Abstract
Alternative splicing of pre-mRNAs is a major contributor to both proteomic diversity and control of gene expression levels. Splicing is tightly regulated in different tissues and developmental stages, and its disruption can lead to a wide range of human diseases. An important long-term goal in the splicing field is to determine a set of rules or “code” for splicing that will enable prediction of the splicing pattern of any primary transcript from its sequence. Outside of the core splice site motifs, the bulk of the information required for splicing is thought to be contained in exonic and intronic cis-regulatory elements that function by recruitment of sequence-specific RNA-binding protein factors that either activate or repress the use of adjacent splice sites. Here, we summarize the current state of knowledge of splicing cis-regulatory elements and their context-dependent effects on splicing, emphasizing recent global/genome-wide studies and open questions.
Keywords: context dependence, pre-mRNA splicing, splicing code, splicing factor, splicing regulation
PRE-MRNA SPLICING
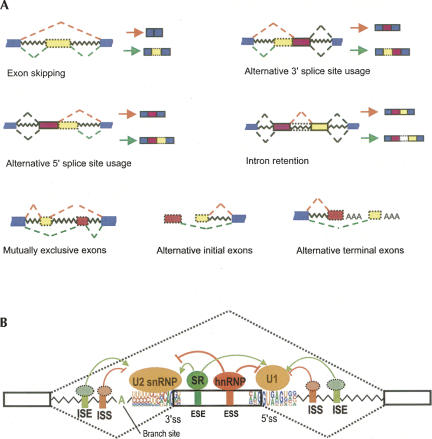
Because human genes typically contain multiple introns, the process of pre-mRNA splicing is an essential step in the expression of most genes. A majority of human genes undergo alternative splicing (AS), generating multiple splicing isoforms containing different combinations of exons (Johnson et al. 2003). The effects of AS on protein products can be dramatic, e.g., producing soluble versus membrane-bound forms of the Fas receptor that have opposing effects on apoptosis (Cascino et al. 1995), or producing isoforms of the Drosophila fruitless protein that act to specify sexual orientation (Demir and Dickson 2005). The major forms of AS are summarized in Figure 1A. Splicing regulation has been comprehensively described in several recent reviews (Black 2003; Konarska and Query 2005; Matlin et al. 2005; Blencowe 2006; House and Lynch 2008). Here, we briefly summarize some aspects of splicing specificity and regulation before turning to our main topic of splicing regulatory elements and the rules governing their activity.
FIGURE 1.
(A) Major forms of alternative splicing. In many cases, these common forms can be combined to generate more complicated alternative splicing events. (B) A schematic of regulated splicing. (Open boxes) Exons, (jagged lines) introns, (brackets) splice sites (ss). The consensus motifs of ss are shown in pictogram, and the branch point adenosine is indicated. (Dashed lines) Two alternative splicing pathways, with the middle exon either included or excluded. Splicing is regulated by cis-elements (ESE, ESS, ISS, and ISE) and trans-acting splicing factors (SR proteins, hnRNP, and unknown factors).
The sequential phosphodiester transfer reactions involved in splicing are catalyzed by large ribonucleoprotein complexes known as spliceosomes. Containing more than 100 core proteins and five small nuclear RNAs (snRNAs U1, U2, U4, U5, and U6), spliceosomes may be the most complex machines in the cell (Zhou et al. 2002; Jurica and Moore 2003; Nilsen 2003). In addition to these core factors, additional regulatory proteins participate in the splicing of particular pre-mRNAs. Splicing of most introns is thought to occur cotranscriptionally with fairly extensive interactions between splicing factors and the core transcription machinery (Das et al. 2006; Hicks et al. 2006). Splicing mechanisms have also been well reviewed recently (Hertel and Graveley 2005; Konarska and Query 2005).
CORE SPLICING SIGNALS
Three sites, the 5′ splice site (5′ss), the 3′ splice site (3′ss), and the branch point sequence (BPS), participate in the splicing reaction and are present in every intron, and thus are known as the core splicing signals. These signals are recognized multiple times during spliceosome assembly, with the 5′ss interacting initially with U1 and later with U6 snRNP, and the BPS interacting with SF1/mBBP and later with U2 snRNP (Fig. 1B; Will and Lührmann 2007). Because the 5′ss and 3′ss can be mapped precisely by aligning cDNA or expressed sequence tag (EST) sequences to the genome, large datasets of accurately annotated exon/intron structures can be obtained, permitting development of statistical models of 5′ss and 3′ss motifs that capture second-order statistical interactions between positions to more accurately predict splice site locations (Yeo and Burge 2004). However, only a few dozen mammalian BPSs have been mapped. The limited size of the available BPS data set and the low information content of this motif make it difficult to derive a reliable sequence model to predict BPSs in introns. A recent study used comparative genomics to improve BPS prediction (Kol et al. 2005), and more human BPS sequences were identified recently by sequencing lariat RT-PCR products (GAO et al. 2008).
EXON DEFINITION AS A KEY STEP IN MAMMALIAN SPLICING REGULATION
A typical human gene contains relatively short exons (typically, 50–250 base pairs [bp] in length) separated by much larger introns (typically, hundreds to thousands of base pairs or more) that on average account for >90% of the primary transcript. This transcript geometry, and the predominant exon-skipping phenotype of splice site mutations, are consistent with the idea that in mammals splice sites are predominantly recognized in pairs across the exon through “exon definition” (Robberson et al. 1990; Nakai and Sakamoto 1994; Sterner et al. 1996). Exon definition involves initial interaction across the exon between factors recognizing the 5′ss and the upstream 3′ss, whereas in the alternative model, intron definition, interactions occur first across the intron between factors recognizing the 5′ss and the downstream 3′ss (for review, see Berget 1995). Recent analyses of the coevolution of the 5′ss and 3′ss have detected predominant cross-exon interactions in human and mouse, but cross-intron interactions in invertebrates, plants, and fungi, with pufferfishes representing an intermediate state, supporting the primacy of exon definition in mammals and intron definition in most other metazoans (Xiao et al. 2007). Because exon definition occurs early during splicing and may lead to commitment of the exon to splicing, this step is critical for splicing regulation and specificity. For example, polypyrimidine tract binding protein (PTB/hnRNP I) can inhibit exon definition complex formation by binding to an ESS sequence causing skipping of Fas exon 6 (Izquierdo et al. 2005); this same factor can also inhibit the spliceosome assembly across introns (intron definition) in repressing splicing of the c-src N1 exon (Sharma et al. 2005). Following initial splice site recognition in exon definition, a series of sequential structural rearrangements is required to activate the spliceosome, and commitment to alternative splice site pairing may occur after initial splice site recognition and E complex formation (Lim and Hertel 2004).
