversity at the S-locus in natural populations of this species (Charlesworth et al. 2006; Hagenblad et al. 2006).
MATERIALS AND METHODS
Study system and rationale:
Leavenworthia is thought to have diverged from the common ancestor of Arabidopsis ∼13–19 MYA and from the common ancestor of Brassica ∼16–21 MYA (Mitchell-Olds 2001; Beilstein et al. 2006). There are eight extant species of Leavenworthia, and within the group there have been at least three independent derivations of self-fertilization (Rollins 1963; Beck et al. 2006). Leavenworthia is endemic to the southeastern United States and is found under a restricted range of ecological conditions (Rollins 1963). A long history of work in this group points to a role of reproductive assurance in the evolution of self-compatibility (Lloyd 1965; Busch 2005a) and the loss of genetic variation in self-compatible populations (Liu et al. 1998, 1999). Leavenworthia alabamica and L. crassa possess considerable intraspecific variation in the presence or absence of SI and are therefore interesting models for the study of S-locus variation.
Plant growth, diallele crosses, and pollen tube visualization:
Seeds were collected from two natural SI populations of L. alabamica during the spring of 2005; these are referred to as the “Isbell” and “Waco” populations (see Busch 2005b for geographic locations). Plants were raised from seed originally collected from 20 distinct plants in these populations. Seeds were germinated according to established protocols (Busch 2005b), and the plants were grown to flowering in the McGill University Phytotron greenhouse under a combination of natural and supplemental light. Plants collected from the same population were paired haphazardly to generate 10 full-sib families; the parents of families 1–5 were of “Waco” origin, whereas the parents of families 6–10 were collected from “Isbell.”
Sixteen or more offspring from each of the 10 full-sib families were germinated in March of 2006 and raised to flowering for the diallele crosses. Within each diallel, replicates of three or more reciprocal crosses were conducted for each parental combination. To carry out a cross, flowers were emasculated, anthers were removed with forceps, and pollen was placed directly on the stigmas of newly opened flowers. Forceps were cleaned with a 70% ethanol solution between individual crosses. Seeds were collected 1 month later as they ripened. Individual plants were considered compatible if the majority of crosses between them yielded large fruits (with four or more seeds). Single-seeded fruits were considered to reflect an incompatible cross with some degree of leakiness (Schierup et al. 2001). Two- and three-seeded fruits were also sometimes produced; these were enumerated separately. Individuals within a diallele cross were placed in the same phenotypic group when they exhibited identical patterns of compatibility.
For visualization of the SI reaction at the level of pollen tube growth, compatible or incompatible pollen was applied to stigmas. Stigmas were harvested after 24 hr and fixed in acetic acid–ethanol for 1 week, then rinsed, cleared in NaOH, and stained using a 0.1% aniline blue solution (Kho and Baer 1968). Fluorescence microscopy with a DAPI filter and a digital camera was used to document the SI reaction.
Cloning and sequencing of SRK gene family members:
Leaves were collected from parents and offspring and flash frozen in liquid nitrogen. DNA was extracted from leaves using DNeasy plant mini kits (QIAGEN, Mississauga, ON, Canada). Degenerate primers that amplify SRK in Arabidopsis and Brassica were used to amplify the S-domain of SRK-like sequences (Charlesworth et al. 2000; Schierup et al. 2001; Mable et al. 2003; supplemental Table 1). Primers were also designed to amplify conserved regions of the kinase domain, which are less variable than the extracellular S-domain (Charlesworth et al. 2003a; kinase 1F: RCTTCARCAYATHAAYCTTG; kinase 1R: TCTTGGTGAAGRTAYARAAG; kinase 2F: ACGGGTGTGTGTATTTGGACTGGA; kinase 2R: AGAATATY CCRTCCATYGC). Kinase 1F and kinase 1R are found at the 3′-end of exon 4 and the middle of exon 5, respectively; kinase 2F and kinase 2R were also designed from the middle of exon 5 and the 5′-end of exon 6. PCRs were conducted at low stringency under the following conditions: 94° for 5 min with 32 cycles of a 30-sec denaturation step at 94°, annealing at 40° for 1 min, and extension at 72° for 1 min, followed by a final 10-min extension step at 72°. Amplicons were cloned using TOPO TA cloning kits (Invitrogen, San Diego) and grown overnight on LB–agarose plates. At least eight clones were randomly selected from each transformed sample. Plasmid DNA was isolated with standard mini-prep procedures. Insert sizes of clones were quantified by digestion with EcoRI and gel electrophoresis. All positive clones near the expected size were sequenced in both the forward and reverse direction using universal M13 primers. DNA templates were purified and cycle sequenced using the v. 3.1 BigDye terminator ready reaction mix and analyzed using an ABI 3730 automated sequencer (Applied Biosystems, Foster City, CA). The GenBank accession numbers for sequence data are EU394446–EU394520. Sequences were aligned using Codon Code Aligner (Codoncode). Nucleotide sequences and their translated protein sequences were screened against known SRK sequences found in closely related species by using the BLAST algorithm (Altschul et al. 1990).
Phylogenetic analyses, sequence diversity estimates, and detecting selection:
Within each family of DNA sequences, alignments were manually edited on the basis of published amino acid alignments of S-domain genes. Phylogenetic relationships among sequences were studied using the neighbor-joining algorithm with Jukes–Cantor correction across all sites. Synonymous and nonsynonymous nucleotide diversities were estimated using DNAsp v. 4.0 (Rozas et al. 2003). Nucleotide diversity among Lal1 sequences was estimated ignoring pseudogenes. The seven longest Lal2 DNA fragments were translated into the predicted amino acid sequences and aligned, and a sliding window of nucleotide variability was calculated using DNAsp. This sliding window provided an aid for nucleotide sequence alignments, with a window size of 50 nucleotides and a step size of one nucleotide. The nucleotide diversities of SRK alleles sampled within A. lyrata, C. grandiflora, R. sativus, and Brassica species were also calculated for purposes of comparison.
To analyze patterns of selection at the codon level in the Lal2 sequences, we used the phylogenetically based, codon-level analysis of nonsynonymous and synonymous substitution (Nielsen and Yang 1998; Yang et al. 2005). This approach calculates posterior probabilities of positive selection (as determined from the ratio of nonsynonymous substitution per nonsynonymous site to synonymous substitution per synonymous site) for individual codons, using an empirical Bayesian approach. The maximum-likelihood tree topology for the Lal2 sequences was first obtained using the branch-and-bound algorithm as implemented in PAUP for the Kimura two-parameter model (Kimura 1980; Swofford 2002), and the selection analysis was conducted using the CODEML program of Yang (1997), as decribed by Sainudiin et al. (2005). The number of Lal2 sequences available to us for analysis (10 sequences) was much smaller than those analyzed by Sainudiin et al. (2005), who used the same method to detect specific classes of positive selection (e.g., polar- or volume-changing amino acid substitutions) in SRK sequences of Brassica and Arabidopsis. Hence, our focus was on evidence of any type of positively selected amino acid substitution, regardless of the particular type of property-changing characteristic. The Lal2 codons available for analysis by this method correspond to amino acid residues 154–410 of B. oleracea.
Cosegregation of SRK-like sequences with the SI reaction:
SRK-like sequences were tested for cosegregation with incompatibility alleles determined by diallele crosses; these were identified by analyzing seed set resulting from crosses between full siblings. All incompatibility groups were classified independently and without prior knowledge of sequence data. Sequence-specific primers were designed and used to amplify SRK-like DNA sequences in the offspring of a full-sib family (supplemental Table 2). SRK-like sequences were inferred to have cosegregated with S-alleles if the sequence was found in individuals from the dialleles that were incapable of mating with one another. PCR products were visualized on 1× TBE gels containing 1% agarose and verified by DNA sequencing.
