Abstract
The conjecture that the deleterious effects of mutations are amplified by stress or interaction with one another remains unsatisfactorily tested. It is now possible to reapproach this problem systematically by using genomic collections of mutants and applying stress-inducing conditions with a well-recognized impact on metabolism. We measured the maximum growth rate of single- and double-gene deletion strains of yeast in several stress-inducing treatments, including poor nutrients, elevated temperature, high salinity, and the addition of caffeine. The negative impact of deletions on the maximum growth rate was relatively smaller in stressful than in favorable conditions. In both benign and harsh environments, double-deletion strains grew on average slightly faster than expected from a multiplicative model of interaction between single growth effects, indicating positive epistasis for the rate of growth. This translates to even higher positive epistasis for fitness defined as the number of progeny. We conclude that the negative impact of metabolic disturbances, regardless of whether they are of environmental or genetic origin, is absolutely and relatively highest when growth is fastest. The effect of further damages tends to be weaker. This results in an average alleviating effect of interactions between stressful environment and gene deletions and among gene deletions.
RECENT experiments suggest that the genomic rate of spontaneous deleterious mutation is high (Denver et al. 2004; Haag-Liautard et al. 2007). Spontaneous mutagenesis must be countered by purging selection—that is, the enhanced mortality or reduced fecundity of bearers of mutations—or offset by compensatory mutations (Silander et al. 2007). It has been repeatedly proposed that a harsh environment, or stress, is likely to aid selection by imposing demands unbearable for individuals weakened by mutations. This simple and intuitively appealing assumption is supported by the results of some experiments demonstrating that the negative effects of random mutations are higher under adverse physical conditions or severe competition (Kondrashov and Houle 1994; Shabalina et al. 1997; Korona 1999; Vassilieva et al. 2000; Szafraniec et al. 2001; Yang et al. 2001; Fry and Heinsohn 2002). However, not all studies confirm this expectation (Fry et al. 1996; Martin and Lenormand 2006) and an opposite effect has also been described (Kishony and Leibler 2003). Moreover, the reported cases of aggravation of deleterious effects in harsh environments are difficult to interpret. Earlier studies often involved organisms with large and unknown numbers of mutations. It is thus unclear whether stress exposes more mutations or increases their average effects (Szafraniec et al. 2001). Finally, it is possible that environmental stress may promote negative (lowering fitness) genetic interactions among deleterious mutations. This direction of epistasis probably does not dominate under normal environmental conditions (De Visser and Elena 2007; Kouyos et al. 2007; Jasnos and Korona 2007). However, it is unsure whether the average effect of epistasis can change under stress (You and Yin 2002; Kishony and Leibler 2003; Cooper et al. 2005; Killick et al. 2006). In sum, the basic characteristics of deleterious mutations are insufficiently recognized, especially when the changeability of the environment is taken into account. It therefore remains unclear whether the accumulation of deleterious mutations can endanger the existence of populations (Kimura and Maruyama 1966; Crow and Kimura 1979; Schultz and Lynch 1997) and whether the widespread occurrence of genetic recombination and sex is an evolutionary response to this threat (Otto and Lenormand 2002).
We chose the organism, mutations, and environments specifically to overcome or reduce the problems typically met in earlier studies. We used isogenic strains of the budding yeast with none, one, or two gene deletions. The use of gene deletions guaranteed that each introduced alteration meant the complete loss of a protein in all studied environments. Another advantage of the laboratory yeast strains is the wealth of information about genetics and cell physiology provided by traditional work as well as recent genomewide studies of gene function and expression (Scherens and Goffeau 2004). Finally, in most previous studies the molecular basis of stress reaction was poorly known. It was defined mostly in gross terms, that is, as the occurrence of additional energetic costs or simply as a decrease in fitness (Martin and Lenormand 2006). In yeast, it is possible to select environments that are known to elicit extensive, specific, and functionally interpretable reactions of cellular metabolism (Bahn et al. 2007).
