Abstract
Pre-harvest sprouting (PHS) or vivipary in cereals is an important agronomic trait that results in significant economic loss. A considerable number of mutations that cause PHS have been identified in several species. However, relatively few viviparous mutants in rice (Oryza sativa L.) have been reported. To explore the mechanism of PHS in rice, we carried out an extensive genetic screening and identified 12 PHS mutants (phs). Based on their phenotypes, these phs mutants were classified into three groups. Here we characterize in detail one of these groups, which contains mutations in genes encoding major enzymes of the carotenoid biosynthesis pathway, including phytoene desaturase (OsPDS), ζ-carotene desaturase (OsZDS), carotenoid isomerase (OsCRTISO) and lycopene β-cyclase (β-OsLCY), which are essential for the biosynthesis of carotenoid precursors of ABA. As expected, the amount of ABA was reduced in all four phs mutants compared with that in the wild type. Chlorophyll fluorescence analysis revealed the occurrence of photoinhibition in the photosystem and decreased capacity for eliminating excess energy by thermal dissipation. The greatly increased activities of reactive oxygen species (ROS) scavenging enzymes, and reduced photosystem (PS) II core proteins CP43, CP47 and D1 in leaves of the Oscrtiso/phs3-1 mutant and OsLCY RNAi transgenic rice indicated that photo-oxidative damage occurred in PS II, consistent with the accumulation of ROS in these plants. These results suggest that the impairment of carotenoid biosynthesis causes photo-oxidation and ABA-deficiency phenotypes, of which the latter is a major factor controlling the PHS trait in rice.
Keywords: rice, pre-harvest sprouting, photo-oxidation, carotenoid biosynthesis, abscisic acid
Introduction
The phenomenon of germination of cereal grains in the ear or panicle, usually under wet conditions shortly before harvest, is termed pre-harvest sprouting (PHS) or vivipary. Pre-harvest sprouting of cereal grains not only causes reduction of grain yield but also affects the quality of the grain. In Southeast Asia, PHS frequently occurs in rice due to the long spell of rainy weather in early summer and autumn (Wan et al., 2006). In south China alone, heavy PHS sometimes occurs in >6% of the rice acreage, which could be up to 20% for hybrid rice (Guo et al., 2004).
Previous work demonstrated that mutants impaired in abscisic acid (ABA) biosynthesis or responsiveness, such as maize viviparous (vp), Arabidopsis ABA-deficient (aba) and ABA-insensitive (abi) mutants, often produce precociously germinating seeds (McCarty, 1995). At least 10 viviparous mutants have been identified from maize (Zea mays L.), most of which (vp2, vp5, vp7, vp9, w3, y3, and y9) were blocked in biosynthesis of the carotenoid precursors for de novo ABA synthesis (Figure S1; Singh et al., 2003). In the early steps of carotenoid biosynthesis, the head-to-head condensation of C20 geranylgeranyl diphosphate (GGPP) molecules to produce a C40 carotenoid phytoene (colorless) is mediated by a soluble enzyme called phytoene synthase (PSY), which is the first committed step in carotenoid synthesis. Subsequently the phytoene undergoes four desaturation reactions with the production of lycopene (Cunningham and Gantt, 1998). Then, a series of steps including cyclization and hydroxylation reactions take place to yield α-carotene, β-carotene, lutein, xanthophyll, and zeaxanthin. The C40 carotenoid precursor is cleaved and followed by a two-step conversion of the intermediate xanthoxin to ABA via ABA aldehyde (Schwartz et al., 2003; Xiong and Zhu, 2003).
The maize Vp5 gene was found to encode a phytoene desaturase (PDS), and transgenic rice plants harboring the PDS–RNAi construct showed a clear albino phenotype (Hable et al., 1998; Miki and Shimamoto, 2004). The maize vp9 mutant and the non dormant-1 (nd-1) mutant of sunflower (Helianthus annuus L.) have mutations in the gene coding for zeta-carotene desaturase (ZDS; Conti et al., 2004; Matthews et al., 2003). The carotenoid isomerase (CRTISO) gene was cloned from the tangerine mutant of tomato and ccr2 mutant of Arabidopsis (Isaacson et al., 2002; Park et al., 2002). The Vp7/Ps1 gene encodes a lycopene-β- cyclase and is necessary for the accumulation of both ABA and carotenoid zeaxanthin in mature maize embryos; the mutant is easily discernible as it has pink kernels because of lycopene accumulation (Singh et al., 2003). These mutants containing defects in carotenoid precursor synthesis exhibit pleiotropic phenotypes, such as albino or pale green, non-viable seedlings and vivipary, due to deficiencies of carotenoid and ABA.
Within the thylakoid membranes of chloroplasts, carotenoids are found to be bound to specific protein complexes of photosystem I (PS I) and photosystem II (PS II), where they augment the light-harvesting capacity by absorbing light in the blue–green range of the visible spectrum (450–550 nm) and transferring the energy to chlorophylls (Holt et al., 2005; Moise et al., 2005). Carotenoid deficiency often causes aberrations in plastid ultrastructure, such as etioplasts from the cotyledon of dark-grown ccr2 mutants (carotenoid isomerase, CRTISO mutation) which lack prolamellar bodies (PLBs; Park et al., 2002). In addition, carotenoids play an essential role in photoprotection in plants. During photosynthesis, the excess absorbed energy can be eliminated as heat by de-exciting 1Chl through the process of non-photochemical quenching of chlorophyll fluorescence (NPQ), minimizing the generation of harmful reactive oxygen species (ROS; Ma et al., 2003; Niyogi, 1999; Tracewell et al., 2001).
Besides, as accessory pigments in photosynthesis and photoprotectors preventing photo-oxidative damage, carotenoids can also be the precursors to the hormone ABA. Analysis of mutant and transgenic plants has provided strong evidence that ABA biosynthesis and responses to this phytohormone are clearly involved in the onset and maintenance of dormancy and inhibition of PHS (Bewley, 1997). Abscisic acid biosynthetic mutants fail to induce seed dormancy and exhibit a vegetative wilty phenotype. These phenotypes have been used to define and prove the function of ABA in seed dormancy and water relations. The seeds of ABA-deficient mutants of tomato (Solanum lycopersicum L., e.g. sitiens), Arabidopsis (e.g. aba) and maize treated with fluridone (a carotenoid biosynthesis inhibitor) germinate readily when placed in water and sometimes show vivipary (Fong et al., 1983; Groot and Karssen, 1992; Leon-Kloosterziel et al., 1996a).
Another phytohormone, gibberellin (GA), is also involved in the release from dormancy of various species. It has been shown that GA-deficient mutants of Arabidopsis and tomato are dependent on exogenous GA for germination (Koornneef et al., 2002). Further work in maize showed that inhibition of GA biosynthesis mimics the effect of exogenous ABA in suppressing vivipary (White and Rivin, 2000; White et al., 2000). These results suggested that the ABA/GA ratio and not the absolute hormone content controls germination (Finch-Savage and Leubner-Metzger, 2006).
In contrast to the intensive molecular and genetic studies of seed dormancy in maize and Arabidopsis, the molecular mechanism of seed dormancy in rice is poorly understood, mainly because of the lack of available mutants with reduced dormancy. So far most research has focused on the quantitative trait locus (QTL) analysis of the natural differences in seed dormancy characteristics (Gonzalez-Guzman et al., 2004; Gu et al., 2004, 2006; Takeuchi et al., 2003; Wan et al., 2006). Only a few reports have been published on phs mutants and only one gene (OsABA1) has been cloned which was involved in ABA biosynthesis (Agrawal et al., 2001).We performed a large-scale screening of a rice T-DNA/Tos17 insertion mutant population to identify the genes involved in PHS. The warm and damp weather, a good elicitor of PHS, during the harvest season in Zhejiang Province on the southeast coast of China, enables us to isolate phs mutants efficiently. In an intensive screening of approximately 16 000 rice T1 mutant lines, we obtained 12 viviparous mutants. In this paper, four genes involved in carotenoid precursors of ABA biosynthesis were cloned. Our results suggested that the impairment in synthesis of the carotenoid precursors of ABA leads to photo-oxidation and PHS in rice, which will definitely be helpful for elucidating the molecular mechanisms of PHS in other crops such as wheat and barley that are susceptible to PHS.