SPLICING REGULATORY ELEMENTS AND ASSOCIATED FACTORS
Monte Carlo simulations inserting artificial motifs with varying information content into transcripts in place of natural splice sites have been used to estimate that the core human splice site motifs contain only about half of the information required to accurately define exon/intron boundaries, even considering only short introns (Lim and Burge 2001). Large human introns typically contain numerous “decoy” splice sites: sequences with similar score/degree of consensus matching as authentic splice sites. Not infrequently, decoy splice sites occur in pairs as “pseudoexons,” which resemble authentic exons in terms of length and splice site strength, but are very rarely if ever spliced (Sun and Chasin 2000). Despite the large potential for errors, the splicing process appears to occur with very high fidelity, implying the widespread involvement of transcript features besides the core splice signals in splice site selection. This additional information is thought to derive in large part from the presence of numerous cis-regulatory elements that serve as either splicing enhancers or silencers. These elements are conventionally classified as exonic splicing enhancers (ESEs) or silencers (ESSs) if from an exonic location they function to promote or inhibit inclusion of the exon they reside in, and as intronic splicing enhancers (ISEs) or silencers (ISSs) if they enhance or inhibit usage of adjacent splice sites or exons from an intronic location. In general, these splicing regulatory elements (SREs) function by recruiting trans-acting splicing factors that activate or suppress splice site recognition or spliceosome assembly by various mechanisms (Matlin et al. 2005; Chasin 2007).
SREs have most often been identified by directed mutagenesis of alternatively spliced genes and by analysis of naturally occurring mutations that perturb splicing of disease genes. ESEs are very abundant in constitutive exons, but there is ample evidence that silencers play at least as important a role in splicing regulation as enhancers (Fairbrother et al. 2002; Wang et al. 2004; Zhang and Chasin 2004). Based on the selective constraints on constitutive exons for efficient and accurate splicing and on AS exons for regulated splicing, enhancing elements are expected to play predominant roles in constitutive splicing, with silencers being relatively more prominent in control of AS.
EXONIC SPLICING ENHANCERS AND SILENCERS
It is now well established that ESEs include a diverse range of sequences, and many if not all exons contain internal ESE sequences (Schaal and Maniatis 1999; Fairbrother et al. 2002; Cartegni et al. 2003). Most ESEs function by recruiting members of the SR protein family (for review, see Graveley 2000). These factors usually regulate splicing by binding ESEs through their N-terminal RRM domains and mediating protein–protein interactions that facilitate spliceosome assembly through C-terminal RS domains (Graveley and Maniatis 1998). However, at least for in vitro splicing, the RS domains may not always be required for ESE-dependent splicing activation (Shaw et al. 2007), and RS domains can interact with RNA at the BPS and 5′ss during splicing complex assembly, at least in some cases (Shen et al. 2004; Shen and Green 2006).
ESSs are often bound by splicing repressors of the hnRNP class, a diverse group of proteins containing one or more RNA-binding domains and sometimes splicing inhibitory domains such as glycine-rich motifs (Pozzoli and Sironi 2005). hnRNPs function by a wide variety of mechanisms. For example, PTB (hnRNP I) can block essential interactions between U1 and U2 snRNPs (Izquierdo et al. 2005; Sharma et al. 2005), whereas hnRNP A1 can inhibit splicing by binding on either side and “looping out” exons or by directly displacing snRNP binding (Zhu et al. 2001; Nasim et al. 2002). Other splicing inhibitory mechanisms may also be used by these or other repressors.
Beyond molecular genetics, global approaches, including both computational and experimental methods, have been developed to identify ESEs and ESSs on a large scale (for review, see Chasin 2007). ESEs have been identified experimentally by in vitro and in vivo SELEX approaches (Tian and Kole 1995; Coulter et al. 1997; Liu et al. 1998, 2000). ESEs have also been computationally identified based on their enrichment in authentic exons versus introns and in exons with weak splice sites (Fairbrother et al. 2002), and by their enrichment in authentic exons versus pseudoexons and 5′ UTRs of intronless genes (Zhang and Chasin 2004). These studies have generated comprehensive lists of ESE oligonucleotides that help to predict the splicing phenotypes of exonic mutations (Cartegni et al. 2002; Fairbrother et al. 2002; Pfarr et al. 2005). ESSs have also been predicted based on the assumption that ESSs are depleted from authentic exons (Zhang and Chasin 2004). We developed a cell-based fluorescence-activated screen (FAS) and used it to identify 133 ESS decanucleotides active in human cells from a library of random sequences (Wang et al. 2004). Further analyses of these FAS–ESS sequences suggested that ESSs play an important role in distinguishing authentic exons from pseudoexons and in regulating alternative splice site usage and intron retention (Wang et al. 2004, 2006).
Additional exonic SREs have been predicted based on sequence conservation (Goren et al. 2006), but the activities of these elements as either splicing enhancers or silencers or neutral sequences were observed to depend heavily on their exonic context. It is not clear whether the context dependence observed was due to the flexible activity of the sequences tested, or resulted from alterations to endogenous SREs in the reporters used. For example, all of the sequences inserted into the SXN minigene promoted exon skipping, which could be explained by disruption of an unknown ESE in the site where the foreign sequences were inserted (Goren et al. 2006). Other studies of ESSs have found much greater consistency of function in different exonic contexts (Wang et al. 2004, 2006).
INTRONIC SPLICING ENHANCERS AND SILENCERS
A number of intronic SREs are also known (for reviews, see Ladd and Cooper 2002; Zheng 2004), but fewer large-scale screens have been conducted for intronic elements, and many more intronic elements likely remain to be identified. One well characterized ISE is the G triplet (GGG) or G run (Gn; n ≥ 3), which often occur in clusters and can enhance recognition of adjacent 5′ss or 3′ss (McCullough and Berget 1997, 2000). This ISE is common in GC-rich introns and is conserved between human and mouse (Yeo et al. 2004). Intronic CA repeats in several cases can enhance splicing of upstream exons, probably through binding of hnRNP L (Hui et al. 2005; Hung et al. 2007). UGCAUG hexanucleotides or slight variations often occur downstream of neuron-specific exons and function as ISEs by binding to the brain- and muscle-specific splicing factors Fox-1/Fox-2 (Brudno et al. 2001; Jin et al. 2003; Minovitsky et al. 2005; Nakahata and Kawamoto 2005; Underwood et al. 2005). Pairs of YCAY motifs (Y = C or U) are recognized by the neuron-specific Nova family of splicing factors to regulate a large number of splicing events in the brain (Jensen et al. 2000; Ule et al. 2003). Interestingly, depending on their relative location in pre-mRNA, YCAY pairs can also function as either ESSs or ISSs (Hui et al. 2005; Ule et al. 2006). Such context dependence will be discussed in more detail below.