The laboratory environment of microbial cultures is determined by the applied nutrients, physical conditions, and additional compounds, such as drugs. In this experiment, one benign and four harsh environments were used. The two most often applied yeast nutritional media are YPD, a rich resource of organic compounds including amino acids and nucleotides, and SD, a synthetic medium containing a minimum set of vitamins, salts, and simple sources of nitrogen and organic carbon (Sherman 2002). In this study, YPD represents the benign environment, whereas SD models nutritional stress. Two model environmental stresses applied in this study are high temperature, 37° instead of the standard 30°, and a high salinity of 1 m NaCl. Faced with stress, the yeast cell reacts universally by remodeling of transcriptional activity, protection of existing proteins, and adjustment of carbohydrate metabolism (Marchler et al. 1993; Martinez-Pastor et al. 1996). High temperature results specifically in massive protein misfolding and ultimately tests the cell's ability to synthesize, maintain, and dispose of proteins with high efficiency (Mager and De Kruijff 1995). High salinity primarily stresses the robustness of the cell wall, cytoskeleton, and vacuolar system (Hampsey 1997; Hohmann 2002). The final environment used in this study is 0.8 mm caffeine. This compound alters the activity of PKA, Tor1, and Pkc1, important regulators of cell metabolism, and influences the stability of chromosomes and the trafficking of proteins through Golgi and vacuole biogenesis (Bianchi et al. 2001; Levin 2005; Kuranda et al. 2006). It likely represents an artificial metabolic stress, one that, in contrast to deprivation of nutrients, heat, or salinity, was not routinely encountered by the yeast in its evolutionary past.
In this experiment we focused on genes whose deletion causes detectable deleterious effects under favorable conditions. We assumed that these genes are likely to provide “housekeeping” activities and therefore be important also in other environments. We therefore asked whether the growth defects detectable under the benign environment are amplified by stress. We found that the relative deleterious effect of single mutations was lower under stress than in the benign environment. The joint fitness effect of two deleterious mutations was smaller than expected from their individual effects in both benign and stressful environments, indicating positive epistasis.
MATERIALS AND METHODS
Strains:
We used a collection of single-gene deletions engineered in the laboratory strain BY (Giaever et al. 2002). In Jasnos and Korona (2007), a list of 639 nonlethal gene deletions was compiled. The sole criterion used in selecting the deletions was the annotation that the deletion generated a detectable growth effect in rich medium. The selection was based on results of genomewide phenotypic screens and traditional genetic studies (Jasnos and Korona 2007). In this study, not all of the originally selected gene deletions were used (see below). The omission of some strains was decided a priori as a result of limited access to instruments, not because of the characteristics of the omitted strains.
Media:
Rich medium was composed of standard YPD (1% yeast extract, 2% peptone, 2% glucose) at 30° and at 37°; YPD with salinity at 1 m NaCl added at 30°; and YPD with 8 mm caffeine added at 30°. Minimal medium was composed of standard SD (2% glucose, 0.6% yeast nitrogen base without amino acids) with uracil (20 mg/liter), histidine (20 mg/liter), methionine (20 mg/liter), leucine (100 mg/liter), and lysine (30 mg/liter) at 30°.
Growth assays:
The growth curve of every progeny strain was assayed independently twice. [In the Jasnos and Korona (2007) study, four replicates in YPD were obtained. Of those, only the first two of the relevant crosses were used in this study when comparisons with new estimates were made.] Growth was measured in an automated workstation, Bioscreen C. Growth curves were transformed with a polynomial function to compensate for the nonlinear relation between population density and the OD readings (Warringer and Blomberg 2003) and then log-normally transformed. Regions of linear relation between the doubly transformed readings and time were defined on the basis of the pilot studies. They were used to calculate regression lines whose slope was equivalent to maximum growth rate (MGR) (Jasnos et al. 2005; Jasnos and Korona 2007). In total, 10,520 individual growth curves were analyzed (263 crosses, each giving four progeny strains, five environments, and two replications).
RESULTS
Growth effects of gene deletions:
It was shown in an earlier study that the functional annotations of gene deletions causing growth defects followed a pattern characteristic for the whole genome; that is, they were roughly representative of complete cellular metabolism (Jasnos and Korona 2007). In half of the deletions, a genetic marker present in the deletion cassette (resistance to geneticin, or kan) was exchanged for another one (resistance to nourseothricin, or nat). Differently marked haploid strains of the opposite mating types were randomly matched, mated, and sporulated. From each cross involving two haploid parental strains with single deletions, a tetrad of recombinant haploid strains was derived: unmarked, kan marked, nat marked, and both kan and nat marked. MGR of the progeny strains was then estimated in a favorable environment of rich broth medium (YPD) at 30° (Jasnos and Korona 2007).