Results
Identification and genetic analysis of the rice phs mutants
To identify rice phs mutants, we have screened a T-DNA/Tos17-mutagenized population (Nipponbare background) under field conditions in Hangzhou, downstream of the Yangtze River with a relatively high degree of humidity. Approximately 16 000 transgenic T1 lines were screened prior to harvest by visual inspection in the paddy field, and 27 putative mutants were identified with a viviparous phenotype. A representative mutant is shown in Figure S2(a). T2 seeds of these 27 putative phs mutants were then grown in Beijing with a lower degree of humidity for a second round of screening. From the secondary screening, 12 mutants showing a viviparous phenotype were recovered (Figure S2b), which could be simply categorized into three groups based on phenotypes besides vivipary. Mutants from category I exhibit an albino or photobleaching phenotype (Figure S2c); while mutants from categories II and III do not show an albino or photobleaching phenotype but have an enhanced wilty phenotype under conditions of water stress (category II; Figure S2d) or with embryo/seedling-lethal phenotypes (category III; Figure S2e). We present here a detailed characterization of six mutants which belong to category I (Table 1).
Table 1.
Rice pre-harvest sprouting mutants and corresponding genes
| Category | Original line | Gene | Arabidopsis homolog | Mutation | Chromosome location | Protein (a.a.) | Accession no. |
|---|---|---|---|---|---|---|---|
| I | T01 | OsPDS | AT4G14210 | Insertion | Chr.3, 20.3cM | 566 | LOC_Os030g08570 |
| HF807 | OsZDS | AT3G04870 | Insertion | Chr.7, 41.7cM | 578 | AK065213 | |
| HG4123 | Deletion | ||||||
| T09 | OsCRTISO* | AT1G06820 | Substitution | Chr.11, 84.6cM | 586 | EF417892 | |
| HC2621 | β-OsLYC | AT3G10230 | Deletion | Chr.2, 23.3cM | 493 | OC_Os02g09750 | |
| HD1449 | Deletion |
The nucleotide sequence of OsCRTISO reported in this paper has been submitted to GenBank under accession number EF417892.
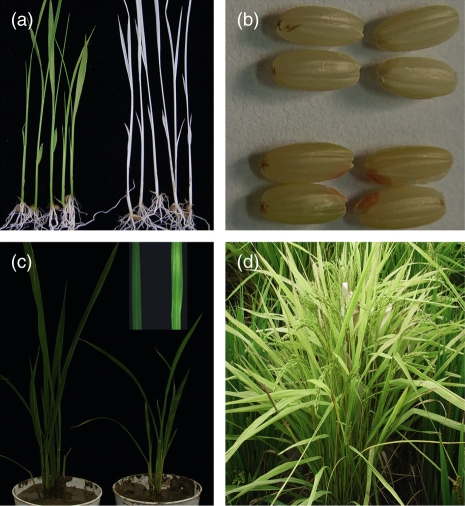
To further characterize these mutants the viviparous seedlings were rescued, and five viviparous mutant lines showed albino seedlings (Figure 1a), these homozygous plants eventually died at about 4 weeks after germination. Interestingly, the homozygous seeds of two mutant lines among them had pink embryos (Figure 1b). Moreover, one of the viviparous mutant lines, T09, developed alternating green and yellow crossbands on the leaf blades at the tillering stage (Figure 1c), like rice zebra mutants previously described (Kusumi et al., 2000), and finally showed pale green on entire plants when mature (Figure 1d).
Figure 1.
- The albino phenotype of phs seedlings.
- The pink-embryo seeds of phs4-1 and phs4-2 lines.
- Phenotypes of the wild type (left) and phs3-1 mutant (right) at the early tillering stage. The inset shows the magnified leaves of the wild type and phs3-1 mutant, respectively.
- The phs3-1 plant shows pale green over the whole leaves at the mature stage in the field.
Except for the T09 mutant, all the other viviparous mutants (T1 plants) are lethal due to lack of pigments, therefore only two genotypes – heterozygous and wild type in the T1 mutant seeds – could germinate after sowing. Statistical analysis of segregation ratios of viviparous and non-viviparous plants demonstrated that the segregation of viviparous and non-viviparous plants is in line with the expected 2:1 ratio (Table S2). Furthermore, in T2 progeny (seeds in the ear) obtained from the viviparous plants, the mutant phenotype segregated in 3:1 (Wt: vivipary, data not shown), indicating that each mutant phenotype was caused by a single recessive mutation. Subsequently genetic analysis suggested that these six mutants could be assigned to four loci and were consecutively designated as phs1 through phs4, respectively, of which two mutant alleles were identified in phs2 (phs2-1 and phs2-2) and phs4 (phs4-1 and phs4-2; see Table 1). In addition, we later identified another two phs3 alleles by the zebra phenotype (see below).
Molecular cloning of PHS1 through PHS4 genes
Genetic analysis suggested that T-DNA was not co-segregated with the mutant phenotype in all these mutants (data not shown). We then made a rough mapping to identify candidate genes in these mutants, and the PHS genes were located to different chromosomes (Table 1 and Figure S5).
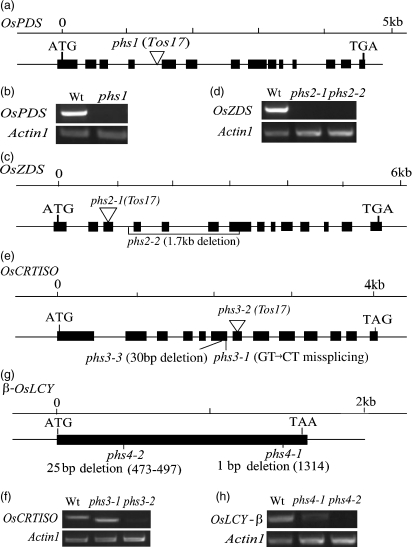
Because only mutants specifically blocked in the carotenoid biosynthetic pathways can manifest as a combination of vivipary with albino phenotype (Wurtzel et al., 2001), we presumed that these mutants may carry lesions in key genes for biosynthesis of carotenoids or ABA. So the predicted amino acid sequences of ABA biosynthesis genes from Arabidopsis were used as probes to screen in silico all available rice databases. In these four regions where mutations phs1 through phs4 mapped, four candidate genes appearing to be orthologs of Arabidopsis genes were found in the carotenoid biosynthesis pathway (Table 1), including a PDS-like gene (phs1), a ZDS-like gene (phs2-1 and phs2-2), a carotenoid isomerase-like gene (phs3-1) and a β-LCY-like gene (phs4-1 and phs4-2). To examine whether these candidate genes carried mutations in the respective mutants, we analyzed the corresponding candidate genes by polymerase chain reaction (PCR) and sequencing; the results revealed that a Tos17 was inserted in intron 4 of the putative OsPDS in phs1 (Figure 2a). As expected, no OsPDS expression was detected by reverse transcription (RT)-PCR analysis in phs1 (Figure 2b). The phs2-1 carried a Tos17 insertion in exon 3 of OsZDS, whereas phs2-2 had a 1.7-kb deletion started from intron 3 spanned to exon 7 (Figure 2c). No OsZDS expression was detectable in either mutant allele (Figure 2d).
Figure 2.
- The position of the mutation in the OsPDS gene of phs1 mutant.
- Expression of the OsPDS gene in the wild type and mutant.
