Abstract
The evolution of self-fertilization can mediate pronounced changes in genomes as a by-product of a drastic reduction in effective population size and the concomitant accumulation of slightly deleterious mutations by genetic drift. In the nematode genus Caenorhabditis, a highly selfing lifestyle has evolved twice independently, thus permitting an opportunity to test for the effects of mode of reproduction on patterns of molecular evolution on a genomic scale. Here we contrast rates of nucleotide substitution and codon usage bias among thousands of orthologous groups of genes in six species of Caenorhabditis, including the classic model organism Caenorhabditis elegans. Despite evidence that weak selection on synonymous codon usage is pervasive in the history of all species in this genus, we find little difference among species in the patterns of codon usage bias and in replacement-site substitution. Applying a model of relaxed selection on codon usage to the C. elegans and C. briggsae lineages suggests that self-fertilization is unlikely to have evolved more than ∼4 million years ago, which is less than a quarter of the time since they shared a common ancestor with outcrossing species. We conclude that the profound changes in mating behavior, physiology, and developmental mechanisms that accompanied the transition from an obligately outcrossing to a primarily selfing mode of reproduction evolved in the not-too-distant past.
LONG-term reductions in effective population size (Ne) will result in pronounced shifts in molecular evolution across the genome. Specifically, a reduction in effective population size causes an increased role of genetic drift relative to selection, leading to the fixation of slightly deleterious mutations that would otherwise be eliminated, or kept at low frequency, by purifying selection (Kimura 1968). Provided that sufficient time has elapsed, the accumulation of such deleterious mutations should manifest in coding sequences as an elevated rate of replacement-site substitution (Popadin et al. 2007) and as a decline in codon usage bias (Akashi 1995; Kreitman and Antezana 1999) and eventually genome degeneration (Mira et al. 2001) or even extinction (Lynch and Gabriel 1990). Small population size might also facilitate intron evolution (Lynch and Conery 2003), and insertion/deletion mutational biases will more strongly influence intron size in small populations (Mira et al. 2001). Genomic features that are shaped by the weakest intensities of selection, such as the selection for translational efficiency and/or accuracy that results in biased usage of synonymous codons, should be most sensitive to changes in population size because they are the most susceptible to shifting into an effectively neutral state. However, because it is genetic drift due to relaxed selection that causes these phenomena, they will occur slowly, at a rate on the order of the mutation rate (Marais et al. 2004a). Here, we investigate the potential for differences in population size, mediated by reproductive mode (selfing vs. outcrossing), to have affected genomic patterns of molecular evolution throughout the nematode genus Caenorhabditis.
The onset of a self-fertilizing mode of reproduction is an example of an evolutionary transition that will cause a reduced effective population size compared to the obligately outcrossing ancestor and close relatives (Pollak 1987). This results as a consequence of the direct effects of homozygosity, induced by selfing, and indirectly through reduced effective recombination (Nordborg 2000; Charlesworth and Wright 2001). The strong linkage disequilibrium within selfing species will cause selective sweeps and purifying selection to reduce genetic diversity across large portions of the genome, rather than just at the targets of selection, which further reduces the effective population size (Maynard Smith and Haigh 1974; Charlesworth et al. 1993). Additional reductions in the effective size of selfing populations can occur if they tend to have greater metapopulation dynamics (Pannell and Charlesworth 1999; Pannell 2003); more generally, the generation of population structure is facilitated by inbreeding and leads to lower effective population sizes locally (Charlesworth et al. 1997). The combined action of these forces can result in drastically depressed effective breeding sizes of selfing populations beyond the simple twofold reduction due to elevated homozygosity, with potentially dramatic consequences for genomic patterns of molecular evolution (Charlesworth and Wright 2001; Charlesworth 2003).
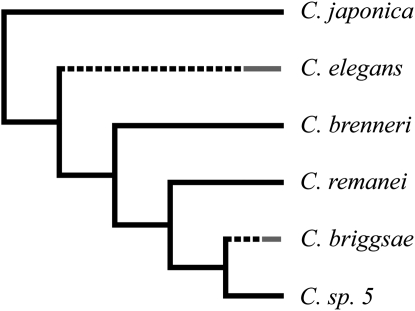
Self-fertilization has arisen independently twice within the nematode genus Caenorhabditis (Kiontke et al. 2004) (Figure 1), but it remains unknown as to how long this mode of reproduction has persisted in these lineages and whether sufficient time has elapsed for the molecular evolutionary consequences of self-fertilization to become apparent. Surveys of population genetic variation show that the selfing species of Caenorhabditis, Caenorhabditis elegans and C. briggsae, do have lower polymorphism levels and effective population sizes than the obligate outcrossing C. remanei (Graustein et al. 2002; Jovelin et al. 2003; Sivasundar and Hey 2003; Barrière and Félix 2005, 2007; Cutter 2006; Cutter et al. 2006a,b), and observed levels of linkage disequilibrium indicate that the effective selfing rate is exceedingly high (Koch et al. 2000; Barrière and Félix 2005; Haber et al. 2005; Cutter 2006; Cutter et al. 2006b). Studies of polymorphism also have determined that weak selection for preferred codons operates in contemporary populations of C. remanei, with the intensity of selection (Nes) estimated to be ∼0.1 on average (Cutter and Charlesworth 2006; Cutter 2008b). For C. elegans, genomic analyses have demonstrated that selection for translational efficiency and/or accuracy has shaped codon usage in its history (Stenico et al. 1994; Duret and Mouchiroud 1999; Duret 2000; Cutter et al. 2006c), although the ∼20-fold smaller effective sizes of present-day C. elegans and C. briggsae populations imply that current selection for preferred codons will effectively be absent. The extent of codon usage bias, the conservation of preferred codon identity, and any association of codon usage bias with reproductive mode is presently uncharacterized in Caenorhabditis generally.
Figure 1.—
Phylogenetic topology of Caenorhabditis that is assumed in calculations of lineage-specific divergence from Kiontke et al. (2004), Kiontke and Sudhaus (2006), and Kiontke et al. (2007). Dashed lines for C. elegans and C. briggsae indicate that self-fertilization (shaded tips) evolved at some point along these branches. Branch lengths are not to scale.
In this study, we test the prediction that selfing species will accumulate slightly deleterious mutations more rapidly than obligately outcrossing relatives in six species of Caenorhabditis. We obtain new coding sequences for three species of Caenorhabditis (C. japonica, C. brenneri, and C. sp. 5 strain JU727) through an expressed sequence tag (EST) effort, yielding 4760 unique nuclear-encoded loci. Using the number of EST hits as a measure of gene expression level, we show that codon bias is stronger in highly expressed genes throughout the genus, identify a set of preferred codons that is conserved across species, and find only modest differences among species in overall biases in codon usage patterns. On the basis of a subset of loci for which orthology could be inferred, we compute lineage-specific substitution rates, but find little evidence for differences in rates of protein evolution among lineages. Application of a model of codon bias evolution suggests that it is unlikely that more than ∼4 million years have elapsed since the onset of relaxed selection on codon usage in C. elegans and C. briggsae and, by implication, the origin of a predominantly self-fertilizing mode of reproduction. We therefore conclude that the novel features involving hermaphroditism and self-fertilization in these species are not ancient in origin.
MATERIALS AND METHODS
Molecular methods:
For each of C. japonica (strain DF5081), C. brenneri (strain CB5161), and C. sp. 5 (strain JU727), cDNA libraries were constructed with the BD Biosciences Clontech SMART cDNA library construction kit as per the manufacturer's instructions. This protocol enriches for full-length cDNAs with complete 5′ ends. Total RNA was extracted from populations of worms of mixed stage and mixed sex with a RNeasy mini kit (QIAGEN, Valencia, CA) from which mRNA was isolated with a QIAGEN Oligotex mRNA mini kit, following manufacturer instructions. The libraries were cloned into the λ-TriplEx2 phage vector and plated with Escherichia coli strain XL1-Blue for blue-white screening of recombinants. Recombinant plaques were picked into buffer, from which an aliquot was used for PCR amplification with primers TW5 (CTCGGGAAGCGCGCCATTGTGTTGGT) and TW3 (AGGCGGCCGACATGTTTTTTTTTTTT), followed by direct sequencing with primer TW5. Sequencing was performed on ABI 3730 automated sequencers by the University of Edinburgh School of Biological Sciences sequencing service and by the University of Arizona Genome and Technology Core sequencing service. EST sequencing was performed on a total of 42 96-well microtitre plates for C. sp. 5, 27 microtitre plates for C. brenneri, and 18 microtitre plates for C. japonica.