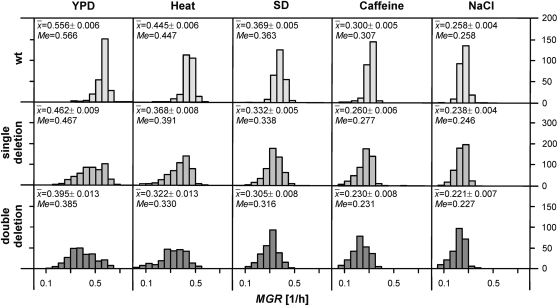
In this experiment, we extended measurements of MGR in four unfavorable environments: minimal medium, high saline, high temperature, and caffeine. In total, we report here 263 random crosses between 526 (263 × 2) parental strains of which 1052 (263 × 4) progeny strains were obtained and assayed for MGR. [We assayed 263 wild-type strains (products of crosses) instead of a single ancestral strain. In this way, we ensured that an average genetic background of the zero-deletion strains was exactly the same as that of the bearers of deletions, including randomly segregating unknown polymorphism.] The new estimates were compared to a relevant sample known from the favorable environment. Figure 1 shows the frequency distributions of MGR for strains with none, one, or two deletions in every environment. The list of gene deletions and estimates of MGR is presented in supplemental Table 1.
Figure 1.—
Frequency distribution of the MGR. Average values with 95% confidence limits and medians are shown. The kan- and nat-marked single deletions are pooled; therefore their samples are twice as numerous as those with none or double deletions.
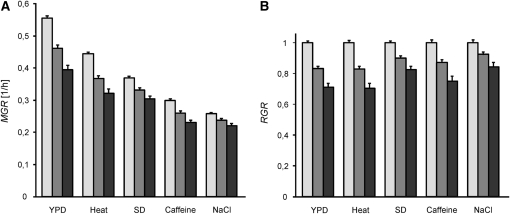
To compare the growth effects of gene deletions in different environments, we converted the absolute values of maximum growth rate to relative growth rates (RGR). Individual MGRs were divided by the average MGR of the 263 strains with no deletion (the progeny wild-type strains). The standardization was done for each environment independently. Figure 2 shows the relation between the average MGR and RGR in all test environments. In general, the average RGR tended to correlate negatively with the average MGR. In other words, the relative damage caused by a gene deletion (1 − RGR) was high when the MGR was high. This means that stress tended to alleviate the negative effect of gene deletion. (Statistical comparisons between pairs of environments are reported below after an ANOVA test.)
Figure 2.—
Average growth rate effects of gene deletions. The average growth rate of strains containing no deletions (open bars), single deletions (shaded bars), and two deletions (solid bars) with 95% confidence limits are shown. Five environments were used: YPD (rich medium), heat (YPD and 37°), SD (minimal synthetic medium), caffeine (8 mm caffeine in YPD), and saline (1 m NaCl in YPD). (A) MGR. (B) RGR.
We then asked whether some deletions tended to be more harmful than others when averaged across all environments. This analysis was restricted to strains bearing single deletions. To be conservative, we excluded deletions that produced the lethal or sublethal phenotype in any of the compared environments (see supplemental Table 1). Results of a two-way ANOVA test carried out for the standardized data, RGRs, are summarized in Table 1. The genetic factor was significant, indicating that some deletions tended to be generally more harmful than others. The negative effect of a deletion was modulated by the environment as evidenced by a statistically significant interaction between gene deletion and environment. The environmental factor was also significant. An a posteriori Tukey's test revealed that there was no difference in the average RGR only between YPD and heat. All other pairs differed significantly, P < 0.0001, yielding four clusters in an increasing order of the average RGR: YPD together with heat, caffeine, SD, and NaCl.
TABLE 1.