- The positions of the mutation in the OsZDS gene of phs2-1 and phs2-2 mutants.
- Expression of the OsZDS gene in the wild type and mutants.
- The positions of the mutation in the OsCRTISO gene of phs3-1, phs3-2/NG0489 and phs3-3 mutants.
- Expression of the OsCRTISO gene in wild type and mutants.
- The positions of the mutation in the β-OsLCY gene of phs4-1 and phs4-2 mutants.
- Expression of the β-OsLCY gene in the wild type and mutant.
Arrowheads indicate the insertion sites of a rice retrotransposon, Tos17. Boxes and lines indicate exons and introns, respectively.
In phs3-1, a G-to-C transition was identified at position 1995 (the putative translation start codon is referred to as +1) of the OsCRTISO gene, which is predicted to be the donor site of intron 6 (Figure 2e). This mutation presumably causes the utilization of a novel splicing donor-like site (GT) 24 bp upstream from the mutated site. Consistent with this speculation, RT-PCR analysis revealed the transcript in phs3-1 was slightly shorter than that in the wild type (Figure 2f), which was confirmed by direct DNA sequencing. In addition, another two phs3 alleles were collected by the zebra phenotype. Sequence analysis revealed that mutations of various types were identified in the candidate gene CRTISO. A Tos17 retrotransposon inserted in exon 7 of phs3-2 (NG0489 from the Rice Tos17 Insertion Mutant database http://tos.nias.affrc.go.jp/~miyao/pub/tos17/index.html.en) completely disrupts the function of CRTISO (Figure 2f). phs3-3 has a 30-bp deletion in exon 6. The deletions in phs3-1 and phs3-3 do not cause a frame-shift in the open reading frame (ORF).
The rice β-OsLCY gene has no intron, and sequence analysis of phs4-1 mutant showed a 1-bp deletion at the 1314 bp starting from ATG, and the phs4-2 allele has a 25-bp deletion in 473–497 bp (the putative translation start codon is referred to as +1). The deletions caused a frame-shift and subsequently created a new stop codon that might result in immature translation (Figure 2g) and disrupt the normal function of the β-LCY gene. Expression analysis also showed that the β-OsLCY transcript was barely detectable in phs4 alleles compared with the wild type (Figure 2h).
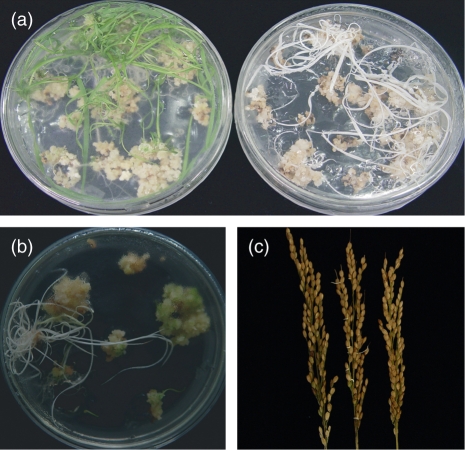
To confirm the identity of these candidate genes, we also performed genetic complementation experiments. Since homozygous mutant seedlings from three genes (OsPDS, OsZDS, and β-OsLCY) are non-viable the homozygous seeds were screened out simply from the heterozygous lines at an early stage of callus induction using mature embryos. The embryos of the homozygous mutant initiated an albino bud after 2 weeks’ culture on Murashige and Skoog medium (Murashige and Skoog, 1962) with addition of 2 mg l−1 2,4-dichlorophenoxyacetic acid (2,4-d) in the light, and their calli derived from homozygous seeds regenerated the albino shoot during later regeneration (Figure 3a). Genomic DNA fragments covering the respective 5′-upstream regions, entire genes and 3′-downstream regions of respective candidate genes were introduced into corresponding mutants by Agrobacterium-mediated transformation. In all cases, the mutant phenotypes were rescued when genomic fragments containing the candidate genes were introduced (Figure 3b,c, and Table S3). In contrast, the transgenic plants containing the empty vectors failed to rescue the phs mutants (data not shown).
Figure 3.
- Plant regeneration from calli derived from embryos of the heterozygous phs4 or wild-type seeds (left) and the homozygous phs4 seeds (right).
- The calli derived from embryos of homozygous phs4 seeds were used for transformation.
- The phenotypes of panicles in mature plants of wt (left), heterozygous mutant (middle) and complementation transgenic plant (right).
Carotenoid profiles of phs mutants
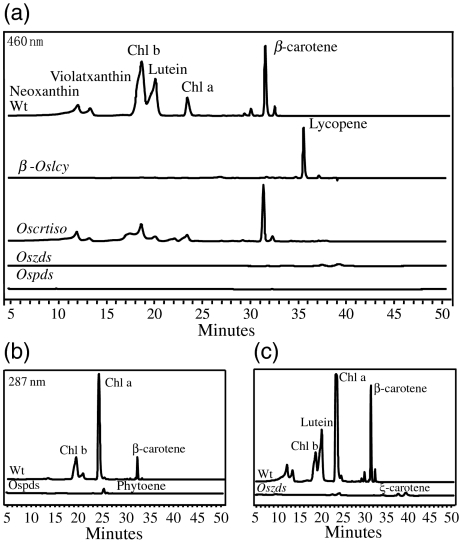
To analyze carotenoid metabolism in the phs mutants, high-performance liquid chromatograph (HPLC) analysis was performed on the extracts prepared from light-grown wild-type and mutant seedlings. As shown in Figure 4a, carotenoids were almost undetectable in phs1 (Ospds mutants) and phs2-1 or phs2-2 (Oszds mutants) seedlings at 460 nm. However, lycopene accumulated in phs4-1 or phs4-2 (β-Oslcy mutants). Carotenoids, especially lutein, were dramatically reduced in light-grown phs3-1 (Oscrtiso mutants) and an all-trans lycopene precursor, prolycopene, was accumulated in dark-grown phs3-1 seedlings (data not shown). The Ospds mutant contained a peak at 287 nm with a retention time of 25.54 min, which was absent in the wild type (Figure 4b). The derived spectrum of the peak showed characteristics of phytoene with three main peaks (data not shown). In phs2 seedlings only small peaks of ξ-carotene and its isomers were detected at 430 nm, which were absent in the wild type (Figure 4c).
Figure 4.
- HPLC chromatograms recorded at 460 nm (most carotenoids).
- HPLC chromatograms recorded at 287 nm (phytoene).
- Chromatograms recorded at 430 nm (ξ-carotene). Chl a, chlorophyll a; Chl b, chlorophyll b.
These results were consistent with the mutated genes in the biosynthetic pathway of carotenoids, and carotenoid biosynthesis is indeed impaired in these mutants. The albino phenotypes of the phs mutants (Ospds, Oszds, and β-Oslcy) were due to chlorophyll and carotenoid deficiency. The pink embryo in phs4 suggested lycopene accumulation, and the phenotype of phs3 (Oscrtiso mutant) seemed to result from photobleaching.
Cellular characters of the phs mutants
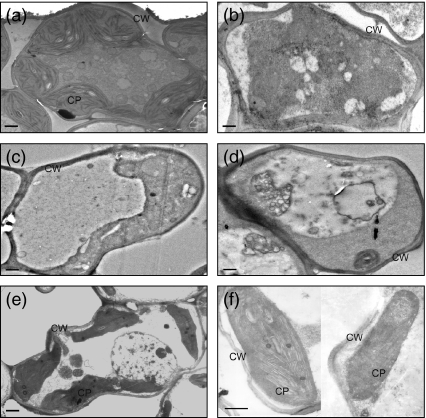
Carotenoids are essential components of photosynthetic systems, which occur in thylakoid membranes in chloroplasts in association with chlorophyll–protein complexes (Green and Durnford, 1996; Moise et al., 2005). To assess the effect of the PHS mutations on chloroplast development, the leaves of 2-week-old seedlings of all phs mutants mentioned above were examined by transmission electron microscopy. In wild types, chloroplasts were well-developed and organized in the mesophyll cells (Figure 5a), in contrast, no plastid- or chloroplast-like structures were found in the mesophyll cells of albino mutants (phs1, phs2, and phs4; Figure 5b–d). In the mesophyll cells of Oscrtiso mutants, irregularly shaped and abnormally developed chloroplasts were observed, with reduced and irregularly organized thylakoid membranes (Figure 5e,f).