Characterized ISSs include binding sites for the splicing repressors PTB and hnRNP A1 (for reviews, see Zheng 2004; Matlin et al. 2005), CA-rich sequences bound by hnRNP L (Hui et al. 2005), specific octamers flanking exon IIIb of the FGFR2 gene (Wagner et al. 2005), and two elements in intron 7 of human SMN2, a therapeutic target for spinal muscular atropy (Singh et al. 2006; Kashima et al. 2007).
Intronic elements (ISS and ISE) are likely of primary importance in regulating AS events, as the intronic regions surrounding alternative exons are far more conserved in mammals than those surrounding constitutive exons, out to a distance of 150 bp or more (Sorek and Ast 2003). Such increased conservation has been used to predict unannotated alternative exons (Sorek et al. 2004; Yeo et al. 2005), and to predict intronic SREs (Voelker and Berglund 2007; Yeo et al. 2007). In the predictions of intronic SREs, both groups identified the Fox-1/2 binding motif UGCAUG as the most conserved sequence near AS exons, and the elements identified by Yeo and colleagues included most known binding motifs associated with tissue-specific splicing factors. Remarkably, not only the Fox-1/Fox-2 splicing factor, but also its highly specific binding motif UGCAUG, are conserved from nematodes to mammals (Kabat et al. 2006), indicating an ancient role in splicing regulation. The analyses by Voelker and colleagues also suggested that the AU-rich motifs are strongly associated with constitutive splicing and may function as ISEs (Voelker and Berglund 2007); rigorous controls for GC content effects and experimental tests will be needed to firmly establish this result.
Splicing enhancers and silencers often function additively, with additional copies increasing their effect on splicing regulation (e.g., Huh and Hynes 1994; McCullough and Berget 1997; Chou et al. 2000; Wang et al. 2004; Zhang and Chasin 2004), either because they increase the affinity of the associated factor (Dominguez and Allain 2006) or because they increase the copy numbers of the factor that are recruited, sometimes in synergistic fashion. Different SREs may also function cooperatively to regulate alternative splicing. For example, exonic UAGG motifs and intronic GGGG motifs overlapping the 5′ss can function cooperatively to silence the brain-specific CI cassette exon (exon 19) of the glutamate NMDA R1 receptor gene (Han et al. 2005); the purine-rich ESE in exon N1 of the c-src gene can cooperate with the downstream G-run ISE to increase exon N1 inclusion (Modafferi and Black 1999)
ELEMENTS THAT REGULATE INTRON RETENTION
Among the major types of alternative splicing, intron retention is unique in that it does not involve a choice between competing pairs of splice sites. Instead, it involves the choice between using a pair of splice sites to excise an intron and bypassing splicing to export intron-containing mRNA to the cytoplasm. Therefore, intron retention is probably regulated both by factors generally involved in splicing—some of which may also play a role in mRNA export—and by dedicated mRNA export factors (for review, see Reed and Cheng 2005).
Many SREs that affect other types of AS can regulate intron retention when properly situated (Sakabe and de Souza 2007). For example, G-run ISEs can promote splicing of a retained intron in human thrombopoietin (Marcucci et al. 2007), and a subset of FAS–ESS elements can activate splicing of a retained intron (Wang et al. 2006). In addition, the CA-rich motifs that bind to hnRNP L can enhance the splicing of multiple retained introns as revealed by splicing microarray analyses following RNAi against hnRNP L (Hung et al. 2007). Another intensely studied case is the Drosophila P-element third intron (INV3), which is retained in somatic cells but is fully spliced in germ cells (Rio 1991). One element that promotes intron retention is a 5′ss-like sequence in the upstream exon (Siebel et al. 1992); similar 5′ss-like sequences have ESS activity in the ATM gene (Pagani et al. 2002) and were identified in the FAS–ESS screen (Wang et al. 2004). Some protein factors that regulate INV3 retention are homologous with vertebrate proteins that regulate splicing by binding to splicing enhancers or silencers (Siebel et al. 1994, 1995; Min et al. 1997). Therefore, it is reasonable to speculate that most general SREs can also regulate intron retention since they usually have a direct effect on splice site activity.
Perhaps the best-studied systems involving regulated intron retention occur in retroviruses (e.g., HIV) that need to balance between splicing of mRNA and transport of unspliced genomic RNA to the cytoplasm for packaging (Fischer et al. 1999). Retroviruses use many general SREs to regulate splicing of their RNA (for review, see Stoltzfus and Madsen 2006) and also use RNA elements to regulate nuclear export of unspliced RNA (for reviews, see Pollard and Malim 1998; Cullen 2003). How HIV RNAs balance between the two pathways is not completely clear, but some common elements might serve as a functional bridge between the two pathways by participating in the regulation of both splicing and RNA export (e.g., an ESS recognized by hnRNP A1 [Asai et al. 2003] and the Rev response elements [Pongoski et al. 2002]).
CONTEXT DEPENDENCE OF SRES
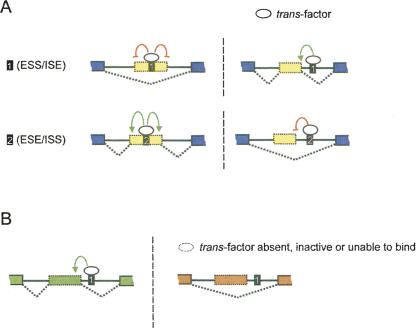
It was realized early in the study of splicing that the activities of SREs may depend on the relative locations of the elements in pre-mRNAs. For example, G triplets commonly enhance splicing from intronic locations (McCullough and Berget 1997), but they function as splicing silencers when located in exons (Chen et al. 1999). A more subtle effect is that the activity of SR proteins to promote splicing depends on the distance between the ESE and the adjacent splice site (Graveley et al. 1998). In some cases, the SREs identified in one AS event fail to regulate splicing when located in a heterologous exon/intron context. These phenomena may be collectively described as the “context dependence” of SREs and can be considered to fall into two categories: (1) location-dependent activity, in which activity varies with relative positions in the pre-mRNA (Fig. 2A); and (2) gene-dependent activity, in which activity observed in one gene is lost when the SRE is moved to another (Fig. 2B).
FIGURE 2.