EST analysis:
The PartiGene software system (Parkinson et al. 2004a) was used to process the raw EST sequence data to trim low quality sequence, remove contaminating vector sequence, and cluster replicate sequences into a single sequence object. Hereafter, we refer to these clustered sequence objects as “genes,” recognizing that in most cases they do not represent full-length coding sequences. The resulting genes were then passed through prot4EST (Wasmuth and Blaxter 2004), a computational pipeline that infers the most appropriate peptide translation, identifies and masks putative insertion/deletion sequencing errors, and identifies putative mitochondrial vs. nuclear-encoded loci. This procedure generated 2320 genes (3857 ESTs) for C. sp. 5, 1405 genes (2449 ESTs) for C. brenneri, and 1073 genes (1906 ESTs, including 218 already present in dbEST) for C. japonica, including mitochondrial loci. Of these, 4760 genes in total are putatively nuclear encoded. The resulting sequences have been deposited in GenBank (C. japonica, FD512256–FD513938; C. brenneri, FD509784–FD512255; and C. sp. 5, FD513939–FD517806) and added to Nembase (Parkinson et al. 2004b).
For the sequenced genomes, we obtained EST sequences from dbEST for C. elegans (346,064), C. remanei (28,205), and C. briggsae (2517). Using a custom Perl script, we associated these ESTs to corresponding predicted coding sequences from WormBase WS170. After applying the restriction that the BLAST alignment must match at least half the length of the EST with ≥90% identity, we associated 280,966 ESTs with 15,207 genes for C. elegans, 15,868 ESTs with 5603 genes for C. remanei, and 1798 ESTs with 765 genes for C. briggsae. Although predefined associations between EST and coding sequence are publicly available for C. elegans, we used the EST hit counts from this procedure to ensure uniform criteria for associating ESTs with genes among these three species.
Analysis of codon usage:
We used the species-specific EST counts for each locus as a measure of the expression level for the corresponding locus in each species, for nuclear-encoded loci only (supplemental Data 1). Following Duret and Mouchiroud (1999) and Cutter et al. (2006c), we partitioned the 26,390 loci described above into two classes: those that had a single EST “hit” and those with EST “hits” numbering greater than or equal to the 90th percentile (n90). Loci in these categories are taken to represent genes with low and high levels of expression. For C. japonica, C. brenneri, and C. sp. 5, n90 = 3. We also associated ESTs in dbEST with the predicted coding sequences of C. elegans, C. remanei, and C. briggsae (see above), and partitioned them into two categories as for the other species, with n90 = 37, 5, and 4, respectively. We then calculated the relative synonymous codon usage (RSCU) separately for the loci in the two expression categories, which for codon j of amino acid i is simply 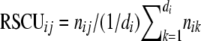 , where d is the degeneracy of the corresponding amino acid and nij is codon frequency.
, where d is the degeneracy of the corresponding amino acid and nij is codon frequency.
To identify those codons that are disproportionately represented in highly expressed genes, termed preferred or optimal codons, we used t-tests to test for significant differences in RSCU between high- and low-expression loci (ΔRSCU) using JMP v.5.01. For this analysis, we restricted the data set to those loci with ≥100 codons. Codons experiencing significantly elevated RSCU in highly expressed genes (optimal codons) were subsequently used to calculate the frequency of optimal codons (Fop) codon usage bias statistic (Ikemura 1985) with a customized optimal codon table in CodonW (J. Peden, http://codonw.sourceforge.net). The effective number of codons (ENC) (Wright 1990) and base composition at fourfold degenerate sites (GC3s) were also calculated for each locus with CodonW and INCA 2.1 (Supek and Vlahovicek 2004). On the basis of these summary statistics, we constructed an ANOVA model to explain variation in codon bias (Fop) as a function of base composition (GC3s), length (log-transformed), and EST hit counts (log-transformed), and their first-order interactions using JMP. A significant positive effect of gene expression (EST hit counts) is consistent with selection having caused codon bias, although this analysis will underestimate the role of selection on codon bias because an association between codon bias and GC3s could result from selection rather than mutational effects. To obtain an overall index of codon usage bias for the species, we report the average of positive ΔRSCU values: 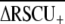 (Cutter et al. 2006c). This metric does not depend strongly on genomic base composition or on the identities of specific optimal codons and therefore represents a useful index for contrasting the strength of selection on codon usage among species (Cutter et al. 2006c), although it is not known how strongly this metric is affected by sample size (and therefore the 90th percentile cutoff level relative to the class of genes with a single EST hit).
(Cutter et al. 2006c). This metric does not depend strongly on genomic base composition or on the identities of specific optimal codons and therefore represents a useful index for contrasting the strength of selection on codon usage among species (Cutter et al. 2006c), although it is not known how strongly this metric is affected by sample size (and therefore the 90th percentile cutoff level relative to the class of genes with a single EST hit).
Divergence:
The coding sequences resulting from the computational procedures applied to ESTs from C. japonica, C. brenneri, and C. sp. 5 were combined with gene predictions for C. elegans, C. briggsae, and C. remanei acquired from WormBase release WS170 to infer putative one-to-one orthologs with OrthoMCL using default parameters (Li et al. 2003). Multiple-sequence alignments of the resulting putative orthologs with ClustalW (Thompson et al. 1994) were followed by calculation of lineage-specific synonymous (dS) and nonsynonymous (dN) site substitution rates with PAML (Yang 1997) assuming the Caenorhabditis phylogenetic topology of Kiontke et al. (2004, 2007) and Kiontke and Sudhaus (2006) (Figure 1), implemented with custom Bioperl-based scripts. Alignments were based on canonical peptide translations and substitution rates used the Goldman–Yang codon-based maximum-likelihood method, permitting branch-specific dN/dS (i.e., model = 1, codon model F3X4) (Yang et al. 1994). Because the EST collections do not fully represent the gene complement of their respective genomes, we eliminated sets of genes for which orthology designation was likely to be inappropriate; specifically, we excluded putative orthologous groups that exhibited excessively high substitution rates along any branch (dN > 0.5 or dS > 5; <3% of genes fit these criteria). This procedure resulted in 63 orthologous groups with representatives in all six species of Caenorhabditis and 1244 orthologous groups with other combinations of three to five species (supplemental Data 2). We exclude from consideration the 6398 orthologous groups specific to the three sequenced genomes (C. elegans, C. briggsae, and C. remanei), because in the six-species context, they provide only lineage-specific data to C. remanei. However, we also followed the above procedure to infer orthologs and calculate divergence for the three sequenced genomes only, excluding the EST-derived sequences, which provided lineage-specific divergence for C. remanei and C. briggsae (7846 orthologous groups) with respect to their common ancestor (using C. elegans as outgroup).
The substitution rate at neutral sites will occur at a rate equal to the mutation rate and independently of population size (Kimura 1968), and therefore can be used to standardize replacement-site substitutions for genomic heterogeneity in mutation rate. Divergence at synonymous sites (dS) is typically used as a neutral reference. However, all of these Caenorhabditis species demonstrate correlations between dS and codon bias (ENC) (Spearman's nonparametric correlations: C. elegans = 0.55, C. brenneri = 0.60, C. remanei = 0.50, C. briggsae = 0.58, C. sp. 5 = 0.51; all P < 0.0001), reflecting selection on codon usage (Sharp and Li 1987). To represent neutral substitution rates, we consequently adjusted the divergence values to correct for selection on codon bias by adding the residuals of least-squares regressions of branch-specific dS on ENC to the value of dS expected at ENC = 61 (where codon bias is not present). These adjusted synonymous-site substitution rates are referred to as dS′, and are used to standardize nonsynonymous substitution rates as dN/dS′. We restrict assessment of substitution rates to those sequences with a Caenorhabditis outgroup, and therefore exclude divergence for the basal C. japonica lineage from consideration (and for C. elegans for the sequenced-genome inference of orthologous groups). This restriction, and the exclusion of loci for which ENC could not be calculated, results in the different sample sizes of genes for each species.
