Abstract
The peptidyl transferase (PT) center of the ribosome catalyzes two nucleophilic reactions, peptide bond formation between aminoacylated tRNA substrates and, together with release factor, peptide release. Structure and function of the PT center are modulated by binding of aminoacyl-tRNA or release factor, thus providing the basis for the specificity of catalysis. Another way by which the function of the PT center is controlled is signaling from the peptide exit tunnel. The SecM nascent peptide induces ribosome stalling, presumably by inhibition of peptide bond formation. Similarly, the release factor-induced hydrolytic activity of the PT center can be suppressed by the TnaC nascent peptide contained in the exit tunnel. Thus, local and long-range conformational rearrangements can lead to changes in the reaction specificity and catalytic activity of the PT center.
Keywords: ribosome; peptidyl transferase center; peptide bond formation; peptide release; SecM, TnaC
INTRODUCTION
The catalytic site of the ribosome, the peptidyl transferase (PT) center on the large 50S ribosomal subunit, facilitates two chemical reactions during synthesis of a protein, formation of peptide bonds and hydrolysis of peptidyl-tRNA (pept-tRNA). During formation of the peptide bond, nucleophilic attack of aminoacyl-tRNA (aa-tRNA) in the A site on the carbonyl carbon of P-site bound pept-tRNA results in A-site bound pept-tRNA that is extended by one amino acid residue and deacylated tRNA in the P site. Catalysis of peptidyl transfer is entropically driven (Sievers et al. 2004) and requires the active participation of the 2′-OH group of the 3′-terminal adenosine residue (A76) of the P-site substrate (Weinger et al. 2004). Hydrolysis of the ester bond in P-site pept-tRNA takes place during termination of protein synthesis. Chemically, the reactions of peptide bond formation and pept-tRNA hydrolysis are similar, as they involve the nucleophilic attack of the A-site substrate (aa-tRNA or H2O, respectively) on the P-site substrate pept-tRNA. However, while peptidyl transfer is performed by the ribosome without the help of accessory factors, hydrolysis of pept-tRNA is induced by release factor (RF), RF1 or RF2 in bacteria, or eRF1 in eukaryotes. The RF appears to make two contributions to catalysis, one leading to a general activation of the catalytic center induced by conformational rearrangements at the active site, and the other resulting in the specific selection of water as nucleophile (Fig. 1; Schmeing et al. 2005b; Shaw and Green 2007).
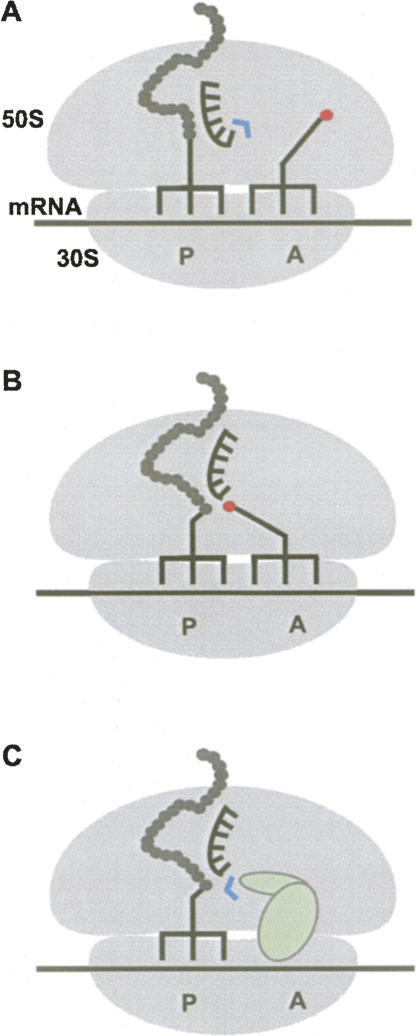
FIGURE 1.