Gene expression of the Lal2-3 allele:
L. alabamica individuals of known genotype were selected for gene expression analysis of the Lal2-3 allele. Tissue was harvested from the leaves, styles, and stigmas. Style tissue was isolated from stigmas and was collected from open flowers. Stigma tissue was harvested from buds 3 days (day −3) and 1 day prior (day −1) to flower opening as well as from open flowers (day 0). Tissue samples from ∼70 flowers harvested from the same individual were pooled and flash frozen in liquid nitrogen. Frozen tissue was ground to a fine powder in liquid nitrogen using a Kontes pestle (VWR, Mississauga, ON, Canada). Total RNA was extracted using the RNeasy plant mini kit (QIAGEN). Extracted RNA was treated with RNAse-free DNAse (Promega, Madison, WI) to eliminate all possible DNA contamination prior to the reverse transcriptase (RT)–PCR) experiments and the enzyme was removed using the cleanup protocol specified for the QIAGEN RNeasy kit. For each sample, 1 μg of RNA was reverse transcribed using oligo(dT) primer (Invitrogen Canada, Burlington, ON, Canada) and Superscript II RT (Invitrogen).
Primers for the Lal2-3 allele (W14F: TCAGCTTGAATACCGTACTGACTTA; W14R: GTCCCTGCTTGTGTTGATGGA) and an actin control (actinF: TATGCACTTCCACATGCTAT; actinR: CTTTGCGATCCACATCTGCTG) were designed. Both of the actin primers were intron spanning to ensure that only cDNAs would be amplified by PCR. PCR products were visualized on a 1% agarose gel by ethidium bromide staining and product identity was also confirmed by sequencing (Genome Quebec, Montreal). No reverse transcriptase controls were used for each sample to confirm the absence of DNA contamination.
RESULTS
Cross-incompatibility and interactions among inferred S-alleles:
Tables 1–5 summarize the results of crosses conducted between full siblings in each of the different diallele crosses. In families 4, 5, and 10, four phenotypic groups were identified on the basis of the patterns of cross-incompatibility. Tables 1 and 2 show that family 4 and family 5 exhibited identical patterns of cross-incompatibility and therefore have identical types of interactions among S-alleles. In particular, in both families, phenotypic group II is incompatible with group I when acting as a maternal parent, while the reciprocal cross successfully produces fruit. This supports the existence of alleles (Si in family 4 and Sm in family 5) that act codominantly in the pistil yet are recessive to some alleles in pollen. Moreover, matings between groups I and III were compatible in both families, suggesting that there are two alleles (Sk in family 4 and So in family 5) that are recessive in both the stigma and pollen. Table 3 shows that, in family 10, most alleles were codominant with one another, but that groups I and III were reciprocally compatible. This pattern suggests the existence of an allele (Ss) with recessive gene action in stigmas and pollen. The overall level of incompatibility between plants sharing S-alleles was lower in family 10, indicating some degree of leakiness in the self-incompatibility mechanism.
TABLE 1.
Summaries of dialleles: family 4
| Recipients
|
||||
|---|---|---|---|---|
| I | II | III | IV | |
| (4) | (7) | (2) | (3) | |
| Donors | SiSk | SiSl | SjSk | SjSl |
| I | 10a/40 | 37b/93 | 25/25 | 38/40 |
| SiSk | (0.18) | (0.20) | (1.0) | (0.95) |
| II | 92/93 | 50c/137 | 39/40 | 18d/69 |
| SiSl | (0.99) | (0.26) | (0.98) | (0.12) |
| III | 28/29 | 44/48 | 0/6 | 4e/16 |
| SjSk | (0.97) | (0.92) | (0.0) | (0.13) |
| IV | 39/40 | 39f/71 | 1g/20 | 0/15 |
| SjSl | (0.98) | (0.27) | (0.0) | (0.0) |
Arabic numerals in parentheses below the roman numeral phenotypic groups indicate the number of full sibs in each category, which are above the inferred genotype for each group at the S-locus. Within each cell is a fraction showing the number of compatible crosses (numerator) and the total number of attempted crosses (denominator). The observed proportion of crosses that yielded fruit with more than three seeds is also shown (in parentheses). Incompatible groups are in italics. S-allele letter designations are used as labels to refer to the alleles inferred within the different crosses (see Table 6 and text).
Three small fruits.
Eighteen small fruits.
Fifteen small fruits.
Ten small fruits.
Two small fruits.
Twenty small fruits.
One small fruit.
TABLE 2.
Summaries of dialleles: family 5
| Recipients
|
||||
|---|---|---|---|---|
| I | II | III | IV | |
| (4) | (2) | (3) | (7) | |
| Donors | SmSo | SmSp | SnSo | SnSp |
| I | 1/38 | 6a/24 | 34/36 | 82/86 |
| SmSo | (0.03) | (0.08) | (0.94) | (0.95) |
| II | 26/26 | 0/5 | 20/20 | 8b/44 |
| SmSp | (1.0) | (0.0) | (1.0) | (0.05) |
| III | 38/38 | 19/21 | 8c/23 | 3/62 |
| SnSo | (1.0) | (0.90) | (0.17) | (0.05) |
| IV | 84/87 | 7d/48 | 30e/83 | 3/117 |
| SnSp | (0.97) | (0.04) | (0.08) | (0.03) |
Four small fruits.
Six small fruits.
Four small fruits.
Five small fruits.
Twenty-three small fruits.
TABLE 3.
Summaries of dialleles: family 10
| Recipients
|
||||
|---|---|---|---|---|
| I | II | III | IV | |
| (6) | (6) | (3) | (2) | |
| Donors | SqSs | SqSt | SrSs | SrSt |
| I | 12a/101 | 65b/113 | 45/51 | 40/41 |
| SqSs | (0.03) | (0.48) | (0.88) | (0.98) |
| II | 27c/121 | 12/97 | 53/54 | 34c/46 |
| SqSt | (0.11) | (0.12) | (0.98) | (0.46) |
| III | 48/51 | 45/54 | 2/14 | 7d/20 |
| SrSs | (0.94) | (0.83) | (0.14) | (0.25) |
| IV | 39/39 | 17e/37 | 1/15 | 0/7 |
| SrSt | (1.0) | (0.35) | (0.07) | (0.0) |
Nine small fruits.
Eleven small fruits.
Thirteen small fruits.
Two small fruits.
Four small fruits.
TABLE 4.
Summaries of dialleles: family 1
| Recipients
|
||
|---|---|---|
| I | II | |
| (8) | (8) | |
| Donors | SaSc,SaSd | SbSc,SbSd |
| I | 25a/164 | 171b/200 |
| SaSc,SaSd | (0.07) | (0.73) |
| II | 183c/197 | 71d/170 |
| SbSc,SbSd | (0.81) | (0.19) |
Thirteen small fruits.
Twenty-five small fruits.
Twenty-four small fruits.
Thirty-nine small fruits.
TABLE 5.
Summaries of dialleles: family 3
| Recipients
|
||
|---|---|---|
| I | II | |
| (8) | (8) | |
| Donors | SeSg,SeSh | SfSg,SfSh |
| I | 28a/147 | 174b/184 |
| SeSg,SeSh | (0.05) | (0.92) |
| II | 184c/187 | 66d/165 |
| SfSg,SfSh | (0.97) | (0.22) |
Twenty-one small fruits.