RESULTS
Selection on codon usage:
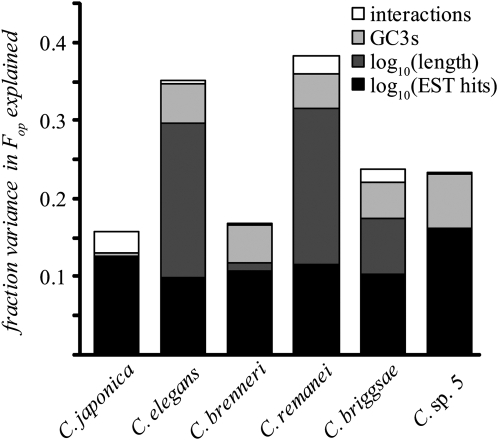
For all six species of Caenorhabditis, we find that codon usage bias is stronger in highly expressed genes (Table 1). In an ANOVA model that explains variation in codon bias (frequency of optimal codons, Fop) (Ikemura 1985) as a function of gene expression (EST hit counts), base composition (GC3s), and sequence length, we show that expression level explains a significant amount of variation in codon bias in each species (Figure 2). These results indicate that selection for translational efficiency and/or accuracy has been a driving force shaping codon usage in the genomes of all Caenorhabditis. This general conclusion is further bolstered by the correlations observed between synonymous site divergence and measures of codon usage bias (see materials and methods), such that loci with stronger codon bias exhibit slower evolution at synonymous sites. The significantly stronger codon bias in short genes that we observe for four of the Caenorhabditis species (Figure 2) is consistent with previous findings for C. elegans (Duret and Mouchiroud 1999) as well as more distantly related nematodes (Cutter et al. 2006c). Intragenic background selection (Loewe and Charlesworth 2007) or Hill–Robertson interference (Comeron and Guthrie 2005) could potentially explain this length effect.
TABLE 1.
Codon usage bias (ENC, Fop, 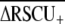 ) and base composition (GC, GC3s) summary statistics
) and base composition (GC, GC3s) summary statistics
| Species | Locia | Mean ENCa | Mean Fopa | Mean GC63b | Mean GC3s63b | Mean ENC63b | Mean 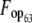 b b
|
Corr[ENC, EST hit count] | 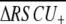 |
|---|---|---|---|---|---|---|---|---|---|
| C. japonica | 1,055 | 47.37 | 0.503 | 0.519 | 0.592 | 40.59 | 0.6617 | −0.350*** | 0.3723 |
| C. elegans | 15,207 | 50.23 | 0.387 | 0.497 | 0.521 | 38.57 | 0.6587 | −0.219*** | 0.3351 |
| C. brenneri | 1,370 | 45.90 | 0.491 | 0.496 | 0.535 | 37.13 | 0.6854 | −0.274*** | 0.4182 |
| C. remanei | 5,603 | 48.62 | 0.432 | 0.497 | 0.529 | 37.35 | 0.6813 | −0.144*** | 0.3643 |
| C. briggsae | 765 | 46.96 | 0.516 | 0.510 | 0.567 | 37.55 | 0.6991 | −0.089*** | 0.3484 |
| C. sp. 5 | 2,295 | 46.27 | 0.508 | 0.512 | 0.583 | 36.70 | 0.7077 | −0.338*** | 0.4977 |
Calculated for loci with ≥1 EST hits.
Calculated for the 63 loci with orthologs in all six species; ***P < 0.0001.
Figure 2.—
Fraction of variance in the frequency of optimal codons (Fop) explained by base composition, gene length, gene expression, and their interactions. Codon usage bias is more extreme in highly expressed genes (Fop × log10[EST hit count] all P < 0.0001) and in shorter genes (Fop × log10[length] P < 0.0001 for C. elegans, C. brenneri, C. remanei, and C. briggsae; P > 0.05 in other species).
Identity of optimal codons:
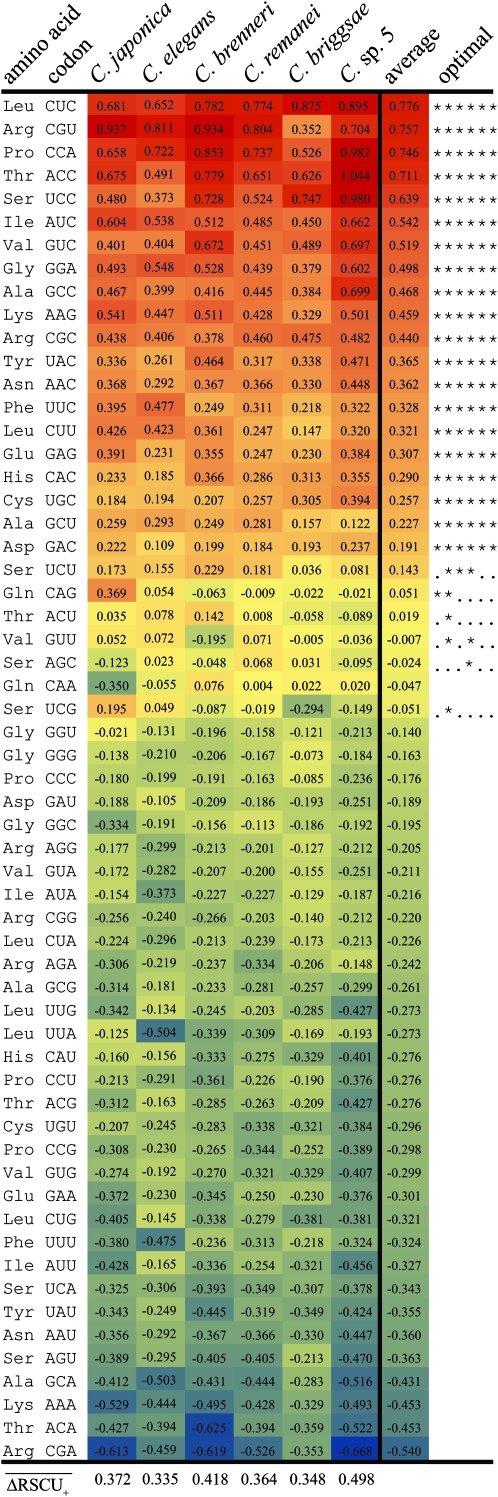
We identified the optimal codon identities for each species on the basis of significant differences in RSCU between highly and lowly expressed loci. Lowly expressed loci are defined as those loci with only a single species-specific EST hit, whereas highly expressed loci have EST hits exceeding the species-specific 90th percentile. This approach identified 20 optimal codons from 17 amino acids that were common to all six species (Figure 3). An additional 6 codons (CAG, UCG, UCU, AGC, ACU, and GUU) from 4 amino acids (glutamine, serine, threonine, and valine) were identified as optimal in some but not all species. It is difficult to discern whether these few variable optimal codons (i) actually are preferred in all taxa, but the current sample is insufficient to define them in some species, (ii) represent gain/loss of optimal codons in some lineages, or (iii) reflect a spurious result. In the case of CAG (Gln) and UCG (Ser), species other than C. japonica and C. elegans show a trend in the opposite direction to what would be expected if this codon were preferred. Consequently, these codons might not truly represent preferred codons or could indicate true losses of preferred codons among the C. brenneri–remanei–briggsae–sp. 5 monophyletic group. Stenico et al. (1994) pointed out that the glutamine CAG codon is used more in highly than lowly expressed genes of C. elegans, despite being underrepresented relative to alternative codons across gene expression classes. Similarly, the threonine ACU codon is significant only for C. elegans, but a positive trend is evident in all species except C. briggsae and C. sp. 5. The valine GUU and serine AGC patterns show no obvious phylogenetic signal, and may be spurious. In contrast, UCU (Ser) is more abundant in highly expressed genes in all taxa, but only significantly so in C. elegans, C. brenneri, and C. remanei. Thus, UCU might represent a preferred codon in all Caenorhabditis, albeit preferred only weakly. tRNA gene copy numbers are characterized only in C. elegans and C. briggsae at present (C. elegans Sequencing Consortium 1998; Stein et al. 2003) (http://lowelab.ucsc.edu/GtRNAdb/); however, the cognate tRNAs do not differ dramatically in abundance between these two species. Because of the ambiguous nature of these six codons and their generally weak effects (Figure 3), we calculate the Fop for individual loci (Ikemura 1985) solely on the basis of the 20 optimal codons that are conserved across Caenorhabditis.