ANOVA for RGR of single-deletion strains
| Effect | d.f. | MS | F | P |
|---|---|---|---|---|
| Environment | 4 | 1.5623 | 245.1916 | <0.0001 |
| Deletion | 438 | 0.1692 | 26.5565 | <0.0001 |
| Interaction | 1752 | 0.0398 | 6.2401 | <0.0001 |
| Error | 2205 | 0.0064 |
MS, mean square.
A possible explanation for the observed alleviating effect of stress on the average effect of deleterious mutations could be that our sample is biased because we selected gene deletions that were previously found deleterious in the benign environment. However, if the deleterious effects were specific to YPD, or even antagonistically pleiotropic, the correlation between YPD and other environments should be absent or negative. Contrary to this expectation, the Pearson correlation coefficient between YPD and the stressful 37°, SD, caffeine, and saline was always positive: 0.511, 0.409, 0.335, and 0.328, respectively. (These statistics were highly significant, as the sample sizes were large: 501, 517, 496, and 505, respectively.) This result is compatible with our assumption that deleterious effects of the selected mutations are not specific for YPD but represent general defects of cellular metabolism.
Epistasis for growth rate and fitness:
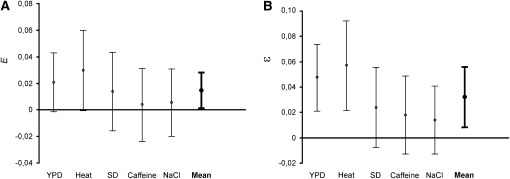
Epistasis is absent when the phenotypic effect of a gene is independent of genes in other loci. Considering deleterious mutations, it is typically assumed that epistasis is absent when the proportional change in a trait caused by an individual mutation is stable, irrespective of the presence of other mutations in the same individual. This leads to a model of multiplicative combination of growth effects caused by gene deletions: RGRwt · RGRkn = RGRk · RGRn, where wt, k, n, and kn denote, respectively, no deletion, kan-marked deletions, nat-marked deletions, and double deletions. Consequently, when growth rate is the considered phenotypic trait, epistasis can be defined as E = RGRwt · RGRkn − RGRk · RGRn. Frequency distributions of E in the five test environments are shown in supplemental Figure 1. Figure 3 shows that the average values of E, although generally small, were positive in every environment. Averaged over all five environments, the positive deviation from zero was statistically significant (Figure 3).
Figure 3.—
Average epistatic effects. The average values and 95% confidence limits of epistasis between two deletions are shown. The confidence limits were calculated using a single (average) value of epistasis for one progeny strain. Sample sizes of the particular environments, beginning from the left, were 257, 238, 254, 235, and 243 strains. The grand mean was calculated as a mean of five environmental means with 4 d.f. (A) Epistasis for growth rate, E. (B) Epistasis for fitness, ɛ.
In population genetics, the status of an individual/clone is identified not by its rate of growth but by its fitness. Fitness, w, is the average number of progeny left by an individual of a given genotype. In a continuously growing population, an equivalent of offspring size is obtained from the expression w = emt, where m is the growth rate and t is generation time (Crow and Kimura 1970). Thus, the growth rate is equal to loge(w) with t set to 1. Consequently, when fitness is the considered phenotypic trait, testing for multiplicity of fitness effects, wwtwnk = wkwn, means testing for the additivity of growth rates, RGRwt + RGRkn = RGRk + RGRn. Epistasis for fitness can be defined as ɛ = (RGRwt + RGRkn) − (RGRk + RGRn) (Jasnos and Korona 2007). Frequency distributions of ɛ in the five environments are shown in supplemental Figure 1. Figure 3 demonstrates that the average ɛ was positive in every environment and significantly >0 when averaged over all environments. Understandably, the values of ɛ were higher than those of E. [The predicted decline in the growth rate of the double mutant is 1 − RGRk − RGRn in the additive model, that is, steeper by RGRkRGRn than in the multiplicative model, (1 − RGRk)(1 − RGRn).]