Five small fruits.
Two small fruits.
Twenty-nine small fruits.
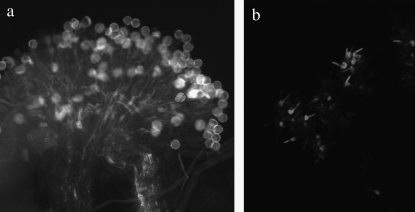
Tables 4 and 5 demonstrate that in families 1 and 3 there were only two phenotypic groups of reciprocal cross-incompatibility. This pattern of cross-incompatibility might occur whenever a parent with two dominant alleles is mated to a parent that is heterozygous or homozygous for recessive S-alleles. The existence of two or four incompatibility groups in dialleles of L. alabamica is consistent with a single-locus system of self-incompatibility in this species. Moreover, the finding of alleles that are recessive either only in pollen or in both stigmas and pollen is consistent with the type of sporophytic system that has been observed in other species in the Brassicaceae. Figure 1 illustrates the characteristic pollen tube germination and growth observed following compatible and incompatible crosses. As seen in other SI members of the Brassicaceae, pollen tube growth of incompatible pollen in L. alabamica is inhibited in the papillae of stigmas.
Figure 1.—
Pollen tube germination and growth after compatible and incompatible crosses. A compatible cross (a) with pollen tube growth into the style and an incompatible cross (b) with pollen tube growth inhibited.
Relationships among SRK-like sequences in L. alabamica and its relatives:
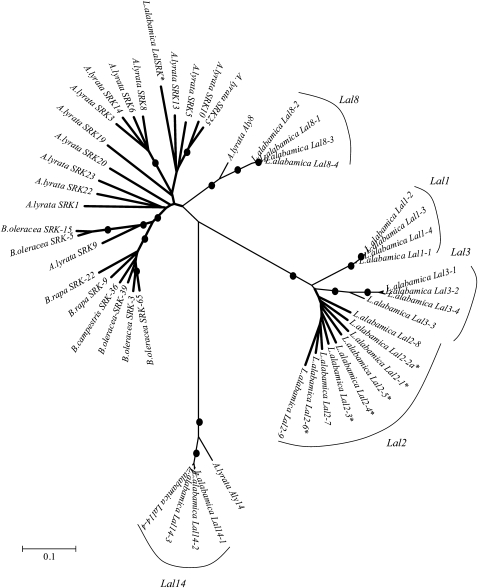
Figure 2 shows a neighbor-joining tree that summarizes the relationships among the sequences amplified in L. alabamica, as well as among SRK gene family members in Brassica and Arabidopsis. In L. alabamica, families of related sequences were identified and given the names LalSRK, Lal1, Lal2, Lal3, Lal8, and Lal14. One of the L. alabamica sequences shares close nucleotide similarity to SRK alleles amplified in A. lyrata and is denoted LalSRK. Of 81 plants collected from natural populations sampled across the geographic range of L. alabamica, we isolated LalSRK with sequence-specific primers from only 1 plant. We amplified two groups of sequences (hereafter termed Lal8 and Lal14) that cluster strongly with their putative orthologs in A. lyrata, Aly8 and Aly14. The large remainder of sequences amplified from L. alabamica fell into one of three closely related clusters that we name Lal1, Lal2, and Lal3.
Figure 2.—
Unrooted neighbor-joining tree with Jukes–Cantor correction of SRK-like sequences amplified in L. alabamica and several relatives. The tree was generated using a pairwise deletion algorithm to handle missing sites. Lal2 and SRK sequences marked with asterisks were shown in this study to be linked to the SI reaction in L. alabamica. Dots denote nodes with ≥95% bootstrap support. The scale denotes the number of substitutions.
Nucleotide BLASTs of Lal1, Lal2, and Lal3 demonstrated highest similarity to alleles of SRK isolated from Arabidopsis, Brassica, and Capsella; to S-locus glycoprotein (SLG) isolated from Brassica; and, to a lesser extent, to the SLR1 locus (Kumar and Trick 1993; Luu et al. 2001), which has been shown to exhibit close identities to SRK in the S-domain region. Most of the Lal1, Lal2, and Lal3 sequences showed high levels of nucleotide identity to alleles of SRK in the conserved 5′ and 3′ regions of the S-domain, which flank the hypervariable portions of SRK. The intervening region of these sequences showed no nucleotide identity to SRK or to any other known plant genes. Sequences of Lal1, Lal2, and Lal3 also share deletions that are 3, 6, and 27 bp in length in the medial portion of the S-domain compared to previously studied sequences of SRK in other mustard species. Protein BLASTS indicate that SRK alleles are the most similar sequences to the predicted amino acid sequences of Lal1, Lal2, and Lal3.
Cosegregation between SRK-like sequences and inferred S-alleles:
Sequence-specific PCR allowed us to amplify six SRK-like sequences in the L. alabamica plants used for diallele analysis of SI (LalSRK, Lal2-1, Lal2-3, Lal2-4, Lal2-5, Lal2-6). Sequences could not be amplified from every plant used in the dialleles, likely because of lack of homology with the PCR primers employed (based mostly on Brassica and Arabidopsis sequence information) and the possibility that Lal2 may not be found in all Leavenworthia S-haplotypes (see below). These six sequences cosegregate with S-alleles inferred on the basis of compatibility patterns in all 113 cases where data were available from crossing studies (Table 6). All Lal2 sequences tested for cosegregation with inferred S-locus alleles were found to segregate in offspring according to Mendelian expectations (χ2 = 4.75, d.f. = 8; P > 0.10; Table 6), as would be expected if these alleles were inherited at a single locus. Given our sample size of n = 97 for tests of cosegregation of Lal2 with inferred S-alleles, we can state with 95% confidence that Lal2 lies at most 3.1 cM from the S-locus in L. alabamica (Stevens 1942).
TABLE 6.
Inferred S-alleles, dominance, and linkage evidence
| Diallele family | Parental genotypes | S-allele interactions | Sequence(s) found | Frequency of sequence in offspring | Evidence sequence is linked to an inferred S-allele |
|---|---|---|---|---|---|
| 1 | Sa,Sb, Sc,Sd | Sa,Sb > Sc,Sd | Lal2-1 | 0.50 | Sequence cosegregates with Sa. |
| Lal2-4 | 0.50 | Sequence cosegregates with Sb. | |||
| Lal2-2a | 0.25 | Recessive allele Sc | |||
| 3 | Se,Sf, Sg,Sh | Se,Sf > Sg,Sh | Lal2-3 | 0.50 | Sequence cosegregates with Se. |
| LalSRK | 0.50 | Sequence cosegregates with Sf. | |||
| Lal2-2b | 0.63 | Recessive allele Sg | |||
| 4 | Si,Sj, Sk,Sl | Sj = Sl = Si > Sk | Lal2-4 | 0.31 | Sequence cosegregates with Sj. |
| Sl > Si in pollen | |||||
| 5 | Sm,Sn, So,Sp | Sn = Sp = Sm > So | Lal2-5 | 0.63 | Sequence cosegregates with Sn. |
| Sp > Sm in pollen | |||||
| 10 | Sq,Sr, Ss,St | Sq = St = Sr > Ss | Lal2-6 | 0.71 | Sequence cosegregates with Sq. |
We amplified a Lal2 sequence (Lal2-3) together with LalSRK from the same individual parent used to generate family 3 (Table 6). Twenty-four offspring of family 3 were genotyped for both of these sequences. Thirteen offspring possessed LalSRK while the other 11 plants possessed Lal2-3; this pattern is consistent with the 1:1 inheritance of these sequences, as would be expected if they were alleles segregating at a single locus or were alleles segregating in repulsion phase at two tightly linked loci.