Figure 5.
Mesophyll cells and chloroplasts from the phs mutants and wild type.
Wild type (a), Ospds (b), Oszds (c), β-Oslcy (d), Oscrtiso (e); the chloroplast from wild type (left) and Oscrtiso mutant (right; f). CP, chloroplast; CW, cell wall; Thy, thylakoid. Scale bar = 1 μm.
OsLCY-RNAi plants and phs mutants suffered from photo-oxidative damages
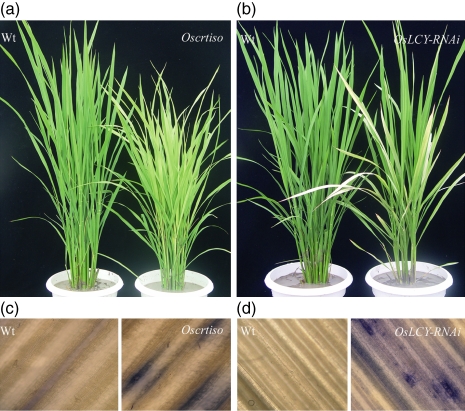
Due to the photoprotective function of carotenoids, plants deprived of these pigments suffered from photobleaching damage, especially under high light conditions (Niyogi, 1999; Sagar et al., 1988). Since the homozygous mutant seedlings from three genes (OsPDS, OsZDS, and β-OsLCY) are nonviable, we generated transgenic rice plants harboring the β-OsLCY- RNAi construct. It was demonstrated that β-OsLCY- RNAi plants and Oscrtiso mutants showed a distinctly photobleached phenotype on leaves (Figure 6a,b), which was in accordance with the accumulation of ROS as revealed by nitro blue tetrazolium (NBT) staining (Figure 6c,d). Furthermore, compared with that in the wild type, higher activities of ROS-scavenging enzymes were found in both β-OsLCY-RNAi plant and phs3-1 mutant (Figure S3), suggesting that these two plants were under oxidative stress.
Figure 6.
Photobleaching phenotype (a, b) and the detection of ROS (c, d) of Oscrtiso and β-LCY-RNAi plants and the corresponding wild types.
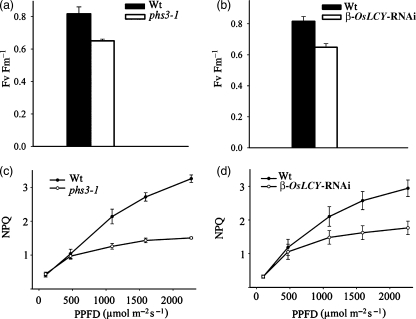
The maximal efficiency of PS II photochemistry (Fv/Fm) is often considered to be an indicator of PS II function (Demmig-Adams and Adams, 1992; Krause and Weis, 1991). Therefore, we further investigated PS II photochemistry in phs3-1/Oscrtiso mutants and β-OsLCY-RNAi plants, as well as the wild type. The Fv/Fm ratios in wild-type plants are above 0.8. However, phs3-1/Oscrtiso mutant and β-OsLCY-RNAi plants show a decrease in the Fv/Fm ratio, which is about 0.65 (Figure 7a,b).
Figure 7.
The maximal efficiency of PS II photochemistry (Fv/Fm; a, b) and non-photochemical quenching of chlorophyll fluorescence (NPQ; c, d) of phs3-1 mutant and β-OsLCY-RNAi plants. Standard deviations were obtained from six measurements.
The phs3-1/Oscrtiso mutants and β-OsLCY-RNAi plants also show different responses of NPQ to increasing photosynthetic photon flux density (PPFD). When PPFD was <500 μmol m−2 sec−1, the phs3-1/Oscrtiso mutants and β-OsLCY-RNAi plants showed almost the same value of NPQ in comparison with the wild type. With further increase in PPFD, NPQ increased more rapidly in wild-type plants than in the Oscrtiso/phs3-1 mutants and β-OsLCY-RNAi plants, leading to much higher values of NPQ than for the latter two plants (Figure 7c,d).
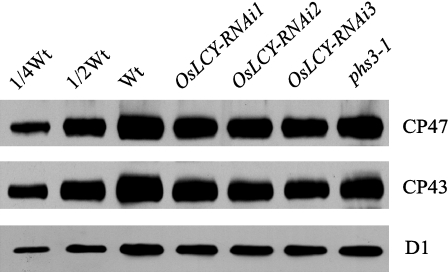
To get further evidence for the photo-oxidative damage occurring in PS II, we examined the levels of essential PS II core proteins CP43, CP47 and D1 (Green and Durnford, 1996), which are the targets of ROS (Niyogi, 1999), in the leaves from the phs3-1 mutant and β-OsLCY-RNAi plants, respectively. Western analysis showed that the level of these proteins in the phs3-1 mutant and β-OsLCY-RNAi plants decreased to 64–77% of that in the wild type, indicating the occurrence of photo-oxidative damage in PS II core proteins (Figure 8).
Figure 8.
Western analysis of the PS II core proteins CP47, CP43 and D1.
ABA and GA synthesis are altered in phs mutants
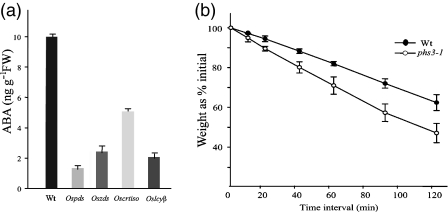
To investigate whether the reduced level of carotenoids in phs mutants causes a decreased level of ABA we measured the ABA content in the respective phs mutants by immunoassay. As shown in Figure 9a, the amount of ABA was reduced in all four mutants, but much more significantly in Ospds, Oszds, and Oslcy mutants compared with that in the wild type. Since ABA is also directly involved in the regulation of stomatal aperture, we examined the water loss characteristics of phs3-1/Oscrtiso plants. Wild-type and mutant plants were grown in the soil under a normal irrigation regime. The water loss analysis was carried out at the tillering stage, and the results revealed a water loss of about 53% of fresh weight within 120 min in phs mutants, whereas wild-type leaves showed only a 38% loss, indicating that the rate of water loss in phs mutants was faster than in wild-type plants (Figure 9b).
Figure 9.
ABA content and water loss assay of phs mutants and wild types. (a) ABA content in wild-type and mutant seedlings. (b) Water loss assays for the leaves of the wild type and Oscrtiso mutant (T09/phs3-1) were performed within 120 min. Standard deviations were obtained from five measurements.
To test whether phs mutant can sense or respond to ABA, we cultured seeds [30 days after pollination (DAP)] from phs3-1 mutant and the wild type in water with or without ABA (of different concentrations) treatment (Figure S4). In all treatments, the phs3-1 mutant and the wild type grew with similar kinetics, indicating that the phs3-1 mutant has a normal response to ABA.