Figure 3.—
Difference in RSCU between highly and lowly expressed genes (ΔRSCU) and inferred optimal codon identity in Caenorhabditis. Codons are sorted by mean ΔRSCU across species. Optimal codons are indicated along the right, with adjacent asterisks indicating statistical significance for different species ordered as for ΔRSCU columns (dots indicate lack of significance for the respective species). Yellow corresponds to ΔRSCU = 0, with shading toward red indicating ΔRSCU > 0 (maximum 1.04) and shading toward blue indicating ΔRSCU < 0 (minimum −0.67).
Codon bias in selfers vs. outcrossers:
We used three approaches to contrast overall codon usage bias among Caenorhabditis species. First, we calculated the average difference in relative synonymous codon usage between highly and lowly expressed genes (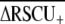 ) across all loci (Cutter et al. 2006c). Second, we calculated the average ENC (Wright 1990) and Fop for the set of 63 orthologous groups of loci that were common to all six taxa (values for the full data set are composed of different sets of loci for each species; Table 1). Because base composition is similar in this group of species (Table 1), it is permissible to contrast mean ENC and Fop across taxa. For all of these metrics, the outcrossing C. sp. 5 exhibits the most extreme value in the direction of strong codon bias (Table 1), although all species of Caenorhabditis demonstrate strong overall codon bias relative to many other nematodes, particularly parasites that have
) across all loci (Cutter et al. 2006c). Second, we calculated the average ENC (Wright 1990) and Fop for the set of 63 orthologous groups of loci that were common to all six taxa (values for the full data set are composed of different sets of loci for each species; Table 1). Because base composition is similar in this group of species (Table 1), it is permissible to contrast mean ENC and Fop across taxa. For all of these metrics, the outcrossing C. sp. 5 exhibits the most extreme value in the direction of strong codon bias (Table 1), although all species of Caenorhabditis demonstrate strong overall codon bias relative to many other nematodes, particularly parasites that have 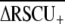 typically ∼0.1 (Cutter et al. 2006c). However, average codon bias did not differ significantly among species (63 orthologous groups, Kruskal–Wallis P > 0.2 for both ENC and Fop). We also tested for a difference in codon bias levels in a pairwise contrast of the >800 orthologs shared between C. briggsae and C. sp. 5 (the two closest relatives). This analysis revealed that codon bias is significantly weaker in the selfing C. briggase (median Fop C. briggsae = 0.461, C. sp. 5 = 0.498, Wilcoxon's P = 0.0022; median ENC C. briggsae = 48.40, C. sp. 5 = 46.67, Wilcoxon's P = 0.0003). Finally, to investigate the correspondence of codon bias in matched orthologous sequences, we performed orthogonal regressions of the frequency of optimal codons for each pair of species for the 63 orthologous groups common to all six species (Figure 4). This third analysis revealed that C. elegans exhibits reduced codon bias relative to most other species of Caenorhabditis, despite the small magnitude of effect. In summary, we find evidence for weaker codon bias in both selfing species, although the magnitude of the reduction is relatively small.
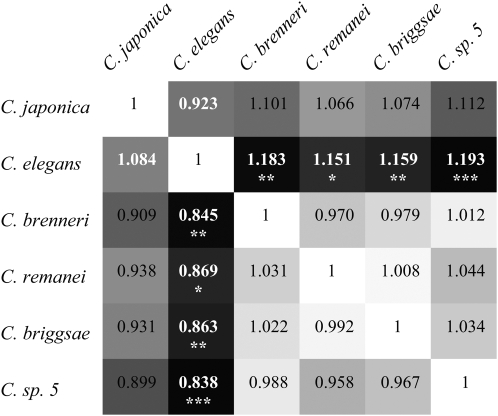
typically ∼0.1 (Cutter et al. 2006c). However, average codon bias did not differ significantly among species (63 orthologous groups, Kruskal–Wallis P > 0.2 for both ENC and Fop). We also tested for a difference in codon bias levels in a pairwise contrast of the >800 orthologs shared between C. briggsae and C. sp. 5 (the two closest relatives). This analysis revealed that codon bias is significantly weaker in the selfing C. briggase (median Fop C. briggsae = 0.461, C. sp. 5 = 0.498, Wilcoxon's P = 0.0022; median ENC C. briggsae = 48.40, C. sp. 5 = 46.67, Wilcoxon's P = 0.0003). Finally, to investigate the correspondence of codon bias in matched orthologous sequences, we performed orthogonal regressions of the frequency of optimal codons for each pair of species for the 63 orthologous groups common to all six species (Figure 4). This third analysis revealed that C. elegans exhibits reduced codon bias relative to most other species of Caenorhabditis, despite the small magnitude of effect. In summary, we find evidence for weaker codon bias in both selfing species, although the magnitude of the reduction is relatively small.
Figure 4.—
Slopes from orthogonal regressions of the frequency of optimal codons (Fop) between pairs of species. Values below the diagonal correspond to species listed along the side as dependent variable; values above the diagonal correspond to species listed along the top as dependent variable. In all comparisons exhibiting a slope >1, the intercept is negative; all comparisons with slope <1 have a positive intercept. The combination of slopes <1 and intercepts >0 for C. elegans as independent variable (second column from left, white text) is indicative of reduced codon bias in this species. Asterisks indicate significant differences from a slope of 1 (*P = 0.017, **P < 0.01, ***P = 0.00023). Darker shades of gray represent slopes with greater deviation from 1.
Modeling changes in codon bias:
Selection for codon bias is very weak, with the selection intensity (γ = 4Nes) estimated to be only ∼0.4 in Caenorhabditis (Cutter and Charlesworth 2006; Cutter 2008b). Consequently, a sharp reduction in effective population size, like that brought on by the evolution of self-fertilization, will result in relaxed selection on codon usage such that its evolution will be governed solely by the neutral processes of mutation and genetic drift. Applying this logic, we draw on the results of Sueoka (1962) and Marais et al. (2004a) to infer the degree of codon bias at time t, pt, following a complete relaxation of selection on codon usage. We begin with the Li–Bulmer (Li 1987; Bulmer 1991) equation for the expected equilibrium frequency of preferred codons in the ancestor that experienced selection for codon bias 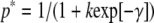 , where k is the ratio of the mutation rate from (u) and to (v) preferred codons and γ is the selection intensity (4Nes). The equilibrium frequency of preferred codons in the absence of selection is simply p** = v/(u + v). Analogous to previous results for the evolution of base composition (Sueoka 1962; Marais et al. 2004a), we find that change in codon bias over time following relaxation of selection can be described by the relation
, where k is the ratio of the mutation rate from (u) and to (v) preferred codons and γ is the selection intensity (4Nes). The equilibrium frequency of preferred codons in the absence of selection is simply p** = v/(u + v). Analogous to previous results for the evolution of base composition (Sueoka 1962; Marais et al. 2004a), we find that change in codon bias over time following relaxation of selection can be described by the relation
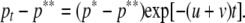 |
(1) |
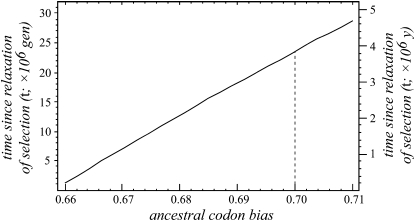
Assuming that C. elegans codon bias has degenerated following the origin of selfing, we can calculate the time over which this process has led to the reduced level of codon bias observed today. We shall focus on the frequency of preferred codons among those 63 highly codon-biased genes that have putative orthologs in all six species of Caenorhabditis, because the change in codon bias should be most apparent for genes with initially high levels of bias. In this case, we shall assume that the ancestral codon bias p* = 0.7 and has degenerated to pt = 0.658 in contemporary C. elegans as a consequence of mutation accumulation (see Fop in Table 1). We take the median frequency of preferred codons in genes with low expression (0.336) as our estimate of p**, which is very close to the value expected under uniform codon usage (20/59 = 0.339) (Stenico et al. 1994). The total per-site single nucleotide mutation rate is estimated to be 9 × 10−9 per generation in C. elegans (Denver et al. 2004). Given that 58.5% of synonymous mutations would result in a change from a preferred to unpreferred codon (or vice versa; so, u + v = 5.26 × 10−9 per site per generation), from our equation describing p**, we can infer that v = 1.89 × 10−9 and u = 3.37 × 10−9. This also assumes that mutational biases have not changed along the lineage. Inserting these values into Equation 1 and numerically solving for t, the model predicts that relaxation of selection upon the origin of selfing began 23.3 × 106 generations ago. Assuming conservatively that C. elegans has a generation time of 60 days in nature, by virtue of spending most of its life in the dauer stage (Barrière and Félix 2005), then this translates to ∼3.8 million years. It is plausible that the generation time in nature is faster than we have assumed, which would imply that relaxation of selection, and selfing, originated more recently (assuming a 14- to 90-day generation time, 8.9 × 105 < t < 5.7 × 106). Likewise, a lower ancestral codon bias would imply a more recent origin than suggested by these calculations (assuming Fop = p* range 0.66–0.71, 1.9 × 105 < t < 4.7 × 106; Figure 5).