In addition to these Lal2 sequences for which there is direct evidence of cosegregation with inferred S-locus alleles, there are three other Lal2 sequences for which crossing data were insufficient to test for cosegregation with inferred SI alleles (Lal2-7, Lal2-8, and Lal2-9), as well as two sequences (Lal2-2a and Lal2-2b) that are 99% identical at the nucleotide level. The 1-bp difference between Lal2-2a and Lal2-2b may be the result of sequencing error, as multiple clones were not sequenced. Lal2-2a and Lal2-2b were amplified from family 1 and family 3, in which cosegregation of incompatibility patterns with genomic sequences cannot be investigated because of recessive gene action. Additional evidence, apart from their nearly complete sequence identity, suggests that Lal2-2a and Lal2-2b either are copies of the same S-allele or are linked in coupling phase to the same S-allele. Lal2-2a and Lal2-2b were found to be reciprocally cross-incompatible and had to be paired with other plants to generate families 1 and 3. Sequence-specific PCR shows that Lal2-2a and Lal2-2b were segregating in both families 1 and 3. Lal2-2a was found in 4 of 16 offspring in family 1 and Lal2-2b in 10 of 16 offspring in family 3. Therefore, both of the originally cross-incompatible parents used to generate families 1 and 3 were heterozygous for these Lal2 alleles. Lal2-2 was the only Lal2 sequence amplified from multiple parental individuals, a finding that is consistent with the hypothesis that recessive S-alleles should equilibrate at higher frequencies in populations (Schierup et al. 1997; Uyenoyama 2000).
Tests of cosegregation with inferred S-locus alleles were also conducted for sequences of Lal3 in family 1. A sequence-specific PCR conducted with the offspring of family 1 successfully amplified the Lal3 variant in all offspring, an inheritance pattern that indicates that Lal3 does not cosegregate with S-locus alleles. Tests for cosegregation with SI were not conducted for Lal1 variants as this locus had relatively low total nucleotide diversity and is therefore not a viable candidate for a S-linked locus, as has been observed for Lal2.
Patterns of nucleotide variability and patterns of selection at the codon level:
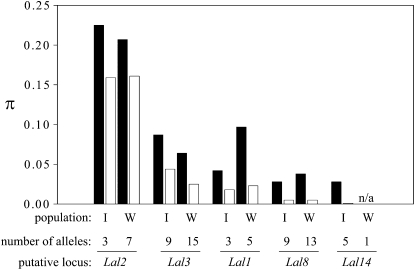
Figure 3 summarizes diversities among alleles at synonymous and nonsynonymous sites of Lal2 and of the other members of this gene family. The Lal2 diversity levels are significantly higher than those observed at all other gene family members for similar lengths of sequence. The average synonymous diversity of Lal2 in two populations of L. alabamica (πs = 0.216) was at least 10 times higher than the average diversity observed at loci in populations of outcrossing plants (Glémin et al. 2006). More modest levels of synonymous diversity were observed at Lal1 (πs = 0.069) and Lal3 (πs = 0.076), although the levels of variability were also high compared to what is expected for loci not experiencing balancing selection. Perhaps most striking were the extremely high estimates of nonsynonymous diversity at the Lal2 locus (πa = 0.160). Lal2 sequences possess synonymous and nonsynonymous diversity levels similar overall to those observed at the SRK locus in Raphanus and Brassica, although they were lower than the levels reported in Arabidopsis and Capsella (Table 7).
Figure 3.—
Estimates of synonymous (solid bars) and nonsynonymous (open bars) nucleotide diversity (π) at five loci studied in two natural populations of L. alabamica.
TABLE 7.
Diversity of Lal2 in comparison to the SRK locus in mustard species
| L. alabamica Lal2 | A. lyrata SRK | C. grandiflora SRK | Brassica spp. SRK | R. sativus SRK | |
|---|---|---|---|---|---|
| πs | 0.20 | 0.73 | 0.71 | 0.39a/0.23b | 0.23 |
| πa | 0.17 | 0.29 | 0.26 | 0.15a/0.12b | 0.13 |
| Amino acid distance | 0.34 | 0.55 | 0.46 | 0.28a/0.24b | 0.26 |
| No. of sequences | 10 | 14 | 6 | 12a/10b | 8 |
| Sequence length | 560 | 654 | 951 | 973a/979b | 978 |
Comparison of genetic diversity at Lal2 and SRK isolated in several mustard species. Synonymous site diversity (πs) and nonsynonymous site diversity (πa) were estimated with Jukes–Cantor correction. Amino acid distances are Poisson-corrected estimates.
Estimate includes both class I and class II (recessive) alleles.
Estimate includes only class I alleles.
We attempted to amplify the kinase domain of Lal2-2b, Lal2-3, and the LalSRK allele found in this study. We successfully amplified a portion of the kinase domain from the same individual from which we amplified the S-domain of LalSRK. Both of these sequences, when used to make independent gene trees with known SRK alleles, cluster with the same A. lyrata SRK alleles (SRK-13 and SRK-5). Our inability to amplify a kinase domain from Lal2 suggests either that Lal2 sequences lack a kinase domain or that there is a marked degree of nucleotide divergence between the Lal2 kinase domain and SRK kinase-domain primers. Although researchers have successfully amplified the kinase domain for many of the SRK alleles studied in A. lyrata (Charlesworth et al. 2003a), it seems possible that divergence from the known allele SRK in the kinase domain of Lal2 could be as pronounced as that observed in the S-domain. Pairwise comparisons demonstrate that LalSRK was most similar to SRK-5 amplified from A. lyrata and that there are a large number of synonymous and nonsynonymous substitutions separating alleles of Lal2 from LalSRK and from SRK alleles amplified in species of Arabidopsis and Brassica (Table 8). The divergence among alleles of Lal2 is on the order of that observed among class I SRK alleles in the genus Brassica (Table 7).
TABLE 8.