Schematic of two types of context dependence for SREs. (A) Location-dependent activity of SREs, in which activity varies with pre-mRNA position. For example, G runs can function as both ESSs and ISEs (upper panel), and some SR protein binding sites can function as both ESEs and ISSs (lower panel). (Yellow) Alternative exons; (blue) constitutive exons. (B) Gene-dependent activity of SREs, in which activity observed in one gene is lost when the SRE is moved to another. Different genes are shown in different colors.
Location-dependent activity reflects the flexibility of splicing regulatory factors in their interactions with core splicing machinery. Given the size and complexity of spliceosomes and their multistep assembly, it is perhaps not surprising that the activities of factors that interact with core spliceosome components would vary depending on their locations relative to these components. For example, the G-run-binding factor hnRNP H can participate in a splicing enhancer complex when G runs are located downstream of the 5′ss (Chou et al. 1999; Caputi and Zahler 2001; Hastings et al. 2001; Schaub et al. 2007) but can inhibit splicing when similar sequences are located in an exon (Chen et al. 1999; Caputi and Zahler 2001).
Although the degree of context dependence of SREs may initially appear chaotic or confusing, there are usually patterns and rules that summarize this activity. For example, the dual activity of G runs as ISEs and ESSs can be conceptually interpreted as defining a region of pre-mRNA as intron rather than exon. We have observed that most ESSs inhibit the intron-proximal 5′ss or 3′ss when located between competing alternative splice sites, and that some ESSs, including G runs, can promote splicing of retained introns when located inside the intron (Wang et al. 2006). The underlying mechanism for these activities of G runs could involve inhibition by hnRNP F/H of exon definition complex formation “across” the site of binding. As noted above, clusters of YCAY motifs that bind the Nova family of neuron-specific splicing factors can function as ESSs, ISEs, or ISSs, depending on their position relative to the regulated exon. An “RNA map” describing these context-dependent activities has been generated that predicts the direction of Nova-dependent splicing regulation with very high accuracy (Ule et al. 2006). This study, including also a dissection of the mechanisms of splicing activation and repression by Nova, represents an important demonstration that the set of splicing regulatory activities of a factor can be rationalized.
Analogous to the common overlap between ESSs and ISEs, some SR proteins can promote splicing when bound to sites (ESEs) in exons, and also inhibit splicing when bound to intronic (ISS) sites (Kanopka et al. 1996; Ibrahim el et al. 2005; Buratti et al. 2007). Results of a FAS-based screen for ISSs suggest that overlap between ISS and ESE activity could be a general phenomenon (Z. Wang, J. Zhang, X. Xiao, and C.B. Burge, unpubl.). A recent report suggested the counterintuitive result that some elements (designated “ESRs”) can function as both ESSs and ESEs depending on exonic context (Goren et al. 2006). However, in our own analyses, large sets of ESEs and ESSs identified using entirely different methods (statistically-based and experimentally-based) were nonoverlapping, suggesting that a single sequence is unlikely to commonly function as both an ESS and an ESE (Fairbrother et al. 2002; Wang et al. 2004).
For intronic elements, the situation is much more complex. For example, Nova binding sequences commonly function as ISEs when located near the splice sites of the intron downstream of the regulated exon and as ISSs when located near the 5′ss of the upstream intron (Ule et al. 2006). Similarly, the Fox-1/2 binding site (UGCAUG) can function as an ISS when located upstream of an exon (Jin et al. 2003) and as an ISE when located downstream of an alternative exon (Huh and Hynes 1994; Lim and Sharp 1998; Jin et al. 2003). CA repeats bound by hnRNP L can function either as ISEs or ISSs depending on their distance to the upstream exon (Hui et al. 2005).
Gene-dependent activity of SREs may often result from factors that determine whether a cis-element can be recognized by the associated factor. Because most splicing regulatory cis-elements are short RNA oligonucleotides, they tend to be very abundant in pre-mRNA. Based on a recent comparative analysis of exonic splicing elements, it was estimated that about 1000 hexamers can function as either ESSs or ESEs (Stadler et al. 2006). Therefore, every pre-mRNA will contain many potential splicing regulatory motifs, only a fraction of which may be recognized by trans-factors. This is analogous to transcriptional regulation in that a search near a promoter region will predict a large number of transcription factor binding sites, only a small fraction of which are functional. Determining which cis-elements will be recognized remains an open problem. One obvious factor is the local structure of the pre-mRNA, which could affect cis-element accessibility. For example, RNA structure in the fibronectin EDA exon appears to affect recognition of an ESE by SR proteins, and this structure accounts for much of the difference between the splicing behavior of the orthologous exons in human and mouse (Buratti et al. 2004). A recent analysis of experimentally determined SREs suggested that these SRE motifs are significantly enriched in single-stranded regions of pre-mRNA (Hiller et al. 2007), suggesting that pre-mRNA secondary structure may play a general role in determining SRE function. Some factors, including Nova and muscle-blind-like splicing factors, recognize their cognate motifs in specific structural contexts (Jensen et al. 2000; Warf and Berglund 2007).
In addition to affecting SRE accessibility, secondary structures may also regulate splicing by directly affecting splice site accessibility. For example, a stem–loop structure at the 5′ss of exon 10 of the human tau gene directly affects the activity of the 5′ss, with stabilization of this structure decreasing exon 10 inclusion and destabilization of this stem–loop increasing exon 10 inclusion (Donahue et al. 2006). Another remarkable example was found in exon 6 of the Drosophila Dscam gene, where the intronic secondary structure ensures that inclusion of 48 alternative exons occurs in a mutually exclusive fashion (Graveley 2005). However, it is unclear whether examples like this represent unusual cases or are a general rule, and even for tau exon 10 the role of structure in splicing regulation is debated (D'Souza and Schellenberg 2002). Spliceosomes contain multiple RNA helicase components that can unwind RNA structures and remodel RNA/protein complexes (for review, see Bleichert and Baserga 2007). Although the primary function of these spliceosome-associated helicases appears to be rearrangement of snRNA/snRNA, snRNA/pre-mRNA, and snRNA/protein interactions in the spliceosome, at least one appears to function at an earlier stage, influencing the alternative splicing of the CD44 pre-mRNA, possibly by remodeling its structure and/or associated protein complexes (Honig et al. 2002; Lee 2002). By analogy to translation, where ribosome-associated helicases alter mRNA structure to facilitate translocation, it is tempting to speculate that spliceosome-associated helicases may also disrupt splicing-inhibitory structure in pre-mRNAs. As the general roles of structure in splicing regulation are still not clearly defined, large-scale measurements of RNA structures should be very valuable. Some recently developed methods like selective 2′-hydroxyl acylation and primer extension (SHAPE) provide new possibilities for high-throughput measurements of RNA structures (Merino et al. 2005), but high-throughput methods capable of assessing structure in vivo still need to be developed.