Figure 5.—
The predicted time since the onset of relaxed selection on codon usage to yield 65.8% optimal codons, as observed for 63 C. elegans genes that are conserved across the genus, as a function of the ancestral level of codon bias. Provided that the evolution of selfing initiated the relaxed selection, this time is equivalent to the duration of selfing in C. elegans. Timescale in years assumes a 60-day generation time in nature; dashed line demarcates the value for ancestral codon bias assumed in point estimation of t.
Divergence:
We observe no significant differences in median values of replacement-site divergence (dN/dS′) (Table 2) for all genes in all lineages considered together (Kruskal–Wallis P > 0.86) and only for the subset of orthologous groups common to all six species of Caenorhabditis (Kruskal–Wallis P > 0.35). These tests do not correct for common ancestry, which would serve to further reduce the likelihood of inferring heterogeneity in substitution rates. When we restrict analysis to a contrast of orthologs of C. briggsae (selfing) and C. sp. 5 (outcrossing), the two closest relatives, again we find no differences in either dN (C. briggsae median = 0.0254, C. sp. 5 median = 0.0213; Wilcoxon's P = 0.075) or dN/dS′ (Wilcoxon's P > 0.73; Table 2). These results generally accord with the small data set that contrasted substitution rates between inbreeding C. briggsae and outbreeding C. remanei (Cutter and Payseur 2003). We also find that divergence at replacement sites correlates with codon bias (Table 2), such that loci with stronger purifying selection on amino acids also exhibit stronger selection to maintain biased usage of codons, as also reported for Drosophila (Betancourt and Presgraves 2002; Marais et al. 2004b; Bierne and Eyre-Walker 2006).
TABLE 2.
Lineage-specific divergence for orthologous groups among six Caenorhabditis species, with C. japonica as outgroup
| All orthologous groups
|
Six-species orthologous groups
|
||||
|---|---|---|---|---|---|
| Species | Loci | Median dN/dS′ | Corr[dN, ENC] | Loci | Median dN/dS′ |
| C. elegans | 356 | 0.0284 | 0.449*** | 63 | 0.0196 |
| C. brenneri | 430 | 0.0272 | 0.373*** | 63 | 0.0169 |
| C. remanei | 1263 | 0.0291 | 0.348*** | 63 | 0.0152 |
| C. briggsae | 822 | 0.0316 | 0.401*** | 63 | 0.0145 |
| C. sp. 5 | 819 | 0.0292 | 0.363*** | 63 | 0.0153 |
P < 0.0001.
For the much larger set of orthologous groups of genes shared between C. elegans–C. remanei–C. briggsae, we also calculated lineage-specific rates of divergence. For these data, the C. briggsae lineage had a significantly higher rate of nonsynonymous site substitution than C. remanei (Wilcoxon's P < 0.0001 for both dN/dS and dN/dS′; Table 3). Despite sharing a common ancestor at the same point in the past, the C. briggsae lineage also exhibits a higher rate of synonymous site substitution than C. remanei, potentially reflecting more generations per unit time in C. briggsae or possibly a higher mutation rate per generation (Baer et al. 2005)(Table 3). In addition, dN is positively correlated with both dS (Spearman's rank correlations: C. remanei = 0.50, P < 0.0001; C. briggsae = 0.53, P < 0.0001) and dS′ (Spearman's rank correlations: C. remanei = 0.43, P < 0.0001; C. briggsae = 0.45, P < 0.0001). Consequently, it becomes difficult to conclude confidently that the difference in replacement-site divergence between the C. remanei and C. briggsae lineages is best explained by a difference in Ne between these species due to selfing. It is plausible that the C. briggsae lineage experiences an elevated mutation rate (causing higher dS and dS′) that is incompletely accounted for in this data set (due to synonymous site saturation), leading to higher replacement-site divergence in the C. briggsae lineage simply as a by-product. Thus, an unambiguous signature of deleterious mutation accumulation in peptide sequences for the selfing C. briggsae lineage also is not found for this larger set of sequences.
TABLE 3.
Lineage-specific divergence for C. elegans–remanei–briggsae orthologs, with C. elegans as outgroup
| Species | Loci | Median dN | Median dN/dS′ | Median dS | Median dS′ |
|---|---|---|---|---|---|
| C. remanei | 7846 | 0.0330 | 0.0350 | 0.635 | 0.904 |
| C. briggsae | 7846 | 0.0446 | 0.0416 | 0.759 | 1.059 |
DISCUSSION
Molecular evolutionary implications for the evolution of selfing:
These results demonstrate that the demographic history of all Caenorhabditis species has enabled their genomes to respond to even very weak natural selection. This is evidenced by the strong patterns of codon usage bias in highly expressed genes that are indicative of selection for translational efficiency and/or accuracy; such selection is very weak, with a selection intensity (Nes) estimated to be ∼0.1 in C. remanei (Cutter and Charlesworth 2006; Cutter 2008b). Therefore, selection on codon usage is expected to be relaxed completely in the present day for those species that reproduce by self-fertilization, by virtue of their ∼20-fold lower effective population sizes (Graustein et al. 2002; Jovelin et al. 2003; Cutter et al. 2006a). However, we report that the overall levels of codon bias are not much reduced in the selfing species, implying that little time has elapsed to permit the accumulation of slightly deleterious mutations in the form of unpreferred codons.
Consistent with the conclusion that the selfing mode of reproduction is not ancient, we also find little evidence for differences in rates of substitution at replacement sites within coding sequences for orthologous groups of genes in these six species of Caenorhabditis. In the large data set of orthologs defined for the three sequenced genomes (C. elegans, C. remanei, and C. briggsae), we do observe an elevation in the rate of replacement substitutions in the selfing C. briggsae lineage. However, the synonymous substitution rate is also higher, perhaps as a consequence of a more rapid turnover of generations in C. briggsae and its recent ancestors or to a higher mutation rate per generation (Baer et al. 2005). Due to the high absolute levels of synonymous site divergence (saturation is observed in pairwise comparisons) and the strong correlation between synonymous and replacement divergence, it is possible that the observed differences in dN and dN/dS′ are due to a more rapid pace of mutation per year rather than to a change in population size coincident with the origin of selfing in C. briggsae's ancestry. The contrast of C. briggsae with the more closely related C. sp. 5 is the more informative comparison, for which no significant difference in replacement-site divergence was detected. It is worth noting that if selection coefficients on replacement sites are sufficiently large, then even a significant reduction in effective population size due to selfing might not lead them to be effectively neutral. However, given a broad distribution of selection coefficients, as observed in Drosophila (Piganeau and Eyre-Walker 2003; Loewe and Charlesworth 2006; Loewe et al. 2006), we would nonetheless expect a reasonable fraction of replacement sites to become effectively neutral with a 20-fold smaller effective population size.