Pairwise nucleotide diversity between alleles in the S-domain of SRK and Lal2
| Sequence | BoSRK3 | BcSRK8 | BrSRK9 | BoSRK5 | BoSRK15 | AlSRK5 | AlSRK1 | AlSRK22 | AlSRK25 | AlSRK6 | LalSRK | Lal2-2a | Lal2-3 | Lal2-4 | Lal2-5 | Lal2-6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BoSRK3 | 0.156 | 0.157 | 0.298 | 0.301 | 0.324 | 0.315 | 0.281 | 0.311 | 0.374 | 0.365 | 0.695 | 0.655 | 0.674 | 0.678 | 0.764 | |
| BcSRK8 | 0.278 | 0.154 | 0.307 | 0.319 | 0.302 | 0.324 | 0.284 | 0.292 | 0.353 | 0.34 | 0.661 | 0.632 | 0.673 | 0.66 | 0.775 | |
| BrSRK9 | 0.275 | 0.247 | 0.296 | 0.302 | 0.317 | 0.314 | 0.317 | 0.288 | 0.354 | 0.365 | 0.65 | 0.623 | 0.7 | 0.677 | 0.762 | |
| BoSRK5 | 0.953 | 0.8 | 0.855 | 0.073 | 0.383 | 0.329 | 0.315 | 0.348 | 0.438 | 0.393 | 0.682 | 0.649 | 0.655 | 0.692 | 0.769 | |
| BoSRK15 | 0.884 | 0.79 | 0.857 | 0.084 | 0.387 | 0.355 | 0.322 | 0.361 | 0.429 | 0.394 | 0.72 | 0.68 | 0.705 | 0.741 | 0.793 | |
| AlSRK5 | 1.006 | 1.036 | 0.914 | 0.842 | 0.86 | 0.315 | 0.344 | 0.194 | 0.296 | 0.249 | 0.593 | 0.63 | 0.615 | 0.639 | 0.72 | |
| AlSRK1 | 0.745 | 0.719 | 0.664 | 0.866 | 0.795 | 0.924 | 0.265 | 0.323 | 0.39 | 0.39 | 0.603 | 0.568 | 0.617 | 0.603 | 0.669 | |
| AlSRK22 | 0.837 | 0.787 | 0.8 | 0.664 | 0.62 | 0.833 | 0.766 | 0.331 | 0.399 | 0.377 | 0.633 | 0.593 | 0.622 | 0.634 | 0.711 | |
| AlSRK25 | 0.991 | 0.813 | 0.93 | 0.729 | 0.742 | 0.381 | 0.758 | 0.815 | 0.338 | 0.25 | 0.664 | 0.693 | 0.69 | 0.668 | 0.779 | |
| AlSRK6 | 0.982 | 0.895 | 0.781 | 1.15 | 1.134 | 0.733 | 1.031 | 0.665 | 0.964 | 0.344 | 0.691 | 0.672 | 0.7 | 0.696 | 0.748 | |
| LalSRK | 1.156 | 1.04 | 1.04 | 0.855 | 0.814 | 0.644 | 0.97 | 0.952 | 0.636 | 0.945 | 0.65 | 0.653 | 0.634 | 0.65 | 0.71 | |
| Lal2-2a | 1.03 | 1.066 | 1.183 | 1.624 | 1.414 | 1.273 | 1.269 | 1.185 | 0.92 | 1.178 | 1.441 | 0.154 | 0.168 | 0.14 | 0.155 | |
| Lal2-3 | 1.134 | 1.107 | 1.222 | 1.87 | 1.58 | 1.429 | 1.252 | 1.462 | 1.206 | 1.285 | 1.725 | 0.151 | 0.147 | 0.146 | 0.14 | |
| Lal2-4 | 1.084 | 1.073 | 1.150 | 1.757 | 1.509 | 1.29 | 1.214 | 1.511 | 1.218 | 1.083 | 1.379 | 0.18 | 0.168 | 0.156 | 0.147 | |
| Lal2-5 | 1.032 | 1.124 | 1.277 | 1.805 | 1.505 | 2.032 | 1.037 | 1.109 | 1.263 | 1.258 | 1.705 | 0.18 | 0.207 | 0.204 | 0.114 | |
| Lal2-6 | 1.335 | 1.107 | 1.185 | 1.385 | 1.298 | 1.541 | 1.236 | 1.126 | 1.172 | 1.256 | 1.37 | 0.161 | 0.205 | 0.201 | 0.19 |
Jukes–Cantor-corrected estimates of nonsynonymous substitutions (Ka) and synonymous substitutions (Ks) are above and below the diagonal, respectively. The levels of synonymous (non-italic type, Ks < 0.5; italic type, 0.5 < Ks < 1.0; and underlining, Ks > 1.0) and nonsynonymous divergence (non-italic type, Ka < 0.3; italic type, 0.3 < Ka < 0.6; and underlining, Ka > 0.6) are shown.
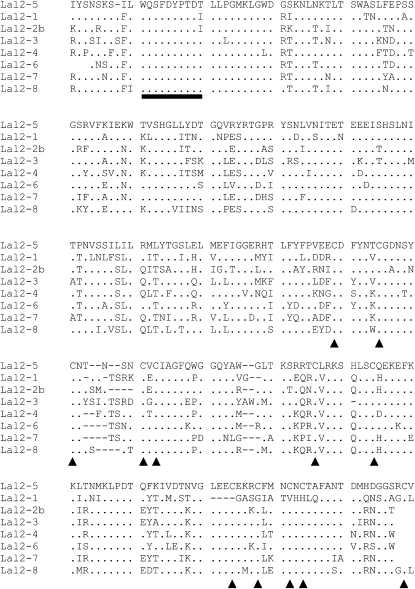
Figure 4 is an alignment of the predicted amino acid sequences for the longest Lal2 variants, showing that that this locus contains a strongly conserved motif (WQSFDYPTDT/I) that is also constrained among other receptor-like protein kinases (Walker 1994). With the exception of Lal2-1, which has a short deletion at the 3′-end of the S-domain, these sequences also possess the predicted cysteine residues that are important to the tertiary structure and function of the S-locus receptor kinases. As a result, Lal2-1 lacks four of the last five conserved cysteines immediately downstream of the deletion, although it retains amino acid identity downstream of the deletion. Overall, the levels of amino acid diversity in Lal2 are on the order of those observed at the SRK locus in other mustard species (Table 7).
Figure 4.—
Alignment of the predicted amino acid sequences of Lal2 alleles. Dots denote amino acid identity with the topmost reference allele. Triangles point to the 12 conserved cysteine residues typically found in S-domain loci. The underlined region of 10 amino acids indicates a strongly conserved motif characteristic of receptor-like kinases. The Lal2-1 allele does not possess four of the last five conserved cysteine residues.
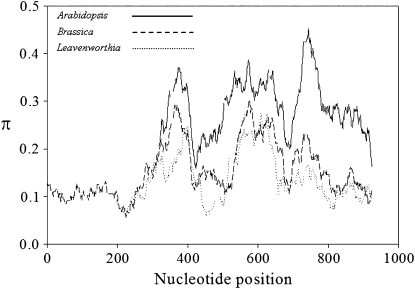
The Lal2 sequences were examined for evidence of balancing selection and, in particular, evidence of “hypervariable” regions that have been proposed to harbor sites controlling S-allele recognition (Sainudiin et al. 2005). Figure 5 shows a sliding-window analysis of nucleotide variability calculated for the alignment of long Lal2 sequences with alleles of SRK sampled in Arabidopsis and class I alleles in Brassica. Overall, Arabidopsis SRK alleles were the most variable, whereas SRK alleles of Brassica and the L. alabamica Lal2 locus exhibited similarly high levels of diversity. Importantly, there were peaks of diversity in Lal2 in the same regions where Brassica alleles exhibit the highest local diversity (sites 350–400 and 550–650), although the peak of diversity at the 3′-end of Lal2 (sites 700–775) was modest in comparison to the elevated diversity observed in this region of SRK.
Figure 5.—
Sliding-window analysis of nucleotide diversities in the S-domain of SRK and Lal2. The nucleotide diversities (π) of Arabidopsis (solid line) and Brassica (dashed line) SRK were estimated using 8 and 11 alleles, respectively. Leavenworthia Lal2 diversity (stippled line) was estimated using seven sequences. The window size is 50 nucleotides with a step size of 1 nucleotide.
The Bayesian empirical inference of amino acid sites under positive selection detected 17 such sites, and an alignment of the Lal2 sequences with SRK60 of B. oleracea shows that many of the positively selected codons correspond closely or exactly with those of the latter species. A Fisher exact test conducted for a 2 × 2 table of sites positively selected in both Brassica and Leavenworthia, positively selected in only one group, or positively selected in neither returns a probability value of 0.034 for the two-tailed null hypothesis test of no association; i.e., the pattern of selection at codons in the two sets of taxa appears to be significantly correlated.