Principles of modulating the activity of the PT center. (A) When the A site on the 50S subunit is empty, for example prior to accommodation of aa-tRNA, water (blue) is prevented from nucleophilic attack on the pept-tRNA by a specific conformation of active-site residues (black spiked element). (B) Upon accommodation of aa-tRNA, the PT center rearranges into an active conformation and positions pept-tRNA for the nucleophilic attack of aa-tRNA (nucleophile is indicated red). (C) Hydrolysis of pept-tRNA during termination of protein synthesis is catalyzed by release factor (pale green), which may induce an active conformation of the PT center similar to that brought about by aa-tRNA. Additionally, the release factor orients and/or activates the hydrolytic water molecule.
PEPTIDE BOND FORMATION AND PEPTIDE RELEASE
Bacterial 50S subunits are composed of two rRNA molecules, 23S rRNA and 5S rRNA, and more than 30 proteins. Based on cross-linking studies, the PT center was identified in domain V of 23S rRNA (Noller 1991). High-resolution crystal structures of archaeal ribosomes revealed that the PT center is composed of RNA only (Nissen et al. 2000), which lent support to earlier biochemical evidence on the key role of rRNA, rather than proteins, in the catalysis of peptide bond formation (Noller et al. 1992). The acceptor ends of A- and P-site tRNAs are located in a cleft of the 50S subunit interface side (Yusupov et al. 2001; Hansen et al. 2002). Their universally conserved 3′-terminal CCA residues are oriented and held in place in the A and P sites by interactions with 23S rRNA. The conserved bases A2451, U2506, U2585, C2452, and A2602 are at the core of the PT center (Bashan et al. 2003; Schmeing et al. 2005a,b). The oxyanion of the tetrahedral reaction intermediate seems to be stabilized by a water molecule that is positioned by interactions with nucleotides A2602 and U2584 (Schmeing et al. 2005a). Generally, the rRNA backbone in the PT center is found in very similar conformations in 50S subunits from different organisms and in different ribosomal complexes. However, small-scale reorientations do occur, and the movement of some particularly flexible nucleotides may have implications for the catalytic mechanism.
Peptide bond formation on the ribosome is fast, with an estimated rate of >300 sec−1 (Bieling et al. 2006). Studying the enzymology of peptidyl transfer revealed the contribution of different catalytic strategies and the catalytic role of ribosomal groups at the active site (for review, see Beringer and Rodnina 2007b). The combined evidence strongly supports the notion that catalysis is predominantly entropic, suggesting an important contribution of such strategies as substrate positioning in the active site, desolvation, and electrostatic shielding (Sievers et al. 2004; Trobro and Åqvist 2005). The 23S rRNA bases or other ionizing groups of the ribosome are not utilized as general acids/bases to any significant extent (Bieling et al. 2006). The presence of the 2′-OH of A76 of pept-tRNA in the P site is crucial for the reaction (Weinger et al. 2004). It is the only group found within hydrogen-bonding distance of the attacking nucleophile in the transition state analogs (Schmeing et al. 2005a). Hydrogen bonding between the 2′-OH of A76 and the nucleophilic α-NH2 group may help to position the nucleophile. Another attractive explanation for the importance of 2′-OH of A76 of the P-site tRNA is its participation in a concerted proton shuttle that bridges the attacking α-NH2 group and the leaving 3′ oxygen of the P-site tRNA (Das et al. 1999; Dorner et al. 2002; Changalov et al. 2005; Schmeing et al. 2005a; Trobro and Åqvist 2005) The main role of ribosomal residues in the active site appears to provide a preorganized hydrogen-bond network that stabilizes the charged transition state (Schmeing et al. 2005a; Trobro and Åqvist 2005). Additionally, they seem to protect pept-tRNA from premature hydrolysis by water when the A site is not filled by aa-tRNA (Fig. 1A; Schmeing et al. 2005b). Crystallographic analysis indicates that only upon interaction of the CCA end of the A-site substrate with the ribosomal A loop do residues G2583, U2506, and U2585 of 23S rRNA rearrange and allow the efficient attack of the nucleophile (Fig. 1B; Schmeing et al. 2005b).