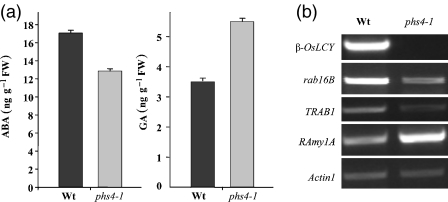
Abscisic acid and another important phytohormone, GA, which can directly antagonize ABA signaling during induction of dormancy, are two important regulators of seed dormancy. Therefore, we measured the ABA and GA content in homozygous phs4-1/β-Oslcy seeds 30 DAP (phs4 homozygous seeds were easily discernible by their pink embryo). As expected, the ABA level in phs4-1/β-Oslcy was lower than that in the wild type; however, GA in phs4-1/β-Oslcy is higher than that in the wild type (Figure 10a). We further examined the expression of two ABA-regulated genes, rab16B and TRAB1, as well as a GA-induced gene, the α-amylase gene RAmy1A (AK073487), which were involved in embryo development and seed maturation (Hobo et al., 1999; Mitsui et al., 1996; O'Neill et al., 1990; Ono et al., 1996). The results demonstrated that the transcripts of rab16B and TRAB1 were significantly reduced in phs4-1/β-Oslcy seeds while the transcript of RAmy1A was increased (Figure 10b), suggesting that the ABA/GA ratio in phs4-1/β-Oslcy seeds is probably a switch for PHS.
Figure 10.
Hormone contents and expression of hormone-regulated genes. (a) ABA and GA content in wild-type and β-Oslcy seeds at 30 days after pollination (DAP). (b) Expression of rab16B (LEA protein gene), TRAB1 (a bZIP factor gene) and RAmy1A (α-amylase gene) genes in wild-type and β-Oslcy seeds at 30 DAP.
Discussion
Analysis of phs mutants in rice
Under normal circumstances, germination in the ear of developing cereal grain is prevented by physiological mechanisms that include dormancy. Only under exceptional circumstances (e.g. rain late in grain maturation) may PHS occur. Since dormancy is a complex trait which is controlled by a large number of genes, not only with strong intergenic interactions but also strong interactions between genes and the environment. Thus, QTL analysis has often been the tool of choice for the dissection of dormancy and germination, and a large number of the QTLs controlling seed dormancy and germination have been identified in Arabidopsis, rice, wheat, and barley (Clerkx et al., 2004; Li et al., 2004; Prada et al., 2004). Comparative genomics approaches were also used to identify candidate gene(s) controlling dormancy and PHS based on the availability of the rice genome sequence, a large number of barley and wheat ESTs, and also the high co-linearity among rice, wheat, and barley (Li et al., 2004; Wilkinson et al., 2002).
Since PHS is a complex trait controlled by both environmental and genetic factors, we carried out intensive screening of the rice mutant population in south China, where long spells of rainy weather often occur in early summer and autumn and successfully isolated 27 rice phs mutants in the paddy fields.
In this study, we focused on phs mutants which belong to category I; using both map-based cloning and comparative genetics approaches, we have identified that the mutated genes in category I mutants encode major enzymes (OsPDS, OsZDS, OsCRTISO, and β-OsLCY) in the carotenoid biosynthesis pathway. The candidate genes were all confirmed via functional complementation. Further work with these rice phs mutants will provide novel insights into the role of these PHS genes in dormancy and germination.
Carotenoid metabolism, plastid development and photo-oxidation in phs mutants
In plant chloroplasts, carotenoids serve important roles in the photosynthetic apparatus as structural components and photosynthetic and photoprotective pigments (Green and Durnford, 1996; Niyogi, 1999; Niyogi et al., 2001). Just as expected, according to the enzymatic steps blocked, phs1 and phs2 accumulated phytoene and ξ-carotene in the light-grown seedlings, respectively. The results are in accordance with maize vp5 and vp9 mutants with mutations in PDS and ZDS (Hable et al., 1998; Matthews et al., 2003). In case of phs4, the mutation in lycopene β-cyclase leads to high lycopene accumulation in both seedlings and embryos from homozygous phs4 seeds. The accumulation of lycopene may be explained by the presumption that lycopene ε-cyclase and lycopene β-cyclase constitute an enzyme complex in the thylakoid membranes of plant chloroplasts; in the absence of β-cyclase, the complex is destabilized, resulting in the loss of both ε- and β-cyclase activity and so lycopene accumulates (Cunningham and Gantt, 1998). The results are consistent with 2-(4-chlorophenylthio)-triethylamine hydrochloride (CPTA)-treated barley seedlings (CPTA is a specific inhibitor of the cyclization of lycopene, especially lycopene β-cyclase; La Rocca et al., 2007). In addition, PSY mRNA is virtually absent in rice endosperm, which does not provide the substrate for these downstream enzymes and is consequently unable to form downstream products (Schaub et al., 2005). Therefore, the phs4 mutant accumulates lycopene (showing a pink color) in the embryo but not in the endosperm.
Unlike phs1, phs2, and phs4 mutants, phs3 mutant show a non-lethal and ‘variegated’ phenotype. The mutation in the PHS3 gene results in a dramatic reduction of lutein in light-grown leaves and accumulation of prolycopene in etiolated seedlings. This result was in agreement with other crtiso mutants from tomato and Arabidopsis (Isaacson et al., 2002; Park et al., 2002). Interestingly, the CRTISO activity could partially be substituted by light, i.e. photoisomerization, which may account for the survival of the phs3 mutant (Isaacson et al., 2002; Park et al., 2002).
Since carotenoids are located in the photosynthetic membrane in the form of chlorophyll–carotenoid–protein complexes and some carotenogenic enzymes are membrane-associated (Cunningham and Gantt, 1998; Green and Durnford, 1996), changes in carotenoid composition or the carotenogenic enzyme itself always lead to abnormal plastid development (Park et al., 2002; Welsch et al., 2000). It has been demonstrated that the location of PSY protein influenced the formation of PLBs (Welsch et al., 2000), CRTISO is required in PS II assembly in cyanobacteria, and loss of CRTISO activity results in the absence of PLB in the ccr2 Arabidopsis mutant (Masamoto et al., 2004; Park et al., 2002). All the phs mutants (phs1 through phs4) showed dramatic changes in carotenoid composition and had abnormal chloroplasts or even none at all, suggesting the importance of these enzymes for plastid development.
Carotenoids could also act as antioxidants to quench excessive free radicals and ROS generated from photo-oxidation (Hirayama et al., 1994; Niyogi, 1999; Niyogi et al., 1998; Woodall et al., 1997). The increasing activities of ROS-scavenging enzymes in β-OsLCY RNAi plants and Oscrtiso mutants suggested that these two plants suffered from oxidative damage. Non-photochemical quenching of chlorophyll fluorescence is one of the major indicators which are closely associated with the onset of harmless dissipation of the excess energy present in the pigment of light-harvesting complexes as heat (Gilmore, 1997; Niyogi, 1999). The significant decrease of NPQ in β-OsLCY RNAi plants and Oscrtiso mutants indicated that excessive absorbed light could not be efficiently dissipated as heat and the excessive energy might result in ROS generation. Nitro blue tetrazolium staining further confirmed that considerable superoxide accumulated in both β-OsLCY RNAi plants and Oscrtiso mutants. The value of Fv/Fm is normally in the range of 0.8–0.85 in healthy leaves independent of plant species, and a lower value indicated that a proportion of PS II reaction centers were damaged (Demmig-Adams and Adams, 1992). The decreased Fv/Fm indicated that photoinhibition occurred in the β-OsLCY RNAi plants and Oscrtiso mutants under normal conditions. Moreover, the decrease of PS II core proteins such as CP43, CP47, and D1 in β-OsLCY RNAi plants and Oscrtiso mutants may provide further evidence for the occurrence of photo-oxidative damage in PS II. Taken together, these results further confirm that the investigated PHS genes are involved in not only synthesis of the carotenoid precursors of ABA, but also chloroplast development and photoprotection. However, in the dark or in weak light the mechanism by which carotenoid deficiency influences plastid development needs to be further elucidated.