Expression of Lal2 in floral and vegetative tissues:
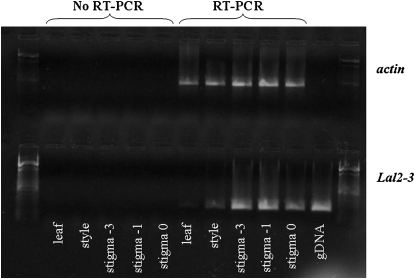
The Lal2-3 allele is expressed at low levels in leaves, at an intermediate level in the styles of mature flowers, and at higher levels in stigmatic tissue (Figure 6). The expression of Lal2-3 at low levels in leaves does not necessarily rule out this locus as being orthologous to SRK, as work in A. lyrata has shown that a class of SRK alleles is transcribed in vegetative tissue (Prigoda et al. 2005). Lal2 transcripts are most prevalent in stigmatic tissue immediately prior to anthesis but decline following anthesis, a pattern that is consistent with the temporal expression of SRK in Brassica (Stein et al. 1996; Kusaba et al. 2001). The decline in Lal2 expression in open flowers is most likely caused by these products being translated into functional proteins during or immediately prior to anthesis. These results suggest that the gene is expressed predominantly in stigmatic tissue, where SRK and SLG have been shown to interact with SCR to block the growth of self-pollen grains (Kachroo et al. 2001; Kusaba et al. 2001; Takayama et al. 2001).
Figure 6.—
Expression of Lal2-3 in vegetative and floral tissues. Samples were harvested 1 (−1) and 3 (−3) days before flowering and on the day the flowers opened (0). PCR amplified actin and Lal2-3 bands are from cDNAs generated by RT–PCR; gDNA stands for a positive control of genomic DNA with the Lal2-3 sequence. RT–PCR controls demonstrate that samples were not contaminated with genomic DNA.
DISCUSSION
The observations made in this study support the hypothesis of sporophytic SI in L. alabamica, as reported in the closely related species L. crassa and several other mustard species (Bateman 1954, 1955; Thompson 1957; Lloyd 1967; Sampson 1967: Schierup et al. 2001). At the molecular level, we amplified a single sequence (LalSRK) linked to the S-locus with strong homology to alleles of SRK that have been sampled from species of Arabidopsis, Brassica, Capsella, and Raphanus (Hinata et al. 1995; Schierup et al. 2001; Castric and Vekemans 2004; Okamoto et al. 2004; Paetsch et al. 2006). In addition, we identified sequences of Lal2 that are S-linked and share many sequence-level characteristics with the known alleles of SRK. Lal2 alleles are most highly expressed in stigmatic tissue, are extremely polymorphic at the nucleotide and amino acid level, and appear to have been subject to selective pressures similar to SRK. Given these findings, we posit that sequences of Lal2 either are divergent SRK alleles or are the result of recent gene duplication in the S-locus region.
Our finding of one allele of SRK and many sequences of Lal2 that are less similar to SRK implies either that (1) successful amplification of more than one L. alabamica SRK was not achieved because of marked divergence between SRK-specific primers (based on sequences from Arabidopsis and Brassica) and their target sites in L. alabamica or (2) Lal2 sequences are, in fact, divergent albeit functional S-alleles and are therefore orthologous to SRK. Experiments involving gain of function may help to establish whether Lal2 is necessary and sufficient to cause SI in L. alabamica (Takasaki et al. 2000; Takayama et al. 2001). For example, experiments whereby self-compatible plants are transformed with an intact Lal2-SCR haplotype can be used to determine whether Lal2 codes for SI in L. alabamica (Nasrallah et al. 2002, 2004).
If Lal2 sequences are functional S-domain receptors affecting SI in L. alabamica, their divergence from SRK alleles would demand explanation. It is possible that a proto-Lal2 sequence may have been captured from another receptor kinase locus, generating divergence between the proto-Lal2 sequence and the SRK sequences in the population. If this gene conversion event caused a bottleneck in S-allele number, the diversification of Lal2 would then simultaneously generate novel S-alleles and explain the similar diversity characteristics of Lal2 and SRK if the same amino acids influence specificity. The relatively high Ka/Ks ratio among Lal2 sequences is compatible with a recent and rapid divergence among Lal2 sequences in their specificities following a bottleneck in S-allele number caused by gene conversion. A scenario such as this has been invoked to explain the ancient divergence between Brassica and Arabidopsis SRK sequences (Kusaba et al. 2001) and the evolution of a divergent clade of closely related SRK alleles in A. lyrata (e.g., SRK3, SRK6, SRK8, and SRK14) that are expressed in leaves (Prigoda et al. 2005). This group of SRK alleles exhibits relatively shallow genetic divergence in both the S and kinase domains, consistent with a recent radiation (Charlesworth et al. 2003a).
Another possibility is that Lal2 is a recently duplicated locus linked to SRK. The finding of a highly polymorphic SRK-like gene linked to SRK would suggest the existence of a locus analogous to SLG of Brassica (Stein et al. 1991). SLG is a S-domain protein lacking a kinase domain that is tightly linked to SRK (Boyes and Nasrallah 1993), facilitates the incompatibility reaction (Suzuki et al. 2000; Takasaki et al. 2000), and is found in most S-haplotypes investigated to date in Brassica (Suzuki et al. 2000). SLG exhibits signatures of balancing selection and patterns of constraint similar to SRK even though it is not required for SI (Hinata et al. 1995; Kusaba et al. 1997), most likely because of gene conversion between SRK and SLG of the same S-haplotype (Watanabe et al. 1994; Fujimoto et al. 2006). There are also other S-domain loci that play no known role in SI that are embedded in the gene-dense S-locus regions of mustard species (Suzuki et al. 1999). In particular, there are at least three S-linked receptor kinases that are not required for the SI response in Brassica (Stein et al. 1991; Suzuki et al. 1997), and a receptor kinase (Aly8) with high homology to SRK is linked to the S-locus in A. lyrata (Charlesworth et al. 2003b). If Lal2 is the product of recent gene duplication at the S-locus, the elevated diversity of Lal2 and its similar diversity characteristics to SRK may involve infrequent gene conversion with SRK or parallel evolutionary constraints on the S-domain portion of the molecule.
Although it is not yet known with certainty whether Lal2 sequences are orthologous to SRK, the work reported here broadens the diversity of mustard species in which the diversity of the S-locus has been studied at the sequence level. Moreover, that Lal2 is S-linked means that it may be employed to infer S-locus genotypes and to investigate selection at the S-locus in L. alabamica. There are several loci closely linked to SRK in A. lyrata that can, in theory, be used to type S-alleles within populations, given the restricted recombination in this region (Charlesworth et al. 2006; Kamau et al. 2007). However, it should also be mentioned that attempts to exploit the close linkage of Aly8 to SRK in A. lyrata to infer S-haplotypes have been complicated by the presence of a tightly linked paralog of Aly8, which causes S-haplotypes to harbor more than one Aly8-like sequence (Hagenblad et al. 2006). If Lal2 is to be applied to infer dynamics at the S-locus in L. alabamica, it will be important to test each Lal2 sequence for linkage with an S-haplotype inferred through controlled dialleles and to continue tests of Mendelian single-locus inheritance.