The activities of SREs are of course dependent on the presence and activity of the associated trans-factors, and this dependence is probably responsible for most of the tissue- or cell-type-specific splicing. SRE activity can also respond to external signals that alter the expression or activity of specific trans-factors (for review, see Shin and Manley 2004). The mechanisms by which inducible splicing responds to different stimuli can be very diverse, including, but not limited to, neuronal depolarization through CaM kinase (Xie and Black 2001; Lee et al. 2007), heat shock response through SR protein dephosphorylation (Shin and Manley 2002; Shin et al. 2004), response to T cell activation through an inducible ESS bound by hnRNP L (Rothrock et al. 2003, 2005), and response to neuronal excitation through a UAGG motif recognized by hnRNP A1 (Han et al. 2005).
GLOBAL ANALYSES OF SPLICING REGULATION
A number of new technologies have been developed recently for genome-wide analysis of AS. Several microarray platforms have been designed to distinguish between different splicing isoforms and to detect AS at a genomic scale. “Exon junction arrays” represent one early design, with high-density oligonucleotide probes targeted to the junctions between consecutive exons (Johnson et al. 2003). Other designs include the use of probe sets to target bodies and junctions of constitutive and alternative exons (Clark et al. 2002; Pan et al. 2004; Blanchette et al. 2005; Sugnet et al. 2006), as well as the use of bead-based fiber-optic microarray platforms with high detection sensitivity (Yeakley et al. 2002). These designs have facilitated analyses of genome-wide alternative splicing in human, mouse, and chimp (Srinivasan et al. 2005; Boutz et al. 2007; Calarco et al. 2007; Ip et al. 2007; Ni et al. 2007), as well as detection of the global impact of specific splicing factors or environmental stimuli on splicing regulation (Park et al. 2004; Hung et al. 2007; Makeyev et al. 2007; Pleiss et al. 2007a,b). A very high-density “exon array” containing probes in essentially all known and predicted human exons was recently developed by Affymetrix (Gardina et al. 2006). Generally speaking, these array platforms appear able to identify a subset of AS events with high confidence, but have an unknown and probably substantial rate of false negatives.
The systematic identification of RNA targets for different trans-factors can also be achieved by some new approaches such as a cross-linking/immunoprecipitation (CLIP) (Ule et al. 2003), RNP immunoprecipitation (RIP) (Keene et al. 2006; Townley-Tilson et al. 2006), and genomic SELEX (Lorenz et al. 2006). These analyses have the potential to identify regulatory targets of a factor and can be applied genome-wide when coupled with microarray or high-throughput sequencing technologies. Analysis of the target sequences will help to define the sequence determinants of binding, and may also help to identify cooperative or antagonistic relationships between different factors. Since only a subset of binding events confers regulatory activity, it is important to also have evidence of regulation. Such evidence can be obtained from knockout/knockdown or overexpression of the factor, which can be also applied on a genome-wide scale when coupled with isoform-specific microarrays (Blanchette et al. 2005), or with high-throughput sequencing.
SPLICING SIMULATION: PUTTING THE PIECES TOGETHER
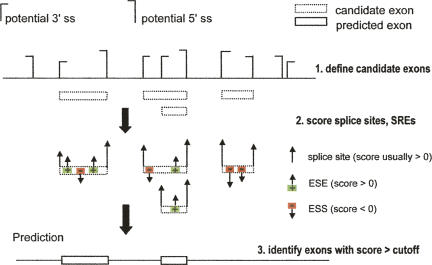
An important long-term goal in the splicing field is to determine a “splicing code:” a set of rules that can predict the splicing pattern of any primary transcript sequence (Fu 2004; Matlin et al. 2005). As large “parts lists” of splicing regulatory cis-elements are identified, and identities and functions of associated trans-acting factors are determined, a natural next step is to assemble the available information into a predictive framework to simulate the recognition of exons and introns that occurs during splicing. An initial approach to this very challenging problem was the development of the general splicing simulation algorithm, ExonScan (Fig. 3; Wang et al. 2004). The goal of splicing simulation is to predict the splicing patterns of transcripts based only on the information that is accessible and known (or strongly implicated) to be recognized by the nuclear pre-mRNA splicing machinery. Thus, in addition to predictive accuracy, faithfulness to the in vivo mechanism is important in splicing simulation, and an important application of splicing simulation is to evaluate how different sequence features contribute to the determination of splicing specificity. This philosophy is different from that of most gene prediction/gene finding algorithms, whose goal is generally to make the most accurate prediction of exon and gene locations using whatever information is available. For example, most current gene finders use reading frame consistency of exons and/or cross-species conservation in their predictions, information that is inaccessible to the spliceosome.
FIGURE 3.
Schematic diagram of a general splicing simulator, ExonScan. (Green boxes) Splicing enhancers, (red boxes) silencers. For simplicity, only ESSs and ESEs are indicated. Step 1, Identify the exon candidate as a pair of splice site (shown as brackets) that is 50–250 bp apart. Step 2, Score the splice sites and splicing enhancers and silencers, and sum up for the score of each exon candidate. Step 3, Determine the threshold and make the final prediction.
The ExonScan algorithm, illustrated in Figure 3, involves scanning the pre-mRNA for pairs of nearby potential 3′ss and 5′ss, and scoring these sites and nearby exonic and intronic SREs. Log-odds scoring is used, which rewards elements based on their statistical enrichment in the relevant location relative to their background frequency in the genome and naturally assigns positive scores to enhancers and negative scores to silencers. The results generated with this algorithm provided important clues to the in vivo function of ESSs (Wang et al. 2004). Such simulation should be improved by use of more refined sets of sequence elements and context-dependent scoring schemes, and by consideration of the functional interactions between different elements, as inferred from experimental assays or from patterns of coevolution (Xiao et al. 2007).
SPLICING REGULATORY NETWORK AS A SUBNETWORK OF GENE REGULATION
The splicing regulatory network is part of a larger network of gene regulation with which it is linked both physically and functionally. Most introns are thought to be spliced cotranscriptionally (de la Mata et al. 2003; Das et al. 2006; Hicks et al. 2006), and SR proteins associate with the C-terminal domain of RNA polymerase II (de la Mata and Kornblihtt 2006; Das et al. 2007). As a downstream event of transcription, AS can be regulated by factors that affect transcription. For example, a mutated RNA polymerase II with a slow elongation rate had been shown to affect the AS of endogenous genes (de la Mata et al. 2003), and the insertion of a transcription-pausing element, MAZ4, into a minigene construct can influence splicing of an alternative exon (Robson-Dixon and Garcia-Blanco 2004). Therefore, it is tempting to speculate that some of the identified SREs could act through effects on transcription, e.g., functioning as a transcription pause site to promote the inclusion of a weak upstream exon. In addition, the whole process of mRNA biosynthesis and processing, including transcription, 5′ capping, splicing, polyadenylation, and transport is extensively coupled, with a number of factors involved in more than one step of this process (Hieronymus and Silver 2004; Kornblihtt et al. 2004). The splicing regulation network should therefore be viewed as a specialized subnetwork of a more general gene regulatory netwok.