Using a model that describes the change in codon bias over time due to relaxed selection, we estimate that selfing likely evolved in the C. elegans lineage within the last ∼4 million years, assuming coincident onset of relaxed selection with the evolution of selfing. If a divergence time of ∼18 million years separating C. elegans from its closest relatives is roughly correct (Cutter 2008a), then this implies that the lineage leading to C. elegans spent <25% of the time since the common ancestor in a selfing state. These calculations could overestimate the date for the origin of selfing for two reasons. First, if codon bias were not as strong in the outcrossing ancestor of C. elegans as we assume, which is plausible because the outgroup C. japonica also exhibits nominally weaker codon bias than the monophyletic group that is sister to C. elegans (Table 1; Figure 1), then less time would be required to account for the smaller resultant change in codon bias to the present day. For example, taking the level of codon bias in C. japonica as the ancestral state for C. elegans, we would infer an origin of selfing of only ∼2.1 × 106 generations or ∼347 thousand years ago (given an average 60-day generation time in nature). Second, if C. elegans's generation time is more rapid than the conservative rate used in our calculations, then a more recent origin of selfing would be inferred. The lack of difference in nonsynonymous substitution rates among species also is consistent with the conclusion that low population sizes in the selfing taxa have not persisted for very long. C. briggsae demonstrates very little reduction in codon bias relative to its sister species, C. sp. 5, suggesting that selfing might have evolved even more recently in C. briggsae's ancestry than for C. elegans. These findings imply that the evolution of hermaphrodite mating behavior (Chasnov and Chow 2002; Garcia et al. 2007; Kleemann and Basolo 2007), chemo-attraction (Chasnov et al. 2007), sperm production (Cutter 2004; Geldziler et al. 2006), and developmental mechanisms (Hill et al. 2006) likely do not have ancient origins.
Additional circumstantial support for the notion of a recent origin of selfing in Caenorhabditis comes from the lack of hermaphrodite-only clades comprising multiple species (Kiontke et al. 2004, 2007), although further sampling of biodiversity in this genus could refute this line of reasoning. Selfing hermaphroditism appears to arise regularly in Rhabditid nematodes, but occurs primarily among tip-taxa (Kiontke and Fitch 2005), suggesting that extinction of hermaphroditic lineages likely outpaces speciation. In addition, to the extent that relaxed selection on male function and hermaphrodite attractiveness to males is occurring in C. elegans and C. briggsae, or active selection against these traits, insufficient time has elapsed for the complete loss of a functional male phenotype. According to the model of male maintenance by Chasnov and Chow (2002) (Equation 13), it would seem that too many male-specific genes occur in the C. elegans genome to permit male maintenance via a balance between degeneration and mating (see also Cutter et al. 2003). Such a balance could maintain males if there were fewer than ∼62 male genes, given an average per gene deleterious mutation rate each generation of 0.48/20,000 = 2.4 × 10−5 (Denver et al. 2004), an average X chromosome nondisjunction rate of 0.0033 per generation (Teotonio et al. 2006), and an average male mating-ability parameter of 0.18 (Teotonio et al. 2006)—yet there are likely >100 male genes in the genome (Kim et al. 2001; Reinke et al. 2004; Cutter and Ward 2005). Consequently, this suggests that male genes are expected to degenerate, which is consistent with phenotypic observations of strains in nature with genetically sterile males (Hodgkin and Doniach 1997; Lints and Emmons 2002) and generally poor male C. elegans mating ability relative to gonochoristic males (Chasnov and Chow 2002). The lack of significantly elevated rates of sequence evolution in male genes (Cutter and Ward 2005) therefore is consistent with a relatively recent origin of the extreme form of selfing currently inferred for C. elegans.
Contrasts with other taxa:
How does codon usage bias in Caenorhabditis compare with other eukaryotes? Differences in background base composition make it infeasible to use average ENC or Fop as a general comparative tool, whereas the 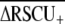 statistic (the average difference in RSCU between highly and lowly expressed genes) provides a useful index for comparison among species with very different GC content (Cutter et al. 2006c). The consistently strong selection-mediated codon bias among Caenorhabditis species is consistent with other free-living nematodes and contrasts with the limited role for selection causing codon bias in parasitic nematodes (Cutter et al. 2006c). For more distant relatives, a reanalysis of Duret and Mouchiroud's (1999) data to calculate
statistic (the average difference in RSCU between highly and lowly expressed genes) provides a useful index for comparison among species with very different GC content (Cutter et al. 2006c). The consistently strong selection-mediated codon bias among Caenorhabditis species is consistent with other free-living nematodes and contrasts with the limited role for selection causing codon bias in parasitic nematodes (Cutter et al. 2006c). For more distant relatives, a reanalysis of Duret and Mouchiroud's (1999) data to calculate 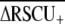 for short proteins indicates that differential selection on codon usage among genes with high vs. low expression has been substantially stronger in C. elegans history (
for short proteins indicates that differential selection on codon usage among genes with high vs. low expression has been substantially stronger in C. elegans history (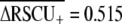 ) than for Drosophila melanogaster (
) than for Drosophila melanogaster (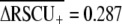 ) or Arabidopsis thaliana (
) or Arabidopsis thaliana (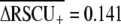 ). The different value for C. elegans
). The different value for C. elegans 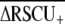 than reported here (0.335; Table 1) stems from their data set containing only short proteins, which experience stronger codon bias and a different definition of highly and lowly expressed genes. Kanaya et al. (2001) also showed that C. elegans exhibits stronger selection on codon usage than D. melanogaster, Xenopus laevis, and human. The substantially higher average frequency of optimal codons observed in D. melanogaster than C. elegans (Duret and Mouchiroud 1999) likely reflects the greater overall GC content of the fly genome relative to the worm, rather than differences in selection on codon usage, because optimal codons tend to be GC rich. However, polymorphism-based inference of codon bias suggests stronger selection intensities for species of Drosophila than Caenorhabditis (reviewed in Cutter and Charlesworth 2006), indicating that further work is necessary to elucidate the conflicting implications of population polymorphism and genomic trends in codon usage for these groups.
than reported here (0.335; Table 1) stems from their data set containing only short proteins, which experience stronger codon bias and a different definition of highly and lowly expressed genes. Kanaya et al. (2001) also showed that C. elegans exhibits stronger selection on codon usage than D. melanogaster, Xenopus laevis, and human. The substantially higher average frequency of optimal codons observed in D. melanogaster than C. elegans (Duret and Mouchiroud 1999) likely reflects the greater overall GC content of the fly genome relative to the worm, rather than differences in selection on codon usage, because optimal codons tend to be GC rich. However, polymorphism-based inference of codon bias suggests stronger selection intensities for species of Drosophila than Caenorhabditis (reviewed in Cutter and Charlesworth 2006), indicating that further work is necessary to elucidate the conflicting implications of population polymorphism and genomic trends in codon usage for these groups.
Breeding system variation analogous to that in Caenorhabditis is found within the plant genus Arabidopsis. Like the results reported here, no differences in substitution rates or codon bias have been observed between the inbreeding A. thaliana and outbreeding A. lyrata (Wright et al. 2002). Overall codon usage bias is much weaker in Arabidopsis than in Caenorhabditis (see above), so an effect of selfing would be more difficult to detect in Arabidopsis because codon usage will be closer to the no-selection equilibrium levels. Furthermore, a recent origin of selfing in A. thaliana (Charlesworth and Wright 2001) or population bottlenecks in A. lyrata might also obscure a detectable effect of selfing on the expected patterns of molecular evolution (Wright et al. 2002, 2003). Analyses of the self-incompatibility locus in Arabidopsis on the basis of patterns of diversity (Shimizu et al. 2004) and linkage disequilibrium (Tang et al. 2007) implicate an origin of selfing of ≤1 million generations ago. This contrasts with the broader plausible range for C. elegans of 0.32 × 106–23.3 × 106 generations ago, on the basis of patterns of nucleotide diversity (Cutter 2006) or the decay of codon bias.
Effects of selfing on other genomic features:
The most straightforward prediction of the smaller effective population sizes induced by selfing is that nucleotide diversity will be reduced genomewide. This prediction is clearly met in Caenorhabditis (Graustein et al. 2002; Jovelin et al. 2003; Cutter et al. 2006a).
We have focused on the potential for accumulation of slightly deleterious mutations in orthologous coding sequences. However, relaxed selection might also impact multigene families disproportionately in selfing lineages, through patterns of divergence and pseudogene production. Such an effect would be important for understanding whether accelerated rates of protein evolution in some gene families result from positive selection or relaxed selection (Stewart et al. 2005; Thomas et al. 2005), making sure to account for past selection on synonymous sites when standardizing by synonymous site divergence.
In addition, the rapid generation of homozygosity produced by self-fertilization might facilitate more rapid fixation of genomic rearrangements, if heterozygous fitness is most strongly affected (Charlesworth 1992). It will be important to determine whether the extensive intrachromosomal rearrangements observed between C. elegans and C. briggsae (Coghlan and Wolfe 2002; Hillier et al. 2007) is a general feature of the genus or whether the rates of translocation, inversion, and transposition are elevated specifically in the selfing lineages.