ABA and pre-harvest sprouting in phs mutants
Seed dormancy and germination are regulated by a wide range of plant hormones, including ABA, ethylene, GA, and brassinosteroids, of which ABA is the primary mediator of seed dormancy (Koornneef et al., 2002). Abscisic acid-deficient or ABA-insensitive Arabidopsis mutants show reduced seed maturation and dormancy (Finch-Savage and Leubner-Metzger, 2006; Koornneef et al., 2002; Leon-Kloosterziel et al., 1996b). Unlike in Arabidopsis, in cereal plants such as maize embryos from ABA-deficient mutants germinate precociously on the ear. However, the absolute level of ABA in the seeds does not always perfectly correlate with the dormancy and germination events, suggesting that other modulating factors are also involved (White et al., 2000). It has been presumed that dormancy released by after-ripening and stratification caused a switch to ABA catabolism resulting in a decrease in ABA content in the embryo and a corresponding increase in inactive ABA metabolites (Gubler et al., 2005).
The amount of GA also plays an important role in controlling seed dormancy and germination. Previous studies revealed that paclobutrazol treatment of the ear suppresses vivipary in ABA-deficient seeds (White and Rivin, 2000). Here, we have demonstrated that the impaired biosynthesis of the carotenoid led to significant reduction of ABA content in the phs mutants. However, these mutants have a normal response to ABA. Interestingly, the amount of GA in phs4-1 seeds was significantly increased; the increased GA might result from a reduced flux of GGPP to carotenogenesis since GGPP is the common precursor for both GA and carotenoid biosynthesis (Rodriguez-Concepcion et al., 2001). It has been demonstrated that the level of GA in PSY-overexpressing plants was reduced due to increasing utilization of GGPP for carotenoid biosynthesis, leading to a dwarf phenotype (Fray et al., 1995). In addition, the ratio of ABA/GA is distinctly reduced in phs4-1 seeds, and the genes involved in seed development were differentially expressed in the seeds of the wild type and mutants. These results suggested that the ABA/GA ratio might play an important role in controlling PHS. However, the contribution of carotenoid synthesis to the regulation of balance between ABA and GA is still elusive.
It was found that some common factors regulated ABA and GA biosynthesis during seed development in Arabidopsis in an opposite manner (Gazzarrini et al., 2004). The viviparous mutants of rice are ideal for elucidating the complex mechanism of germination and dormancy, and a detailed comparison on differential expression patterns of genes between different kinds of phs mutants and wild type by microarray or other technologies will finally help us to identify the major factors controlling PHS and gain more insight into the physiological functions of carotenoids and the molecular mechanism of PHS.
Experimental procedures
Plant materials and growth conditions
Approximately 160 000 rice T-DNA/Tos17 mutant lines (Nipponbare background) were sown in the field in Hangzhou, Zhejiang Province in 2003. The panicles of mature plants were observed in late September to detect the germinated seeds. In 2004, the screened phs mutants (T2) were sown in Beijing for further screening. Segregation ratios of viviparous and non-viviparous plants were investigated in the T2 plants. The other two phs3 alleles were obtained as follows: phs3-2 was obtained from TOS17 retrotransposon insertion lines (http://tos.nias.affrc.go.jp/~miyao/pub/tos17/phenotype/17-NG.html, line number NG 0489) and phs3-3 was derived from a spontaneous mutation.
Map-based cloning of PHS genes
To map the mutated genes, linkage analyses were performed using an F2 population derived from the cross between mutants (Nipponbare, Japonica) and TN1 or Minghui 63 (Indica) varieties. The simple-sequence repeat (SSR) and sequence-tagged-site (STS) markers used to analyze the polymorphisms between Nipponbare and TN1 (or Minghui 63) were obtained from the DNA bank at the National Institute of Agrobiologica Sciences (http://www.nias.affrc.go.jp).
Sequence analysis
The protein sequences of ABA biosynthetic enzymes from Arabidopsis (http://www.arabidopsis.org/) were compared with the rice database of TIGR Rice Databases (http://www.tigr.org), the Rice Genome Research Program (http://rgp.dna.affrc.go.jp/), the Beijing Genomics Institute (http://btn.genomics.org.cn/rice), and the Knowledge-based Oryza Molecular biological Encyclopedia (KOME; http://cdna01.dna.affrc.go.jp/cDNA/) for searching the rice orthologs.
Construction of complementation and RNAi vectors and plant transformation
A 6.5-kb DNA fragment containing a full-length genomic β-OsLCY gene was obtained by digesting the bacterial artificial chromosome (BAC) clone OSJNBb0031B09 with Pst I and Bam HI; the fragment was inserted into a binary vector pCAMBIA2300. Similarly, the complementation vectors for other phs mutants were also constructed, using corresponding BAC clones (Table S3). In addition, empty vectors were transformed into the corresponding phs mutant as the respective controls. For β-OsLCY RNAi construct, an 856-bp fragment from 630 to 1486 bp of the β-OsLCY ORF was inserted as a Bam HI/Sal I fragment in sense orientation downstream of the potato (Solanum tuberosum L.) GA20 oxidase intron into pUC-RNAi (Luo et al., 2006). The same fragment was inserted in antisense orientation into the Bgl II/Xho I sites of pUC-RNAi already carrying the sense fragment. Subsequently, the fragment comprising sense and antisense fragments of β-OsLCY interspersed by potato GA20 oxidase intron was excised from pUC-RNAi using the flanking Pst I and inserted into a pXQAct plasmid between rice actin1 promoter and Ocs terminator, yielding the binary construct. The rice transformation was performed as described (Liu et al., 2007).
Carotenoid analysis
The extraction and analysis of carotenoids was performed as previously described (Fraser et al., 2000). Briefly, a reverse-phase C30, 5 μm column (250 × 4.6 mm) coupled to a 20 × 4.6 mm C30 guard (YMC Inc.; http://www.ymc.co.jp) with a methanol/tert-methyl butyl ether-based mobile phase were used with a HPLC 10Avp system (Shimadzu, http://www.shimadzu.com/). Throughout chromatography, the elution was monitored continuously from 200 to 600 nm by an online Shimadzu SPD-10Avp PDA detector. Carotenoids were identified by their characteristic absorption spectra, typical retention time, and comparison with authentic standards.
Transmission electron microscopic analysis
Rice leaves were cut into 1 mm squares, fixed in 2.5% (v/v) glutaraldehyde in PBS (pH 7.2) for 24 h and post-fixed in 2% OsO4 in PBS (pH 7.2) for 2 h. Following ethanol series dehydration, samples were embedded in Epon 812 (Shell Chemicals; http://www.shellchemicals.com) and polymerized for 24 h at 60°C. Ultrathin sections (50–70 nm) were double stained with uranyl acetate and lead citrate and examined with a transmission electron microscope (TEM; FEI Tecnai 20; http://www.fei.com) at 120 kV.
Histochemical detection of O2−
Superoxide accumulation in rice leaves was visualized by 0.1% NBT staining as described (Fitzgerald et al., 2004). Rice leaves were vacuum-infiltrated and stained with NBT solution. After staining, the chlorophyll was removed by incubating in 96% (v/v) ethanol overnight.
Chlorophyll fluorescence measurements
Chlorophyll fluorescence measurements were carried out with attached leaves in the greenhouse using a PAM 2100 portable fluorometer (Walz, http://www.walz.com/) as described (Lu et al., 2001).
Determination of ABA, GA, and water-loss assay
Determination of ABA and GA with 2-week-old seedlings was performed as previously described (Agrawal et al., 2001; Luo et al., 2006). For the water-loss assay, plants were grown in a paddy field and the detached leaves (300 mg from 10 seedlings) of 1-month-old seedlings were kept on aluminum foil at room temperature (25°C). The weight of leaves was taken at 10-min intervals until 60 min or longer. Each experiment was performed with five replicates.