PERSPECTIVE
Sorting out the complex network that controls constitutive and alternative splicing represents a major challenge for post-genomic biology. A promising route to such global understanding is a “bottom-up” approach, involving systematic identification of the cis-regulatory components of the network, determination of rules—context dependence or otherwise—for their activity and functional interactions with other elements, and integration of these rules into simulation algorithms representing successive approximations to the splicing code. Beyond the challenges discussed above, another important challenge will be to learn how to generalize the cis-elements, factors, and rules identified in one system (e.g., in one cell type or developmental stage) to others. This will undoubtedly require new experimental approaches that can be readily adapted to a variety of systems.
Given the complexity of splicing regulation, the splicing code will not have the simple tabular form of the genetic code. Instead, it may look more like the U.S. tax code (i.e., IRS form 1040), with a variety of tables and subtables that are applicable in different circumstances, and a certain amount of arithmetic required to obtain the correct answer.
ACKNOWLEDGMENTS
We thank Xinshu Xiao for critical reading of our manuscript. Our research is supported by grants from the NIH and NSF (C.B.B.) and from the Damon Runyon Cancer Research Foundation (Z.W.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.876308.
REFERENCES
- Asai, K., Platt, C., Cochrane, A. Control of HIV-1 env RNA splicing and transport: Investigating the role of hnRNP A1 in exon splicing silencer (ESS3a) function. Virology. 2003;314:229–242. doi: 10.1016/s0042-6822(03)00400-8. [DOI] [PubMed] [Google Scholar]
- Berget, S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blanchette, M., Green, R.E., Brenner, S.E., Rio, D.C. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila . Genes & Dev. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert, F., Baserga, S.J. The long unwinding road of RNA helicases. Mol. Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Boutz, P.L., Stoilov, P., Li, Q., Lin, C.H., Chawla, G., Ostrow, K., Shiue, L., Ares M., Jr, Black, D.L. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno, M., Gelfand, M.S., Spengler, S., Zorn, M., Dubchak, I., Conboy, J.G. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29:2338–2348. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti, E., Muro, A.F., Giombi, M., Gherbassi, D., Iaconcig, A., Baralle, F.E. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti, E., Stuani, C., De Prato, G., Baralle, F.E. SR protein-mediated inhibition of CFTR exon 9 inclusion: Molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 2007;35:4359–4368. doi: 10.1093/nar/gkm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco, J.A., Xing, Y., Caceres, M., Calarco, J.P., Xiao, X., Pan, Q., Lee, C., Preuss, T.M., Blencowe, B.J. Global analysis of alternative splicing differences between humans and chimpanzees. Genes & Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi, M., Zahler, A.M. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., Krainer, A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Wang, J., Zhu, Z., Zhang, M.Q., Krainer, A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascino, I., Fiucci, G., Papoff, G., Ruberti, G. Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J. Immunol. 1995;154:2706–2713. [PubMed] [Google Scholar]
- Chasin, L. Searching for splicing motifs. In: Blencowe B.J., Graveley B.R., editors. Alternative splicing in the postgenomic era. Landes Biosciences; Austin, TX: 2007. pp. 85–106. [Google Scholar]
- Chen, C.D., Kobayashi, R., Helfman, D.M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes & Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, M.Y., Rooke, N., Turck, C.W., Black, D.L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, M.Y., Underwood, J.G., Nikolic, J., Luu, M.H., Black, D.L. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- Clark, T.A., Sugnet, C.W., Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Coulter, L.R., Landree, M.A., Cooper, T.A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B.R. Nuclear mRNA export: Insights from virology. Trends Biochem. Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- D'Souza, I., Schellenberg, G.D. Tau exon 10 expression involves a bipartite intron 10 regulatory sequence and weak 5′ and 3′ splice sites. J. Biol. Chem. 2002;277:26587–26599. doi: 10.1074/jbc.M203794200. [DOI] [PubMed] [Google Scholar]
- Das, R., Dufu, K., Romney, B., Feldt, M., Elenko, M., Reed, R. Functional coupling of RNAP II transcription to spliceosome assembly. Genes & Dev. 2006;20:1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, R., Yu, J., Zhang, Z., Gygi, M.P., Krainer, A.R., Gygi, S.P., Reed, R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- de la Mata, M., Kornblihtt, A.R. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat. Struct. Mol. Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- de la Mata, M., Alonso, C.R., Kadener, S., Fededa, J.P., Blaustein, M., Pelisch, F., Cramer, P., Bentley, D., Kornblihtt, A.R. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Demir, E., Dickson, B.J. fruitless splicing specifies male courtship behavior in Drosophila . Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Dominguez, C., Allain, F.H. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: A novel mode of RNA recognition. Nucleic Acids Res. 2006;34:3634–3645. doi: 10.1093/nar/gkl488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue, C.P., Muratore, C., Wu, J.Y., Kosik, K.S., Wolfe, M.S. Stabilization of the tau exon 10 stem–loop alters pre-mRNA splicing. J. Biol. Chem. 2006;281:23302–23306. doi: 10.1074/jbc.C600143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother, W.G., Yeh, R.F., Sharp, P.A., Burge, C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Fischer, U., Pollard, V.W., Luhrmann, R., Teufel, M., Michael, M.W., Dreyfuss, G., Malim, M.H. Rev-mediated nuclear export of RNA is dominant over nuclear retention and is coupled to the Ran-GTPase cycle. Nucleic Acids Res. 1999;27:4128–4134. doi: 10.1093/nar/27.21.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X.D. Towards a splicing code. Cell. 2004;119:736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- Gao, K., Masuda, A., Matsuura, T., Ohno, K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn073. PMID: 18285363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardina, P.J., Clark, T.A., Shimada, B., Staples, M.K., Yang, Q., Veitch, J., Schweitzer, A., Awad, T., Sugnet, C., Dee, S., et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren, A., Ram, O., Amit, M., Keren, H., Lev-Maor, G., Vig, I., Pupko, T., Ast, G. Comparative analysis identifies exonic splicing regulatory sequences—The complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R., Maniatis, T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R., Hertel, K.J., Maniatis, T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, K., Yeo, G., An, P., Burge, C.B., Grabowski, P.J. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings, M.L., Wilson, C.M., Munroe, S.H. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. RNA. 2001;7:859–874. doi: 10.1017/s1355838201002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel, K.J., Graveley, B.R. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem. Sci. 2005;30:115–118. doi: 10.1016/j.tibs.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Hicks, M.J., Yang, C.R., Kotlajich, M.V., Hertel, K.J. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4:e147. doi: 10.1371/journal.pbio.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus, H., Silver, P.A. A systems view of mRNP biology. Genes & Dev. 2004;18:2845–2860. doi: 10.1101/gad.1256904. [DOI] [PubMed] [Google Scholar]