We applied two-way ANOVA to test how variation in epistasis is modulated by the environmental and genetic (pair of deletions) factors. We excluded crosses that included deletions producing the (sub)lethal phenotype in any of the compared environments. Table 2 reports results for ɛ. All three determinants—environmental, genetic, and the interaction between them—were statistically significant. An analogous analysis for E yielded qualitatively similar results, that is, high significance of all three factors (not shown). Inspection of Figure 3 suggests that the dependence of E and ɛ on environment followed a pattern similar to that observed for the relative effect of a single mutation (1 − RGR). That is, the epistatic effect tended to be higher in environments in which growth was faster, although the differences were not statistically significant.
TABLE 2.
ANOVA for epistasis for fitness, ɛ
| Effect | d.f. | MS | F | P |
|---|---|---|---|---|
| Environment | 4 | 0.1756 | 10.4381 | <0.0001 |
| Deletion | 199 | 0.2650 | 15.7464 | <0.0001 |
| Interaction | 796 | 0.0670 | 4.0391 | <0.0001 |
| Error | 1000 | 0.0168 |
MS, mean square.
DISCUSSION
The interactions of deleterious mutations with the environment and with one another are typically weak, making them difficult to predict, estimate, and interpret (Remold and Lenski 2004; Demuth and Wade 2006; Martin and Lenormand 2006; De Visser and Elena 2007; Kouyos et al. 2007). To circumvent these difficulties, we used a model system of yeast gene deletions assayed in standardized laboratory environments. Our recent (Jasnos and Korona 2007) and present results show a consistent pattern: the interactions of deleterious mutations with a stress-inducing environment and with one another are definitely not of the aggravating (synergistic) type. Instead, consistent bias toward alleviation is seen. We found that the average proportional effect of deletion on growth (1 − RGR) under heat stress was close to that measured in the benign YPD environment. In the nutritionally poorer synthetic medium, and especially in saline and caffeine, the average 1 − RGR was clearly lower than in YPD, indicating that mutations became less deleterious in these media. We deliberately chose environments known to induce different functional reactions to increase the chances of exposing the variability of interactions. However, our results hint that the growth rate itself, rather than the specificities of cellular metabolism, constituted an important determinant of the phenotypic reaction to mutational damage. When growth was increasingly impeded by adverse external conditions, metabolic defects caused by mutations were decreasingly harmful.
A similar alleviation of deleterious effects of mutation by stress was previously described for Escherichia coli. Bacterial cells carried random point mutations, supposedly one per genome, whose negative impact on the rate of growth decreased after the addition of several antibiotics, simulating a decline in environmental quality (Kishony and Leibler 2003). In this experiment, the alleviation of mutational harm was particularly clear-cut not only in caffeine but also in high salt conditions. The latter is a typical natural stressor known to elicit cellular reactions that are highly evolved and common for many organisms (Hohmann 2002). It is thus remarkable that both prokaryotic and eukaryotic cells faced with a variety of environmental stresses and mutational damages show a similar general pattern of gene–environment interactions.
In our experiment, genetic epistasis was positive on average under both favorable and stressful conditions. More specifically, the growth rate of double-deletion strains was higher than predicted by the multiplicative and especially by the additive model of how mutational effects accumulate. Multiplicity and additivity of growth rate effects are both simplified and arbitrarily chosen expectations of how complex metabolic networks should behave. Multiplicity of growth rates is a dominating model in theoretical and empirical studies aiming to uncover specific functional interactions between genes (Tong et al. 2004; Davierwala et al. 2005; Kelley and Ideker 2005; Collins et al. 2006; Ooi et al. 2006; Boone et al. 2007; Collins et al. 2007; St. Onge et al. 2007). The additive model of growth rate effects is special in the sense that it is equivalent to the multiplicity of fitness defined as the number of progeny (Jasnos and Korona 2007). This definition of fitness is standard in models of population genetics, including the theoretical work on the evolution of genetic recombination and sexual reproduction (Otto and Lenormand 2002). Thus, while the multiplicative model is well suited as the null hypothesis in metabolic experiments, the additive model is appropriate when comparisons with multicellular organisms or tests of population genetics theory are to be made. In this study, the null predictions of both models were rejected in favor of positive (antagonistic, alleviating) epistasis. This result, together with the alleviating effect of interaction between environment and single deletions discussed above, fits into a single general pattern. The first impairment inflicted upon the metabolism of a rapidly growing cell is relatively the most harmful irrespective of whether the insult originates from a decline in quality of the external environment or from the deregulation of the internal environment. This leads to speculation that antagonism is likely to be common in interactions among environment, drugs, and mutations. It may be an evolved adaptation to downregulate growth more strongly when it is rapid because this allows the avoidance of excessive overinvestment. Alternatively, the very construction of a complex cellular system makes it most sensitive when operating at its highest rate.