Models of transposable element dynamics in selfers vs. outcrossers depend critically on the relative importance of deleterious insertion vs. ectopic exchange (unequal crossing-over caused by repetitive elements) as selective agents against transposable element proliferation (Nuzhdin 1999; Morgan 2001). A dominant effect of deleterious insertion could cause mobile elements to be eliminated completely from highly selfing lineages. Consequently, if it is generally true that outcrossing Caenorhabditis have higher mobile element loads than the selfing species, as appears to be the case for retroelements in C. remanei (Z. Bao and N. Jiang, personal communication), then it might support a primary role for the deleterious effects of element insertion in controlling copy numbers, in contrast to the prevailing view for Drosophila (Petrov et al. 2003). The greater abundance of transposable elements in the genome of C. briggsae than in C. elegans (Stein et al. 2003) therefore might be a consequence of a more recent origin of selfing in C. briggsae. It is also important to note that differences among species in the effectiveness of an RNA-interference response to limit transposition could play a fundamental role in transposable element activity and genomic copy number (Sijen and Plasterk 2003).
Conclusions:
Transitions in breeding system from obligately outcrossing to predominantly self-fertilizing will shift genomic patterns of molecular evolution, through the accumulation of slightly deleterious mutations by genetic drift due to the concomitant reduction in effective population size. However, the extent of change resulting from relaxed selection depends on the mutation rate and the time since the origin of selfing and will be more apparent for genomic features that are more susceptible to becoming selectively neutral, such as codon usage bias. We have shown that Caenorhabditis genomes have been shaped by weak selection in their history, yet selfing species do not differ greatly from outcrossing ones in terms of the magnitude of codon bias or replacement-site substitution rate. As a result, a model of relaxed selection suggests that it is unlikely that selfing evolved much longer than ∼4 million years ago. We therefore conclude that evolution over a modest timescale resulted in the novel features of hermaphrodites relative to females, in such traits as mate searching and avoidance (Lipton et al. 2004; Kleemann and Basolo 2007), activity during mating (Garcia et al. 2007), loss of pheromone attraction (Chasnov et al. 2007), self-sperm production, activation, and numerical optimization (Cutter 2004; Geldziler et al. 2006), and the sex-determination pathway (Hill et al. 2006). Additional consequences of selfing for other aspects of genome architecture, such as chromosomal rearrangements, intron size, and gene family and transposable element dynamics, will be much informed by comparisons among the ongoing genome sequencing projects in this group.
Acknowledgments
We thank M. A. Felix and K. Kiontke for sharing strains used in EST sequencing, and we are grateful to B. Charlesworth for discussing the model of codon bias evolution. The manuscript was improved by the comments of several anonymous reviewers. This research was supported by grants to A.D.C. from the National Science Foundation (Doctoral Dissertation Improvement grant and International Research Fellowship Program grant 0401897) and by startup funds from the Department of Ecology and Evolutionary Biology at the University of Toronto.
References
- Akashi, H., 1995. Inferring weak selection from patterns of polymorphism and divergence at silent sites in Drosophila DNA. Genetics 139 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer, C. F., F. Shaw, C. Steding, M. Baumgartner, A. Hawkins et al., 2005. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc. Natl. Acad. Sci. USA 102 5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière, A., and M. A. Félix, 2005. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 15 1176–1184. [DOI] [PubMed] [Google Scholar]
- Barrière, A., and M. A. Félix, 2007. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics 176 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt, A. J., and D. C. Presgraves, 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99 13616–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne, N., and A. Eyre-Walker, 2006. Variation in synonymous codon use and DNA polymorphism within the Drosophila genome. J. Evol. Biol. 19 1–11. [DOI] [PubMed] [Google Scholar]
- Bulmer, M., 1991. The selection-mutation-drift theory of synonymous codon usage. Genetics 129 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282 2012–2018. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1992. Evolutionary rates in partially self-fertilizing species. Am. Nat. 140 126–148. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., M. Nordborg and D. Charlesworth, 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70 155–174. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., 2003. Effects of inbreeding on the genetic diversity of populations. Phil. Trans. R. Soc. Lond. B Biol. Sci. 358 1051–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., and S. I. Wright, 2001. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 11 685–690. [DOI] [PubMed] [Google Scholar]
- Chasnov, J. R., and K. L. Chow, 2002. Why are there males in the hermaphroditic species Caenorhabditis elegans? Genetics 160 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasnov, J. R., W. K. So, C. M. Chan and K. L. Chow, 2007. The species, sex, and stage specificity of a Caenorhabditis sex pheromone. Proc. Natl. Acad. Sci. USA 104 6730–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan, A., and K. H. Wolfe, 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 12 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J. M., and T. B. Guthrie, 2005. Intragenic Hill-Robertson interference influences selection intensity on synonymous mutations in Drosophila. Mol. Biol. Evol. 22 2519–2530. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., 2004. Sperm-limited fecundity in nematodes: How many sperm are enough? Evolution 58 651–655. [PubMed] [Google Scholar]
- Cutter, A. D., 2006. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics 172 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., 2008. a Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 25 778–786. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., 2008. b Multilocus patterns of polymorphism and selection across the X-chromosome of Caenorhabditis remanei. Genetics 178 1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., and B. Charlesworth, 2006. Selection intensity on preferred codons correlates with overall codon usage bias in Caenorhabditis remanei. Curr. Biol. 16 2053–2057. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., and B. A. Payseur, 2003. Rates of deleterious mutation and the evolution of sex in Caenorhabditis. J. Evol. Biol. 16 812–822. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., and S. Ward, 2005. Sexual and temporal dynamics of molecular evolution in C. elegans development. Mol. Biol. Evol. 22 178–188. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., L. Avilés and S. Ward, 2003. The proximate determinants of sex ratio in C. elegans populations. Genet. Res. 81 91–102. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., S. E. Baird and D. Charlesworth, 2006. a High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics 174 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., M. A. Felix, A. Barriere and D. Charlesworth, 2006. b Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., J. Wasmuth and M. L. Blaxter, 2006. c The evolution of biased codon and amino acid usage in nematode genomes. Mol. Biol. Evol. 23 2303–2315. [DOI] [PubMed] [Google Scholar]
- Denver, D. R., K. Morris, M. Lynch and W. K. Thomas, 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430 679–682. [DOI] [PubMed] [Google Scholar]
- Duret, L., 2000. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16 287–289. [DOI] [PubMed] [Google Scholar]
- Duret, L., and D. Mouchiroud, 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 96 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, L. R., B. LeBoeuf and P. Koo, 2007. Diversity in mating behavior of hermaphroditic and male-female Caenorhabditis nematodes. Genetics 175 1761–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldziler, B., I. Chatterjee, P. Kadandale, E. Putiri, R. Patel et al., 2006. A comparative study of sperm morphology, cytology and activation in Caenorhabditis elegans, Caenorhabditis remanei and Caenorhabditis briggsae. Dev. Genes Evol. 216 198–208. [DOI] [PubMed] [Google Scholar]
- Graustein, A., J. M. Gaspar, J. R. Walters and M. F. Palopoli, 2002. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics 161 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, M., M. Schungel, A. Putz, S. Muller, B. Hasert et al., 2005. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol. Biol. Evol. 22 160–173. [DOI] [PubMed] [Google Scholar]
- Hill, R. C., C. Egydio de Carvalho, J. Salogiannis, B. Schlager, D. Pilgrim et al., 2006. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev. Cell 10 531–538. [DOI] [PubMed] [Google Scholar]
- Hillier, L. W., R. D. Miller, S. E. Baird, A. Chinwalla, L. A. Fulton et al., 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5 e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., and T. Doniach, 1997. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura, T., 1985. Codon usage and transfer-RNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2 13–34. [DOI] [PubMed] [Google Scholar]
- Jovelin, R., B. C. Ajie and P. C. Phillips, 2003. Molecular evolution and quantitative variation for chemosensory behaviour in the nematode genus Caenorhabditis. Mol. Ecol. 12 1325–1337. [DOI] [PubMed] [Google Scholar]
- Kanaya, S., Y. Yamada, M. Kinouchi, Y. Kudo and T. Ikemura, 2001. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 53 290–298. [DOI] [PubMed] [Google Scholar]
- Kim, S. K., J. Lund, M. Kiraly, K. Duke, M. Jiang et al., 2001. A gene expression map for Caenorhabditis elegans. Science 293 2087–2092. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1968. Evolutionary rate at molecular level. Nature 217 624–626. [DOI] [PubMed] [Google Scholar]
- Kiontke, K., and D. H. A. Fitch, 2005. The phylogenetic relationships of Caenorhabditis and other rhabditids, in WormBook, edited by The C. Elegans Research Community. WormBook, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Kiontke, K., and W. Sudhaus, 2006. Ecology of Caenorhabditis species, in WormBook, edited by D. H. A. Fitch and The C. Elegans Research Community. WormBook, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Kiontke, K., N. P. Gavin, Y. Raynes, C. Roehrig, F. Piano et al., 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke, K., A. Barrière, I. Kolotuev, B. Podbilewicz, R. Sommer et al., 2007. Trends, stasis, and drift in the evolution of nematode vulva development. Curr. Biol. 17 1925–1937. [DOI] [PubMed] [Google Scholar]
- Kleemann, G. A., and A. L. Basolo, 2007. Facultative decrease in mating resistance in hermaphroditic Caenorhabditis elegans with self-sperm depletion. Anim. Behav. 74 1339–1347. [Google Scholar]
- Koch, R., H. G. A. M. van Luenen, M. van der Horst, K. L. Thijssen and R. H. A. Plasterk, 2000. Single nucleotide polymorphisms in wild isolates of Caenorhabditis elegans. Genome Res. 10 1690–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman, M., and M. Antezana, 1999. The population and evolutionary genetics of codon bias, pp. 82–101 in Evolutionary Genetics: From Molecules to Morphology, edited by R. S. Singh and C. B. Krimbas. Cambridge University Press, New York.