Germination and ABA response assay
Seeds (30 days after pollination) from phs mutants, the wild type and complementation transgenic plants were sampled for germination ratio and ABA response assay. Seeds were cultured in a growth chamber at 30°C with a 16-h/8-h cycle for 3 days for the germination assay and 11 days for the ABA response assay. Shoots from seeds without ABA treatment were measured regularly, while shoots from seeds treated with different concentrations of ABA were measured at the 11th day after growth.
Expression analysis
Total RNAs were isolated from various organs of rice as described (Luo et al., 2006). The RT-PCR was performed with DNase-treated total RNAs using the RT-for-PCR Kit (Promega, http://www.promega.com/). The primers used for each gene are listed in Table S1. The products of PCR amplification were separated by electrophoresis on 1.5% (w/v) agarose gels and stained with ethidium bromide.
Western blot analysis
Thylakoid were extracted from the first leaves of β-OsLCY-RNAi plants, phs3-1 mutant and the wild type when β-OsLCY-RNAi plants and phs3-1 mutant showed a photobleaching phenotype at the tillering stage. Thylakoid membrane preparation and immunodetection of the PS II core proteins CP43, CP47 and D1 were carried out as described (Peng et al., 2006).
Enzyme activity assay
All enzyme activities were measured from 0.5 g first leaves of β-OsLCY-RNAi plants and the phs3-1 mutant as well as the wild type when β-OsLCY-RNAi plants and the phs3-1 mutant showed a photobleaching phenotype at the tillering stage. The enzyme activities of SOD, CAT, APX, MDAR, DHAR, and GR were measured as described previously (Chen and Gallie, 2004; Gonzalez et al., 1998; Jiang and Zhang, 2002; Knöraer et al., 1996).
Acknowledgments
We are grateful to Dr Jianru Zuo of IGDB, CAS for critically reading the manuscript. This work was supported by grants from the Chinese Academy of Sciences (KSCX2-YW-N-010) and Ministry of Science and Technology of China (2002CCA03200, 2002CB111301, and 2005CB120806) and also the National Natural Sciences Foundation of China (3062100, 30425034, 30710103903).
Supplementary Material
The following supplementary material is available for this article online:
Carotenoid and abscisic acid biosynthetic pathway.
The phenotypes of the rice pre-harvest sprouting mutants.
Activities of ROS-scavenging enzymes of leaves from OsLCY-RNAi plant, phs3-1, and wild type at tillering stage.
Abscisic acid responsiveness test of phs3-1 mutant.
Map-based cloning of PHS genes.
The primers used for expression analysis via RT-PCR.
Segregation ratios of viviparous and non-viviparous plants in the T2 generation.
Constructions for genetic complementation of phs mutants and plant transformation events.
This material is available as part of the online article from http://www.blackwell-synergy.com.
Please note: Blackwell publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Agrawal GK, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H. Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol. 2001;125:1248–1257. doi: 10.1104/pp.125.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gallie DR. The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell. 2004;16:1143–1162. doi: 10.1105/tpc.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkx EJ, El-Lithy ME, Vierling E, Ruys GJ, Blankestijn-De Vries H, Groot SP, Vreugdenhil D, Koornneef M. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 2004;135:432–443. doi: 10.1104/pp.103.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti A, Pancaldi S, Fambrini M, Michelotti V, Bonora A, Salvini M, Pugliesi C. A deficiency at the gene coding for zeta-carotene desaturase characterizes the sunflower non dormant-1 mutant. Plant Cell Physiol. 2004;45:445–455. doi: 10.1093/pcp/pch052. [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW. Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992;43:599–626. [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald HA, Chern MS, Navarre R, Ronald PC. Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol. Plant Microbe Interact. 2004;17:140–151. doi: 10.1094/MPMI.2004.17.2.140. [DOI] [PubMed] [Google Scholar]
- Fong F, Smith JD, Koehler DE. Early events in maize seed development: 1-methyl-3-phenyl-5-(3-[trifluoromethyl]phenyl)-4-(1H)-pyridinone induction of vivipary. Plant Physiol. 1983;73:899–901. doi: 10.1104/pp.73.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Pinto ME, Holloway DE, Bramley PM. Technical advance: application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 2000;24:551–558. doi: 10.1046/j.1365-313x.2000.00896.x. [DOI] [PubMed] [Google Scholar]
- Fray RG, Wallace A, Fraser PD, Valero D, Hedden P, Bramley PM, Grierson D. Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J. 1995;8:693–701. [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell. 2004;7:373–385. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Gilmore AM. Mechanistic aspects of xanthophyll cycle-dependent photoprotection in higher plant chloroplasts and leaves. Physiol. Plant. 1997;99:197–209. [Google Scholar]
- Gonzalez A, Steffen KL, Lynch JP. Light and excess manganese. Implications for oxidative stress in common bean. Plant Physiol. 1998;118:493–504. doi: 10.1104/pp.118.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Abia D, Salinas J, Serrano R, Rodriguez PL. Two new alleles of the abscisic aldehyde oxidase 3 gene reveal its role in abscisic acid biosynthesis in seeds. Plant Physiol. 2004;135:325–333. doi: 10.1104/pp.103.036590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BR, Durnford DG. The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:685–714. doi: 10.1146/annurev.arplant.47.1.685. [DOI] [PubMed] [Google Scholar]
- Groot SP, Karssen CM. Dormancy and germination of abscisic acid-deficient tomato seeds: studies with the sitiens mutant. Plant Physiol. 1992;99:952–958. doi: 10.1104/pp.99.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XY, Kianian SF, Foley ME. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa) Genetics. 2004;166:1503–1516. doi: 10.1534/genetics.166.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XY, Kianian SF, Foley ME. Dormancy genes from weedy rice respond divergently to seed development environments. Genetics. 2006;172:1199–1211. doi: 10.1534/genetics.105.049155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV. Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 2005;8:183–187. doi: 10.1016/j.pbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Guo L, Zhu L, Xu Y, Zeng D, Wu P, Qian Q. QTL analysis of seed dormancy in rice. Euphytica. 2004;140:155–162. [Google Scholar]
- Hable WE, Oishi KK, Schumaker KS. Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol. Genet. Genomics. 1998;257:167–176. doi: 10.1007/s004380050636. [DOI] [PubMed] [Google Scholar]
- Hirayama O, Nakamura K, Hamada S, Kobayasi Y. Singlet oxygen quenching ability of naturally occurring carotenoids. Lipids. 1994;29:149–150. doi: 10.1007/BF02537155. [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl Acad. Sci. USA. 1999;96:15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt NE, Zigmantas D, Valkunas L, Li XP, Niyogi KK, Fleming GR. Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science. 2005;307:433–436. doi: 10.1126/science.1105833. [DOI] [PubMed] [Google Scholar]
- Isaacson T, Ronen G, Zamir D, Hirschberg J. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell. 2002;14:333–342. doi: 10.1105/tpc.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Zhang J. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 2002;53:2401–2410. doi: 10.1093/jxb/erf090. [DOI] [PubMed] [Google Scholar]
- Knöraer OC, Durner J, Böger P. Alterations in the antioxidative system of suspension-cultured soybean cells (Glycine max) induced by oxidative stress. Physiol. Plant. 1996;97:388–396. [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Curr. Opin. Plant Biol. 2002;5:33–36. doi: 10.1016/s1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- Krause GH, Weis E. Chlorophyll fluorescence and photosynthesis: the basics. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991;42:313–349. [Google Scholar]