Results of earlier studies often appear incompatible with the model suggested above. In particular, it was found in yeast that the effect of multiple mutations on the growth rate was more deleterious in stressful than in benign environments (Korona 1999; Szafraniec et al. 2001). However, in these studies random mutageneis was applied and therefore the exact number, location, and functional aspects of mutations were not known. Substitutions of amino acids, a common basis of random mutation, are ambiguous in their metabolic effect. They often result in only partial destabilization of protein structures, which does not impair protein function under normal conditions but is critically exacerbated under stress (Pakula and Sauer 1989). Thus, mutations without phenotypic penetration under favorable conditions can emerge as new phenotypic defects under stress (Hampsey 1997; Szafraniec et al. 2001). This does not occur when a known number of null mutations (here gene deletions) is considered. As the proportion of partial and null mutations is often unknown under random mutagenesis, the results of previous experiments are difficult to interpret. Another question is why earlier studies did not unambiguously detect antagonism among deleterious mutations. Experiments involving unicellular microorganisms usually showed that most epistatic effects were close to zero. Notably, in leading examples of such studies, fitness was defined as the relative rate of growth and consequently the multiplicative model of interaction was used (Elena and Lenski 1997; Segre et al. 2005). Redefining the fitness as the number of progeny and applying the additive model would likely shift the results toward positive values. Thus, we believe that the postulated general alleviating effect of interactions between mutations and environment and among mutations are not contradicted by results of previous work.
Evolutionary biologists are concerned with the role of epistasis in the evolution of genetic recombination. The mutational deterministic hypothesis predicts that not only sporadic genetic recombination but also obligatory sexual reproduction would be convincingly explained if deleterious mutations interacted synergistically (Feldman et al. 1980; Kondrashov 1988; but see also Kouyos et al. 2006). Empirical support for this assumption is generally weak if not absent (De Visser and Elena 2007; Kouyos et al. 2007). This study provides further evidence against synergistic epistasis by including stressful environments. Some current theories of sex and recombination reject the assumption of synergistic epistasis and rely on stochastic explanations. Briefly, unfavorable combinations of new mutations and genetic backgrounds tend to be overly frequent even in relatively large populations. Recombination alleviates this effect and therefore its spread and maintenance is advantageous. However, the predicted benefit of genetic recombination is relatively small or even very small (Hill and Robertson 1966). It may be insufficient if the average epistasis for fitness is positive and not exceedingly small (Keightley and Otto 2006). Here, epistasis was relatively high—higher than the difference between the multiplicative and additive combination of log fitness. Thus, positive (antagonistic, alleviating) epistasis is at odds with both deterministic and stochastic models explaining the evolution of recombination and sex (Barton 1995). It is therefore highly desirable to test whether antagonistic epistasis applies to traits unrelated to fast growth and whether it extends to multicellular organisms.
Acknowledgments
This work was supported by a grant from the State Committee for Scientific Research of Poland (0684/P01/2006/30).
References
- Bahn, Y. S., C. Xue, A. Idnurm, J. C. Rutherford, J. Heitman et al., 2007. Sensing the environment: lessons from fungi. Nat. Rev. Microbiol. 5 57–69. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., 1995. A general model for the evolution of recombination. Genet. Res. 65 123–145. [DOI] [PubMed] [Google Scholar]
- Bianchi, M. M., S. Ngo, M. Vandenbol, G. Sartori, A. Morlupi et al., 2001. Large-scale phenotypic analysis reveals identical contributions to cell functions of known and unknown yeast genes. Yeast 18 1397–1412. [DOI] [PubMed] [Google Scholar]
- Boone, C., H. Bussey and B. J. Andrews, 2007. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8 437–449. [DOI] [PubMed] [Google Scholar]
- Collins, S. R., M. Schuldiner, N. J. Krogan and J. S. Weissman, 2006. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 7 R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham et al., 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 806–810. [DOI] [PubMed] [Google Scholar]
- Cooper, T. F., R. E. Lenski and S. F. Elena, 2005. Parasites and mutational load: an experimental test of a pluralistic theory for the evolution of sex. Proc. Biol. Sci. 272 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970. An Introduction to Population Genetics Theory. Harper & Row, New York.