- Hiller, M., Zhang, Z., Backofen, R., Stamm, S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig, A., Auboeuf, D., Parker, M.M., O'Malley, B.W., Berget, S.M. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell. Biol. 2002;22:5698–5707. doi: 10.1128/MCB.22.16.5698-5707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House, AE., Lynch, KW. Regulation of alternative splicing: More than just the ABCs. J. Biol. Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- Huh, G.S., Hynes, R.O. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes & Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- Hui, J., Hung, L.H., Heiner, M., Schreiner, S., Neumuller, N., Reither, G., Haas, S.A., Bindereif, A. Intronic CA-repeat and CA-rich elements: A new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, L.H., Heiner, M., Hui, J., Schreiner, S., Benes, V., Bindereif, A. Diverse roles of hnRNP L in mammalian mRNA processing: A combined microarray and RNAi analysis. RNA. 2007;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim el, C., Schaal, T.D., Hertel, K.J., Reed, R., Maniatis, T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl. Acad. Sci. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip, J.Y., Tong, A., Pan, Q., Topp, J.D., Blencowe, B.J., Lynch, K.W. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo, J.M., Majos, N., Bonnal, S., Martinez, C., Castelo, R., Guigo, R., Bilbao, D., Valcarcel, J. Regulation of fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Jensen, K.B., Musunuru, K., Lewis, H.A., Burley, S.K., Darnell, R.B. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl. Acad. Sci. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y., Suzuki, H., Maegawa, S., Endo, H., Sugano, S., Hashimoto, K., Yasuda, K., Inoue, K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., Shoemaker, D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Jurica, M.S., Moore, M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kabat, J.L., Barberan-Soler, S., McKenna, P., Clawson, H., Farrer, T., Zahler, A.M. Intronic alternative splicing regulators identified by comparative genomics in nematodes. PLoS Comput. Biol. 2006;2:e86. doi: 10.1371/journal.pcbi.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka, A., Muhlemann, O., Akusjarvi, G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- Kashima, T., Rao, N., Manley, J.L. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc. Natl. Acad. Sci. 2007;104:3426–3431. doi: 10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, J.D., Komisarow, J.M., Friedersdorf, M.B. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Kol, G., Lev-Maor, G., Ast, G. Human–mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum. Mol. Genet. 2005;14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M., Query, C.C. Insights into the mechanisms of splicing: More lessons from the ribosome. Genes & Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- Kornblihtt, A.R., de la Mata, M., Fededa, J.P., Munoz, M.J., Nogues, G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd, A.N., Cooper, T.A. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3:reviews0008.1–0008.16. doi: 10.1186/gb-2002-3-11-reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.G. RH70, a bidirectional RNA helicase, co-purifies with U1snRNP. J. Biol. Chem. 2002;277:39679–39683. doi: 10.1074/jbc.C200337200. [DOI] [PubMed] [Google Scholar]
- Lee, J.A., Xing, Y., Nguyen, D., Xie, J., Lee, C.J., Black, D.L. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L.P., Burge, C.B. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl. Acad. Sci. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S.R., Hertel, K.J. Commitment to splice site pairing coincides with A complex formation. Mol. Cell. 2004;15:477–483. doi: 10.1016/j.molcel.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Sharp, P.A. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol. Cell. Biol. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.X., Zhang, M., Krainer, A.R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes & Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.X., Chew, S.L., Cartegni, L., Zhang, M.Q., Krainer, A.R. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 2000;20:1063–1071. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, C., von Pelchrzim, F., Schroeder, R. Genomic systematic evolution of ligands by exponential enrichment (Genomic SELEX) for the identification of protein-binding RNAs independent of their expression levels. Nat. Protoc. 2006;1:2204–2212. doi: 10.1038/nprot.2006.372. [DOI] [PubMed] [Google Scholar]
- Makeyev, E.V., Zhang, J., Carrasco, M.A., Maniatis, T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci, R., Baralle, F.E., Romano, M. Complex splicing control of the human Thrombopoietin gene by intronic G runs. Nucleic Acids Res. 2007;35:132–142. doi: 10.1093/nar/gkl965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin, A.J., Clark, F., Smith, C.W. Understanding alternative splicing: Toward a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- McCullough, A.J., Berget, S.M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough, A.J., Berget, S.M. An intronic splicing enhancer binds U1 snRNPs to enhance splicing and select 5′ splice sites. Mol. Cell. Biol. 2000;20:9225–9235. doi: 10.1128/mcb.20.24.9225-9235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino, E.J., Wilkinson, K.A., Coughlan, J.L., Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- Min, H., Turck, C.W., Nikolic, J.M., Black, D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes & Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- Minovitsky, S., Gee, S.L., Schokrpur, S., Dubchak, I., Conboy, J.G. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modafferi, E.F., Black, D.L. Combinatorial control of a neuron-specific exon. RNA. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata, S., Kawamoto, S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., Sakamoto, H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- Nasim, F.U., Hutchison, S., Cordeau, M., Chabot, B. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA. 2002;8:1078–1089. doi: 10.1017/s1355838202024056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, J.Z., Grate, L., Donohue, J.P., Preston, C., Nobida, N., O'Brien, G., Shiue, L., Clark, T.A., Blume, J.E., Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen, T.W. The spliceosome: The most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- Pagani, F., Buratti, E., Stuani, C., Bendix, R., Dork, T., Baralle, F.E. A new type of mutation causes a splicing defect in ATM. Nat. Genet. 2002;30:426–429. doi: 10.1038/ng858. [DOI] [PubMed] [Google Scholar]
- Pan, Q., Shai, O., Misquitta, C., Zhang, W., Saltzman, A.L., Mohammad, N., Babak, T., Siu, H., Hughes, T.R., Morris, Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Park, J.W., Parisky, K., Celotto, A.M., Reenan, R.A., Graveley, B.R. Identification of alternative splicing regulators by RNA interference in Drosophila . Proc. Natl. Acad. Sci. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr, N., Prawitt, D., Kirschfink, M., Schroff, C., Knuf, M., Habermehl, P., Mannhardt, W., Zepp, F., Fairbrother, W., Loos, M., et al. Linking C5 deficiency to an exonic splicing enhancer mutation. J. Immunol. 2005;174:4172–4177. doi: 10.4049/jimmunol.174.7.4172. [DOI] [PubMed] [Google Scholar]