- Kusumi K, Komori H, Satoh H, Iba K. Characterization of a zebra mutant of rice with increased susceptibility to light stress. Plant Cell Physiol. 2000;41:158–164. doi: 10.1093/pcp/41.2.158. [DOI] [PubMed] [Google Scholar]
- La Rocca N, Rascio N, Oster U, Rudiger W. Inhibition of lycopene cyclase results in accumulation of chlorophyll precursors. Planta. 2007;225:1019–1029. doi: 10.1007/s00425-006-0409-7. [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996a;10:655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, van de Bunt GA, Zeevaart JA, Koornneef M. Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 1996b;110:233–240. doi: 10.1104/pp.110.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ni P, Francki M, et al. Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Funct. Integr. Genomics. 2004;4:84–93. doi: 10.1007/s10142-004-0104-3. [DOI] [PubMed] [Google Scholar]
- Liu X, Bai X, Wang X, Chu C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 2007;164:969–979. doi: 10.1016/j.jplph.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Lu C, Lu Q, Zhang J, Kuang T. Characterization of photosynthetic pigment composition, photosystem II photochemistry and thermal energy dissipation during leaf senescence of wheat plants grown in the field. J. Exp. Bot. 2001;52:1805–1810. doi: 10.1093/jexbot/52.362.1805. [DOI] [PubMed] [Google Scholar]
- Luo A, Qian Q, Yin H, et al. EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 2006;47:181–191. doi: 10.1093/pcp/pci233. [DOI] [PubMed] [Google Scholar]
- Ma YZ, Holt NE, Li XP, Niyogi KK, Fleming GR. Evidence for direct carotenoid involvement in the regulation of photosynthetic light harvesting. Proc. Natl Acad. Sci. USA. 2003;100:4377–4382. doi: 10.1073/pnas.0736959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamoto K, Hisatomi S, Sakurai I, Gombos Z, Wada H. Requirement of carotene isomerization for the assembly of photosystem II in Synechocystis sp. PCC 6803. Plant Cell Physiol. 2004;45:1325–1329. doi: 10.1093/pcp/pch144. [DOI] [PubMed] [Google Scholar]
- Matthews PD, Luo R, Wurtzel ET. Maize phytoene desaturase and zeta-carotene desaturase catalyse a poly-Z desaturation pathway: implications for genetic engineering of carotenoid content among cereal crops. J. Exp. Bot. 2003;54:2215–2230. doi: 10.1093/jxb/erg235. [DOI] [PubMed] [Google Scholar]
- McCarty DR. Genetic control and integration of maturation and germination pathways in seed development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995;46:71–93. [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Mitsui T, Yamaguchi J, Akazawa T. Physicochemical and serological characterization of rice alpha-amylase isoforms and identification of their corresponding genes. Plant Physiol. 1996;110:1395–1404. doi: 10.1104/pp.110.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moise AR, von Lintig J, Palczewski K. Related enzymes solve evolutionarily recurrent problems in the metabolism of carotenoids. Trends Plant Sci. 2005;10:178–186. doi: 10.1016/j.tplants.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- Niyogi KK. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Bjorkman O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell. 1998;10:1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Shih C, Soon Chow W, Pogson BJ, Dellapenna D, Bjorkman O. Photoprotection in a zeaxanthin- and lutein-deficient double mutant of Arabidopsis. Photosynth. Res. 2001;67:139–145. doi: 10.1023/A:1010661102365. [DOI] [PubMed] [Google Scholar]
- O'Neill SD, Kumagai MH, Majumdar A, Huang N, Sutliff TD, Rodriguez RL. The alpha-amylase genes in Oryza sativa: characterization of cDNA clones and mRNA expression during seed germination. Mol. Genet. Genomics. 1990;221:235–244. doi: 10.1007/BF00261726. [DOI] [PubMed] [Google Scholar]
- Ono A, Izawa T, Chua NH, Shimamoto K. The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol. 1996;112:483–491. doi: 10.1104/pp.112.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell. 2002;14:321–332. doi: 10.1105/tpc.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L. LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell. 2006;18:955–969. doi: 10.1105/tpc.105.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada D, Ullrich SE, Molina-Cano JL, Cistue L, Clancy JA, Romagosa I. Genetic control of dormancy in a Triumph/Morex cross in barley. Theor. Appl. Genet. 2004;109:62–70. doi: 10.1007/s00122-004-1608-x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Ahumada I, Diez-Juez E, Sauret-Gueto S, Lois LM, Gallego F, Carretero-Paulet L, Campos N, Boronat A. 1-Deoxy-d-xylulose 5-phosphate reductoisomerase and plastid isoprenoid biosynthesis during tomato fruit ripening. Plant J. 2001;27:213–222. doi: 10.1046/j.1365-313x.2001.01089.x. [DOI] [PubMed] [Google Scholar]
- Sagar AD, Horwitz BA, Elliott RC, Thompson WF, Briggs WR. Light effects on several chloroplast components in norflurazon-treated pea seedlings. Plant Physiol. 1988;88:340–347. doi: 10.1104/pp.88.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub P, Al-Babili S, Drake R, Beyer P. Why is golden rice golden (yellow) instead of red? Plant Physiol. 2005;138:441–450. doi: 10.1104/pp.104.057927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JA. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003;131:1591–1601. doi: 10.1104/pp.102.017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Lewis PE, Hardeman K, Bai L, Rose JK, Mazourek M, Chomet P, Brutnell TP. Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell. 2003;15:874–884. doi: 10.1105/tpc.010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Lin SY, Sasaki T, Yano M. Fine linkage mapping enables dissection of closely linked quantitative trait loci for seed dormancy and heading in rice. Theor. Appl. Genet. 2003;107:1174–1180. doi: 10.1007/s00122-003-1364-3. [DOI] [PubMed] [Google Scholar]
- Tracewell CA, Vrettos JS, Bautista JA, Frank HA, Brudvig GW. Carotenoid photooxidation in photosystem II. Arch. Biochem. Biophys. 2001;385:61–69. doi: 10.1006/abbi.2000.2150. [DOI] [PubMed] [Google Scholar]
- Wan JM, Jiang L, Tang JY, Wang CM, Hou MY, Jing W, Zhang LX. Genetic dissection of the seed dormancy trait in cultivated rice (Oryza sativa L.) Plant Sci. 2006;170:786–792. [Google Scholar]
- Welsch R, Beyer P, Hugueney P, Kleinig H, von Lintig J. Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta. 2000;211:846–854. doi: 10.1007/s004250000352. [DOI] [PubMed] [Google Scholar]
- White CN, Rivin CJ. Gibberellins and seed development in maize. II. Gibberellin synthesis inhibition enhances abscisic acid signaling in cultured embryos. Plant Physiol. 2000;122:1089–1097. doi: 10.1104/pp.122.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CN, Proebsting WM, Hedden P, Rivin CJ. Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol. 2000;122:1081–1088. doi: 10.1104/pp.122.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, McKibbin RS, Bailey PC, Flintham JE. Use of comparative molecular genetics to study preharvest sprouting in wheat. Euphytica. 2002;126:27–33. [Google Scholar]
- Woodall AA, Britton G, Jackson MJ. Carotenoids and protection of phospholipids in solution or in liposomes against oxidation by peroxyl radicals: relationship between carotenoid structure and protective ability. Biochim. Biophys. Acta. 1997;1336:575–586. doi: 10.1016/s0304-4165(97)00007-x. [DOI] [PubMed] [Google Scholar]
- Wurtzel ET, Luo RB, Yatou O. A simple approach to identify the first rice mutants blocked in carotenoid biosynthesis. J. Exp. Bot. 2001;52:161–166. [PubMed] [Google Scholar]
- Xiong L, Zhu JK. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003;133:29–36. doi: 10.1104/pp.103.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Carotenoid and abscisic acid biosynthetic pathway.
The phenotypes of the rice pre-harvest sprouting mutants.
Activities of ROS-scavenging enzymes of leaves from OsLCY-RNAi plant, phs3-1, and wild type at tillering stage.
Abscisic acid responsiveness test of phs3-1 mutant.
Map-based cloning of PHS genes.
The primers used for expression analysis via RT-PCR.
Segregation ratios of viviparous and non-viviparous plants in the T2 generation.
Constructions for genetic complementation of phs mutants and plant transformation events.