- Crow, J. F., and M. Kimura, 1979. Efficiency of truncation selection. Proc. Natl. Acad. Sci. USA 76 396–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davierwala, A. P., J. Haynes, Z. Li, R. L. Brost, M. D. Robinson et al., 2005. The synthetic genetic interaction spectrum of essential genes. Nat. Genet. 37 1147–1152. [DOI] [PubMed] [Google Scholar]
- Demuth, J. P., and M. J. Wade, 2006. Experimental methods for measuring gene interactions. Annu. Rev. Ecol. Evol. Sys. 37 289–316. [Google Scholar]
- Denver, D. R., K. Morris, M. Lynch and W. K. Thomas, 2004. High mutation rate and predominance of insertions in the C. elegans nuclear genome. Nature 430 679–682. [DOI] [PubMed] [Google Scholar]
- de Visser, J. A., and S. F. Elena, 2007. The evolution of sex: empirical insights into the roles of epistasis and drift. Nat. Rev. Genet. 8 139–149. [DOI] [PubMed] [Google Scholar]
- Elena, S. F., and R. E. Lenski, 1997. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390 395–398. [DOI] [PubMed] [Google Scholar]
- Feldman, M. W., F. B. Christiansen and L. D. Brooks, 1980. Evolution of recombination in a constant environment. Proc. Natl. Acad. Sci. USA 77 4838–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, J. D., and S. L. Heinsohn, 2002. Environment dependence of mutational parameters for viability in Drosophila melanogaster. Genetics 161 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, J. D., S. L. Heinsohn and T. F. C. Mackay, 1996. The contribution of new mutations to genotype-environment interaction for fitness in Drosophila melanogaster. Evolution 50 2316–2327. [DOI] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard, C., M. Dorris, X. Maside, S. Macaskill, D. L. Halligan et al., 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445 82–85. [DOI] [PubMed] [Google Scholar]
- Hampsey, M. A., 1997. Review of phenotypes in Saccharomyces cerevisiae. Yeast 13 1099–1133. [DOI] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1966. The effects of linkage on the limits to artificial selection. Genet. Res. 8 269–294. [PubMed] [Google Scholar]
- Hohmann, S., 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasnos, L., and R. Korona, 2007. Epistatic buffering of fitness loss in yeast double deletion strains. Nat. Genet. 39 550–554. [DOI] [PubMed] [Google Scholar]
- Jasnos, L., P. Sliwa and R. Korona, 2005. Resolution and repeatability of phenotypic assays by automated growth curve analysis in yeast and bacteria. Anal. Biochem. 344 138–140. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., and S. P. Otto, 2006. Interference among deleterious mutations favours sex and recombination in finite populations. Nature 443 89–92. [DOI] [PubMed] [Google Scholar]
- Kelley, R., and T. Ideker, 2005. Systematic interpretation of genetic interactions using protein networks. Nat. Biotechnol. 23 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick, S. C., A. M. Carlsson, S. A. West and T. J. Little, 2006. Testing the pluralist approach to sex: the influence of environment on synergistic interactions between mutation load and parasitism in Daphnia magna. J. Evol. Biol. 19 1603–1611. [DOI] [PubMed] [Google Scholar]
- Kimura, M., and T. Maruyama, 1966. The mutational load with epistatic gene interactions in fitness. Genetics 54 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishony, R., and S. Leibler, 2003. Environmental stresses can alleviate the average deleterious effect of mutations. J. Biol. 2 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336 435–440. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., and D. Houle, 1994. Genotype-environment interactions and the estimation of the genomic mutation rate in Drosophila melanogaster. Proc. Biol. Sci. 258 221–227. [DOI] [PubMed] [Google Scholar]
- Korona, R., 1999. Genetic load of the yeast Saccharomyces cerevisiae under diverse environmental conditions. Evolution 53 1966–1971. [DOI] [PubMed] [Google Scholar]