Identification of S-linked loci in SI species and their close relatives that are self-compatible should provide insights into the evolution of self-fertilization from the outcrossing condition (Bechsgaard et al. 2006; Liu et al. 2007; Sherman-Broyles et al. 2007), which is a pervasive evolutionary trend in angiosperms (Darwin 1876; Baker 1955; Stebbins 1974; Igic et al. 2006). Studies of S-linked Lal2 variation within this and other species of Leavenworthia may be especially valuable for the insights that they may provide into the population–genetic mechanisms underlying the maintenance and loss of SI in natural populations of flowering plants (Busch and Schoen 2008).
Acknowledgments
We thank V. Koelling for generously providing seed collected in wild populations of L. alabamica, H. Jensen for laboratory assistance, and A-M. L'Heureux and K. Vitale for conducting crosses in the greenhouse. We acknowledge Genome Quebec for conducting sequencing reactions and D. Charlesworth, B. Mable, and J. Nasrallah for helpful discussions. Comments by S. Hoebee, E. Newbigin, and two anonymous reviewers improved the article. D.J.S. acknowledges continuing support from the Natural Sciences and Engineering Research Council of Canada (NSERC). J.S. was supported by NSERC. J.W.B. was supported with funds from McGill University's Tomlinson Fellowship program and by NSERC.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Baker, H. G., 1955. Self-compatibility and establishment after long distance dispersal. Evolution 9 347–349. [Google Scholar]
- Bateman, A. J., 1954. Self-incompatibility systems in angiosperms. II. Iberis amara. Heredity 8 305–322. [Google Scholar]
- Bateman, A. J., 1955. Self-incompatibility systems in angiosperms. III. Cruciferae. Heredity 9 53–68. [Google Scholar]
- Bechsgaard, J. S., V. Castric, X. Vekemans, D. Charlesworth and M. H. Schierup, 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 million years. Mol. Biol. Evol. 23 1741–1750. [DOI] [PubMed] [Google Scholar]
- Beck, J. B., I. A. Al-Shehbaz and B. A. Schaal, 2006. Leavenworthia (Brassicaceae) revisited: testing classic systematic and mating system hypotheses. Syst. Bot. 31 151–159. [Google Scholar]
- Beilstein, M. A., I. A. Al-Shehbaz and E. A. Kellogg, 2006. Brassicaceae phylogeny and trichome evolution. Am. J. Bot. 93 607–619. [DOI] [PubMed] [Google Scholar]
- Boyes, D. C., and J. B. Nasrallah, 1993. Physical linkage of the SLG and SRK genes at the self-incompatibility locus of Brassica oleracea. Mol. Gen. Genet. 236 369–373. [DOI] [PubMed] [Google Scholar]
- Busch, J. W., 2005. a The evolution of self-compatibility in geographically peripheral populations of Leavenworthia alabamica (Brassicaceae). Am. J. Bot. 92 1503–1512. [DOI] [PubMed] [Google Scholar]
- Busch, J. W., 2005. b Inbreeding depression in self-incompatible and self-compatible populations of Leavenworthia alabamica. Heredity 94 159–165. [DOI] [PubMed] [Google Scholar]
- Busch, J. W., and D. J. Schoen, 2008. The evolution of self-incompatibility when mates are limiting. Trends Plant Sci. 13 128–136. [DOI] [PubMed] [Google Scholar]
- Castric, V., and X. Vekemans, 2004. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Mol. Ecol. 13 2873–2889. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., P. Awadalla, B. K. Mable and M. H. Schierup, 2000. Population-level studies of multiallelic self-incompatibility, with particular reference to Brassicaceae. Ann. Bot. 85 227–239. [Google Scholar]
- Charlesworth, D., C. Bartolomé, M. H. Schierup and B. K. Mable, 2003. a Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol. Biol. Evol. 20 1741–1753. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., B. K. Mable, M. H. Schierup, C. Bartolomé and P. Awadalla, 2003. b Diversity and linkage of genes in the self-incompatibility gene family in Arabidopsis lyrata. Genetics 164 1519–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., E. Kamau, J. Hagenblad and C. L. Tang, 2006. Trans-specificity at loci near the self-incompatibility loci in Arabidopsis. Genetics 172 2699–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington, C. D., and K. Mather, 1949. The Elements of Genetics. Allen & Unwin, London.
- Darwin, C., 1876. The Effects of Cross and Self-fertilisation in the Vegetable Kingdom. John Murray, London.
- Fobis-Loisy, I., C. Meige and T. Gaude, 2004. Molecular evolution of the S locus controlling mating in the Brassicaceae. Plant Biol. 6 109–118. [DOI] [PubMed] [Google Scholar]
- Fujimoto, R., T. Sugimura and T. Nishio, 2006. Gene conversion from SLG to SRK resulting in self-compatibility in Brassica rapa. FEBS Lett. 580 425–430. [DOI] [PubMed] [Google Scholar]
- Glémin, S., E. Bazin and D. Charlesworth, 2006. Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc. Biol. Sci. 273 3011–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad, J., J. Bechsgaard and D. Charlesworth, 2006. Linkage disequilibrium between incompatibility locus region genes in the plant Arabidopsis lyrata. Genetics 173 1057–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinata, K., M. Watanabe, S. Yamakawa, Y. Satta and A. Isogai, 1995. Evolutionary aspects of the S-related genes of the Brassica self-incompatibility system: synonymous and nonsynonymous base substitutions. Genetics 140 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscock, S. J., and D. A. Tabah, 2003. The different mechanisms of sporophytic self-incompatibility. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic, B., and J. R. Kohn, 2001. Evolutionary relationships among self-incompatibility RNases. Proc. Natl. Acad. Sci. USA 98 13167–13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic, B., and J. R. Kohn, 2006. The distribution of plant mating systems: study bias against obligately outcrossing species. Evolution 60 1098–1103. [PubMed] [Google Scholar]
- Igic, B., L. Bohs and J. R. Kohn, 2006. Ancient polymorphism reveals unidirectional breeding system shifts. Proc. Natl. Acad. Sci. USA 103 1359–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A., C. R. Schopfer, M. E. Nasrallah and J. B. Nasrallah, 2001. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293 1824–1826. [DOI] [PubMed] [Google Scholar]
- Kamau, E., B. Charlesworth and D. Charlesworth, 2007. Linkage disequilibrium and recombination rate estimates in the self-incompatibility region of Arabidopsis lyrata. Genetics 176 2357–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho, Y. O., and J. Baer, 1968. Observing pollen tubes by means of fluorescence. Euphytica 17 298–302. [Google Scholar]
- Kimura, M., 1980. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111–120. [DOI] [PubMed] [Google Scholar]
- Kumar, V., and M. Trick, 1993. Sequence complexity of the S receptor kinase gene family in Brassica. Mol. Gen. Genet. 241 440–445. [DOI] [PubMed] [Google Scholar]
- Kusaba, M., T. Nishio, Y. Satta, K. Hinata and D. Ockendon, 1997. Striking sequence similarity in inter- and intra-specific comparisons of class I SLG alleles from Brassica oleracea and Brassica campestris: implications for the evolution and recognition mechanism. Proc. Natl. Acad. Sci. USA 94 7673–7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba, M., K. Dwyer, J. Hendershot, J. Vrebalov, J. B. Nasrallah et al., 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13 627–643. [PMC free article] [PubMed] [Google Scholar]
- Lawrence, M. J., 2000. Population genetics of the homomorphic self-incompatibility polymorphisms in flowering plants. Ann. Bot. 85 221–226. [Google Scholar]