- Li, L., C. J. Stoeckert, Jr. and D. S. Roos, 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. H., 1987. Models of nearly neutral mutations with particular implications for nonrandom usage of synonymous codons. J. Mol. Evol. 24 337–345. [DOI] [PubMed] [Google Scholar]
- Lints, R., and S. W. Emmons, 2002. Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 16 2390–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton, J., G. Kleemann, R. Ghosh, R. Lints and S. W. Emmons, 2004. Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. J. Neurosci. 24 7427–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe, L., and B. Charlesworth, 2006. Inferring the distribution of mutational effects on fitness in Drosophila. Biol. Lett. 2 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe, L., and B. Charlesworth, 2007. Background selection in single genes may explain patterns of codon bias. Genetics 175 1381–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe, L., B. Charlesworth, C. Bartolome and V. Noel, 2006. Estimating selection on nonsynonymous mutations. Genetics 172 1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2003. The origins of genome complexity. Science 302 1401–1404. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and W. Gabriel, 1990. Mutation load and the survival of small populations. Evolution 44 1725–1737. [DOI] [PubMed] [Google Scholar]
- Marais, G., B. Charlesworth and S. I. Wright, 2004. a Recombination and base composition: the case of the highly self-fertilizing plant Arabidopsis thaliana. Genome Biol. 5 R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G., T. Domazet-Loso, D. Tautz and B. Charlesworth, 2004. b Correlated evolution of synonymous and nonsynonymous sites in Drosophila. J. Mol. Evol. 59 771–779. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. Hitch-hiking effect of a favorable gene. Genet. Res. 23 23–35. [PubMed] [Google Scholar]
- Mira, A., H. Ochman and N. A. Moran, 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17 589–596. [DOI] [PubMed] [Google Scholar]
- Morgan, M. T., 2001. Transposable element number in mixed mating populations. Genet. Res. 77 261–275. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., 2000. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin, S. V., 1999. Sure facts, speculations, and open questions about the evolution of transposable element copy number. Genetica 107 129–137. [PubMed] [Google Scholar]
- Pannell, J. R., 2003. Coalescence in a metapopulation with recurrent local extinction and recolonization. Evolution 57 949–961. [DOI] [PubMed] [Google Scholar]
- Pannell, J. R., and B. Charlesworth, 1999. Neutral genetic diversity in a metapopulation with recurrent local extinction and recolonization. Evolution 53 664–676. [DOI] [PubMed] [Google Scholar]
- Parkinson, J., A. Anthony, J. Wasmuth, R. Schmid, A. Hedley et al., 2004. a PartiGene - constructing partial genomes. Bioinformatics 20 1398–1404. [DOI] [PubMed] [Google Scholar]
- Parkinson, J., C. Whitton, R. Schmid, M. Thomson and M. Blaxter, 2004. b NEMBASE: a resource for parasitic nematode ESTs. Nucleic Acids Res. 32 D427–D430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D. A., Y. T. Aminetzach, J. C. Davis, D. Bensasson and A. E. Hirsh, 2003. Size matters: non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol. Biol. Evol. 20 880–892. [DOI] [PubMed] [Google Scholar]
- Piganeau, G., and A. Eyre-Walker, 2003. Estimating the distribution of fitness effects from DNA sequence data: implications for the molecular clock. Proc. Natl. Acad. Sci. USA 100 10335–10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak, E., 1987. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics 117 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadin, K., L. V. Polishchuk, L. Mamirova, D. Knorre and K. Gunbin, 2007. Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc. Natl. Acad. Sci. USA 104 13390–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, V., I. S. Gil, S. Ward and K. Kazmer, 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 311–323. [DOI] [PubMed] [Google Scholar]
- Sharp, P. M., and W. H. Li, 1987. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol. Biol. Evol. 4 222–230. [DOI] [PubMed] [Google Scholar]
- Shimizu, K. K., J. M. Cork, A. L. Caicedo, C. A. Mays, R. C. Moore et al., 2004. Darwinian selection on a selfing locus. Science 306 2081–2084. [DOI] [PubMed] [Google Scholar]
- Sijen, T., and R. H. Plasterk, 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426 310–314. [DOI] [PubMed] [Google Scholar]
- Sivasundar, A., and J. Hey, 2003. Population genetics of Caenorhabditis elegans: the paradox of low polymorphism in a widespread species. Genetics 163 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L. D., Z. Bao, D. Blasiar, T. Blumenthal, M. R. Brent et al., 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1 166–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenico, M., A. T. Lloyd and P. M. Sharp, 1994. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 22 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, M. R., N. L. Clark, G. Merrihew, E. M. Galloway and J. H. Thomas, 2005. High genetic diversity in the chemoreceptor superfamily of Caenorhabditis elegans. Genetics 169 1985–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka, N., 1962. On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl. Acad. Sci. USA 48 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek, F., and K. Vlahovicek, 2004. INCA: synonymous codon usage analysis and clustering by means of self-organizing map. Bioinformatics 20 2329–2330. [DOI] [PubMed] [Google Scholar]
- Tang, C., C. Toomajian, S. Sherman-Broyles, V. Plagnol, Y. L. Guo et al., 2007. The evolution of selfing in Arabidopsis thaliana. Science 317 1070–1072. [DOI] [PubMed] [Google Scholar]
- Teotonio, H., D. Manoel and P. C. Phillips, 2006. Genetic variation for outcrossing among Caenorhabditis elegans isolates. Evolution 60 1300–1305. [PubMed] [Google Scholar]
- Thomas, J. H., J. L. Kelley, H. M. Robertson, K. Ly and W. J. Swanson, 2005. Adaptive evolution in the SRZ chemoreceptor families of Caenorhabditis elegans and Caenorhabditis briggsae. Proc. Natl. Acad. Sci. USA 102 4476–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth, J., and M. Blaxter, 2004. prot4EST: translating expressed sequence tags from neglected genomes. BMC Bioinformatics 5 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, F., 1990. The effective number of codons used in a gene. Gene 87 23–29. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., B. Lauga and D. Charlesworth, 2002. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 19 1407–1420. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., B. Lauga and D. Charlesworth, 2003. Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Mol. Ecol. 12 1247–1263. [DOI] [PubMed] [Google Scholar]
- Yang, Z. H., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., N. Goldman and A. Friday, 1994. Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol. Biol. Evol. 11 316–324. [DOI] [PubMed] [Google Scholar]