- Kouyos, R. D., S. P. Otto and S. Bonhoeffer, 2006. Effect of varying epistasis on the evolution of recombination. Genetics 173 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyos, R. D., O. K. Silander and S. Bonhoeffer, 2007. Epistasis between deleterious mutations and the evolution of recombination. Trends Ecol. Evol. 22 308–315. [DOI] [PubMed] [Google Scholar]
- Kuranda, K., V. Leberre, S. Sokol, G. Palamarczyk and J. Francois, 2006. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 61 1147–1166. [DOI] [PubMed] [Google Scholar]
- Levin, D. E., 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager, W. H., and A. J. De Kruijff, 1995. Stress-induced transcriptional activation. Microbiol. Rev. 59 506–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler, G., C. Schuller, G. Adam and H. Ruis, 1993. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 12 1997–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G., and T. Lenormand, 2006. The fitness effect of mutations across environments: a survey in light of fitness landscape models. Evolution 60 2413–2427. [PubMed] [Google Scholar]
- Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis et al., 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15 2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Ooi, S. L., X. Pan, B. D. Peyser, P. Ye, P. B. Meluh et al., 2006. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 22 56–63. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and T. Lenormand, 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3 252–261. [DOI] [PubMed] [Google Scholar]
- Pakula, A. A., and R. T. Sauer, 1989. Genetic analysis of protein stability and function. Annu. Rev. Genet. 23 289–310. [DOI] [PubMed] [Google Scholar]
- Remold, S. K., and R. E. Lenski, 2004. Pervasive joint influence of epistasis and plasticity on mutational effects in Escherichia coli. Nat. Genet. 36 423–426. [DOI] [PubMed] [Google Scholar]
- Scherens, B., and A. Goffeau, 2004. The uses of genome-wide yeast mutant collections. Genome Biol. 5 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, S. T., and M. Lynch, 1997. Mutation and extinction: the role of variable mutational effects, synergistic epistasis, beneficial mutations, and degree of outcrossing. Evolution 51 1363–1371. [DOI] [PubMed] [Google Scholar]
- Segre, D., A. Deluna, G. M. Church and R. Kishony, 2005. Modular epistasis in yeast metabolism. Nat. Genet. 37 77–83. [DOI] [PubMed] [Google Scholar]
- Shabalina, S. A., L. Y. Yampolsky and A. S. Kondrashov, 1997. Rapid decline of fitness in panmictic populations of Drosophila melanogaster maintained under relaxed natural selection. Proc. Natl. Acad. Sci. USA 94 13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., 2002. Getting started with yeast. Methods Enzymol. 350 3–41. [DOI] [PubMed] [Google Scholar]
- Silander, O. K., O. Tenaillon and L. Chao, 2007. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 5 e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Onge, R. P., R. Mani, J. Oh, M. Proctor, E. Fung et al., 2007. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 39 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafraniec, K., R. H. Borts and R. Korona, 2001. Environmental stress and mutational load in diploid strains of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A. H. Y., G. Lesage, G. D. Bader, H. M. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303 808–813. [DOI] [PubMed] [Google Scholar]
- Vassilieva, L. L., A. M. Hook and M. Lynch, 2000. The fitness effects of spontaneous mutations in Caenorhabditis elegans. Evolution 54 1234–1246. [DOI] [PubMed] [Google Scholar]
- Warringer, J., and A. Blomberg, 2003. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast 20 53–67. [DOI] [PubMed] [Google Scholar]
- Yang, H. P., A.Y. Tanikawa, W. A. Van Voorhies, J. C. Silva and A. S. Kondrashov, 2001. Whole-genome effects of ethyl methanesulfonate-induced mutation on nine quantitative traits in outbred Drosophila melanogaster. Genetics 157 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, L., and J. Yin, 2002. Dependence of epistasis on environment and mutation severity as revealed by in silico mutagenesis of phage t7. Genetics 160 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]