- Liu, F., L. Zhang and D. Charlesworth, 1998. Genetic diversity in Leavenworthia populations with different inbreeding levels. Proc. Biol. Sci. 265 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., D. Charlesworth and M. Kreitman, 1999. The effect of mating system differences on nucleotide diversity at the phosphoglucose isomerase locus in the plant genus Leavenworthia. Genetics 151 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., S. Sherman-Broyles, M. E. Nasrallah and J. B. Nasrallah, 2007. A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr. Biol. 17 734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, D. G., 1965. Evolution of self-compatibility and racial differentiation in Leavenworthia (Cruciferae). Contr. Gray Herb. Harv. Univ. 195 3–134. [Google Scholar]
- Lloyd, D. G., 1967. The genetics of self-incompatibility in Leavenworthia crassa Rollins (Cruciferae). Genetica 38 227–242. [Google Scholar]
- Luu, D-T., S. Hugues, E. Passelègue and P. Heizmann, 2001. Evidence for orthologous S-locus related I genes in several genera of Brassicaceae. Mol. Gen. Genet. 264 735–745. [DOI] [PubMed] [Google Scholar]
- Mable, B. K., M. H. Schierup and D. Charlesworth, 2003. Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata. Heredity 90 422–431. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds, T., 2001. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol. Evol. 16 693–700. [Google Scholar]
- Murase, K. H., H. Shiba, M. Iwano, F. S. Che, M. Watanabe et al., 2004. A membrane-anchored protein kinase involved in Brassica self-incompatibility signalling. Science 303 1516–1519. [DOI] [PubMed] [Google Scholar]
- Nasrallah, M. E., P. Liu and J. B. Nasrallah, 2002. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297 247–249. [DOI] [PubMed] [Google Scholar]
- Nasrallah, M. E., P. Liu, S. Sherman-Broyles, N. A. Boggs and J. B. Nasrallah, 2004. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc. Natl. Acad. Sci. USA 101 16070–16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R., and Z. H. Yang, 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamota, S., Y. Sato, K. Sakamoto and T. Nishio, 2004. Distribution of similar self-incompatibility (S) haplotypes in different genera, Raphanus and Brassica. Sex Plant Reprod. 17 33–39. [Google Scholar]
- Paetsch, M., S. Mayland-Quellhorst and B. Nueffer, 2006. Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity 97 283–290. [DOI] [PubMed] [Google Scholar]
- Prigoda, N. L., A. Nassuth and B. K. Mable, 2005. Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol. Biol. Evol. 22 1609–1620. [DOI] [PubMed] [Google Scholar]
- Richards, A. J., 1986. Plant Breeding Systems. Allen & Unwin, London.
- Richman, A. D., and J. R. Kohn, 2000. Evolutionary genetics of self-incompatibility in the Solanaceae. Plant Mol. Biol. 42 169–179. [PubMed] [Google Scholar]
- Richman, A. D., M. K. Uyenoyama and J. R. Kohn, 1996. Allelic diversity and gene genealogy at the self-incompatibility locus in the Solanaceae. Science 273 1212–1216. [DOI] [PubMed] [Google Scholar]
- Rollins, R. C., 1963. The evolution and systematics of Leavenworthia (Cruciferae). Contr. Gray Herb. Harv. Univ. 192 1–98. [Google Scholar]
- Rozas, J., J. C. Sanchez-Delbarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Sainudiin, R., W. S. W. Wong, K. Yogeeswaran, J. B. Nasrallah, Z. H. Yang et al., 2005. Detecting site-specific physiochemical selective pressures: applications to the class IHLA of the human major histocompatibility complex and the SRK of the plant sporophytic self-incompatibility system. J. Mol. Evol. 60 315–326. [DOI] [PubMed] [Google Scholar]
- Sampson, D. R., 1967. Frequency and distribution of self-incompatibility alleles in Raphanus raphanistrum. Genetics 56 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., X. Vekemans and F. B. Christiansen, 1997. Evolutionary dynamics of sporophytic self-incompatibility alleles in plants. Genetics 147 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., B. K. Mable, P. Awadalla and D. Charlesworth, 2001. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer, C. R., M. E. Nasrallah and J. B. Nasrallah, 1999. The male determinant of self-incompatibility in Brassica. Science 286 1697–1700. [DOI] [PubMed] [Google Scholar]
- Sherman-Broyles, S., N. Boggs, A. Farkas, P. Liu, J. Vrebalov et al., 2007. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, G. L., 1974. Flowering Plants: Evolution Above the Species Level. Harvard University Press, Cambridge, MA.
- Stein, J. C., B. Howlett, D. C. Boyes, M. E. Nasrallah and J. B. Nasrallah, 1991. Molecular cloning of a putative receptor kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 88 8816–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, J. C., R. Dixit, M. E. Nasrallah and J. B. Nasrallah, 1996. SRK, the stigma-specific S locus receptor kinase of Brassica, is targeted to the plasma membrane in transgenic tobacco. Plant Cell 8 429–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, W. L., 1942. Accuracy of mutation rates. J. Genet. 43 301–307. [Google Scholar]
- Suzuki, G., M. Watanabe, N. Kai, N. Matsuda, K. Toriyama et al., 1997. Three members of the S multigene family are linked to the S locus of Brassica. Mol. Gen. Genet. 256 257–264. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., N. Kai, T. Hirose, K. Fukui, T. Nishio et al., 1999. Genomic organization of the S locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 153 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., M. Kusaba, M. Matsuthita, K. Okazaki and T. Nishio, 2000. Characterization of Brassica S-haplotypes lacking S-locus glycoprotein. FEBS Lett. 482 102–108. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2002. PAUP*. Phylogenetic Analysis Using Parsimony, Version 4. Sinauer Associates, Sunderland, MA.
- Takasaki, T., K. Hatakeyama, G. Suzuki, M. Watanabe, A. Isogai et al., 2000. The S receptor kinase determines self-incompatibility in Brassica stigmas. Nature 403 913–916. [DOI] [PubMed] [Google Scholar]
- Takayama, S., H. Shimosato, H. Shiba, M. Funato, F. S. Che et al., 2001. Direct ligand-receptor complex interaction controls Brassica self- incompatibility. Nature 413 534–538. [DOI] [PubMed] [Google Scholar]
- Thompson, K. F., 1957. Self-incompatibility in marrow-stem kale, Brassica oleracea var. acephala. I. Demonstration of a sporophytic system. J. Genet. 55 45–60. [Google Scholar]
- Uyenoyama, M. K., 1995. A generalized least-squares estimate for the origin of sporophytic self-incompatibility. Genetics 139 975–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama, M. K., 2000. Evolutionary dynamics of self-incompatibility alleles in Brassica. Genetics 156 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J. C., 1994. Structure and function of the receptor-like protein-kinases of higher-plants. Plant Mol. Biol. 26 1599–1609. [DOI] [PubMed] [Google Scholar]
- Watanabe, M., T. Takasaki, T. Toriyama, S. Yamakawa, A. Isogai et al., 1994. A high degree of homology exists between the protein encoded by SLG and the S receptor domain encoded by SRK in self-incompatible Brassica campestris L. Plant Cell Physiol. 33 1221–1229. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1939. The distribution of self-sterility alleles in populations. Genetics 24 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., W. S. W. Wong and R. Nielsen, 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22 1107–1118. [DOI] [PubMed] [Google Scholar]