- Pleiss, J.A., Whitworth, G.B., Bergkessel, M., Guthrie, C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell. 2007a;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss, J.A., Whitworth, G.B., Bergkessel, M., Guthrie, C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007b;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, V.W., Malim, M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Pongoski, J., Asai, K., Cochrane, A. Positive and negative modulation of human immunodeficiency virus type 1 Rev function by cis and trans regulators of viral RNA splicing. J. Virol. 2002;76:5108–5120. doi: 10.1128/JVI.76.10.5108-5120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzoli, U., Sironi, M. Silencers regulate both constitutive and alternative splicing events in mammals. Cell. Mol. Life Sci. 2005;62:1579–1604. doi: 10.1007/s00018-005-5030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R., Cheng, H. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Rio, D.C. Regulation of Drosophila P element transposition. Trends Genet. 1991;7:282–287. doi: 10.1016/0168-9525(91)90309-E. [DOI] [PubMed] [Google Scholar]
- Robberson, B.L., Cote, G.J., Berget, S.M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson-Dixon, N.D., Garcia-Blanco, M.A. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J. Biol. Chem. 2004;279:29075–29084. doi: 10.1074/jbc.M312747200. [DOI] [PubMed] [Google Scholar]
- Rothrock, C., Cannon, B., Hahm, B., Lynch, K.W. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Rothrock, C.R., House, A.E., Lynch, K.W. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe, N.J., de Souza, S.J. Sequence features responsible for intron retention in human. BMC Genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal, T.D., Maniatis, T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub, M.C., Lopez, S.R., Caputi, M. Members of the heterogeneous nuclear ribonucleoprotein H family activate splicing of an HIV-1 splicing substrate by promoting formation of ATP-dependent spliceosomal complexes. J. Biol. Chem. 2007;282:13617–13626. doi: 10.1074/jbc.M700774200. [DOI] [PubMed] [Google Scholar]
- Sharma, S., Falick, A.M., Black, D.L. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, S.D., Chakrabarti, S., Ghosh, G., Krainer, A.R. Deletion of the N-terminus of SF2/ASF permits RS-domain-independent pre-mRNA splicing. PLoS ONE. 2007;2:e854. doi: 10.1371/journal.pone.0000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Green, M.R. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes & Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Kan, J.L., Green, M.R. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Shin, C., Manley, J.L. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- Shin, C., Manley, J.L. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- Shin, C., Feng, Y., Manley, J.L. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- Siebel, C.W., Fresco, L.D., Rio, D.C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: Multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes & Dev. 1992;6:1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- Siebel, C.W., Kanaar, R., Rio, D.C. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes & Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- Siebel, C.W., Admon, A., Rio, D.C. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes & Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- Singh, N.K., Singh, N.N., Androphy, E.J., Singh, R.N. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Ast, G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Shemesh, R., Cohen, Y., Basechess, O., Ast, G., Shamir, R. A non-EST-based method for exon-skipping prediction. Genome Res. 2004;14:1617–1623. doi: 10.1101/gr.2572604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, K., Shiue, L., Hayes, J.D., Centers, R., Fitzwater, S., Loewen, R., Edmondson, L.R., Bryant, J., Smith, M., Rommelfanger, C., et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–359. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Stadler, M.B., Shomron, N., Yeo, G.W., Schneider, A., Xiao, X., Burge, C.B. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet. 2006;2:e191. doi: 10.1371/journal.pgen.0020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D.A., Carlo, T., Berget, S.M. Architectural limits on split genes. Proc. Natl. Acad. Sci. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus, C.M., Madsen, J.M. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV Res. 2006;4:43–55. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- Sugnet, C.W., Srinivasan, K., Clark, T.A., O'Brien, G., Cline, M.S., Wang, H., Williams, A., Kulp, D., Blume, J.E., Haussler, D., et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Chasin, L.A. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, H., Kole, R. Selection of novel exon recognition elements from a pool of random sequences. Mol. Cell. Biol. 1995;15:6291–6298. doi: 10.1128/mcb.15.11.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley-Tilson, W.H., Pendergrass, S.A., Marzluff, W.F., Whitfield, M.L. Genome-wide analysis of mRNAs bound to the histone stem–loop binding protein. RNA. 2006;12:1853–1867. doi: 10.1261/rna.76006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule, J., Jensen, K.B., Ruggiu, M., Mele, A., Ule, A., Darnell, R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Ule, J., Stefani, G., Mele, A., Ruggiu, M., Wang, X., Taneri, B., Gaasterland, T., Blencowe, B.J., Darnell, R.B. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Underwood, J.G., Boutz, P.L., Dougherty, J.D., Stoilov, P., Black, D.L. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker, R.B., Berglund, J.A. A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing. Genome Res. 2007;17:1023–1033. doi: 10.1101/gr.6017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.J., Baraniak, A.P., Sessions, O.M., Mauger, D., Moskowitz, E., Garcia-Blanco, M.A. Characterization of the intronic splicing silencers flanking FGFR2 exon IIIb. J. Biol. Chem. 2005;280:14017–14027. doi: 10.1074/jbc.M414492200. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Rolish, M.E., Yeo, G., Tung, V., Mawson, M., Burge, C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Xiao, X., Van Nostrand, E., Burge, C.B. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf, M.B., Berglund, J.A. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C., Lührmann, R. Spliceosome structure and function. In: Gesteland R.F., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 369–400. [Google Scholar]
- Xiao, X., Wang, Z., Jang, M., Burge, C.B. Coevolutionary networks of splicing cis-regulatory elements. Proc. Natl. Acad. Sci. 2007;104:18583–18588. doi: 10.1073/pnas.0707349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., Black, D.L. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410:936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- Yeakley, J.M., Fan, J.B., Doucet, D., Luo, L., Wickham, E., Ye, Z., Chee, M.S., Fu, X.D. Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol. 2002;20:353–358. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- Yeo, G., Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Yeo, G., Hoon, S., Venkatesh, B., Burge, C.B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl. Acad. Sci. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, G.W., Van Nostrand, E., Holste, D., Poggio, T., Burge, C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, G.W., Nostrand, E.L., Liang, T.Y. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.H., Chasin, L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes & Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.M. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Mayeda, A., Krainer, A.R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]