The catalytic contribution of individual active-site residues was assessed by introducing mutations at different positions of the PT center and testing the effects of replacements on the rate of peptide bond formation. A number of 23S rRNA bases in the so-called “inner shell” of the active site (positions A2541, U2506, U2585, and A2602) (Polacek et al. 2001; Thompson et al. 2001; Katunin et al. 2002; Hesslein et al. 2004; Youngman et al. 2004; Beringer et al. 2005), or G2447 adjacent to it (Bayfield et al. 2001; Polacek et al. 2001; Thompson et al. 2001; Beringer et al. 2003), and the noncanonical, ionized base-pair A2450–C2063 (Bayfield et al. 2004; Hesslein et al. 2004) were mutated and the effects of the replacements were examined. Strikingly, none of the single-mutant ribosomes showed any defect in the rate of peptide bond formation, provided that the interaction between C75 of the A-site tRNA and G2553 in the A loop was intact (Youngman et al. 2004; Brunelle et al. 2006). In contrast, disruption of the base-pairing between tRNA C75 and G2553 in 23S rRNA, or the use of puromycin as an A-site substrate, which does not form this interaction, leads to sensitivity of the peptidyl transfer rate toward base substitutions in the active site and toward changes in pH (Katunin et al. 2002; Brunelle et al. 2006; Beringer and Rodnina 2007a). This suggests that a predominant part of the overall catalysis arises from the correct positioning of the substrates, mediated by 23S rRNA–tRNA interactions. The functions of mutated active-site residues can be taken over by other nucleotides or water molecules.
Hydrolysis of pept-tRNA during termination of protein synthesis is chemically more challenging than peptidyl transfer, because of the weaker nucleophilicity of water relative to the primary amine of aa-tRNA. The residues at the PT center appear to play a more active role for RF-mediated peptide release, compared to peptidyl transfer. Mutations of inner shell residues lead to large or even dramatic (∼9400-fold) effects in the rate of peptide release (Youngman et al. 2004). RF1 and RF2 each have a universally conserved GGQ sequence (Frolova et al. 1999) that is located in a loop that enters the PT center (Klaholz et al. 2003; Rawat et al. 2003). In the active site, the GGQ sequence of RF2 faces A76 of the P-site tRNA and is surrounded by nucleotides C2063, A2451, U2506, and A2602 of 23S rRNA (Petry et al. 2005). The closest residues are A2451 and A2602, the latter being the nucleotide most essential for hydrolysis, as indicated by mutational data (Polacek et al. 2003; Youngman et al. 2004). In the absence of release factor, the ribosome protects pept-tRNA from hydrolysis by precluding the access of water to positions from where it could attack the ester group (Fig. 1A). The release factors may promote conformational rearrangements of inner-shell nucleotides, in particular U2585 (Schmeing et al. 2005b), leading to a general stimulation of the catalytic activity (Shaw and Green 2007). The rearrangement would move the ester group of pept-tRNA into a more exposed position and allow the attack of a water molecule (Fig. 1C). Such a scenario is consistent with the observation that mutations of U2585 lead to a decreased rate of RF-catalyzed peptide release (Polacek et al. 2003; Youngman et al. 2004). In addition, release factors appear to specifically promote pept-tRNA hydrolysis by positioning the nucleolytic water molecule in the active site via hydrogen bonding to the conserved glutamine in the GGQ motif, thus achieving specificity of hydrolysis versus aminolysis (Song et al. 2000; Shaw and Green 2007; Trobro and Aqvist 2007).
MODULATION OF PEPTIDYL TRANSFERASE ACTIVITY
The PT center has to deal with aa-tRNA and pept-tRNA differing in size, charge, and polarity of the amino acid residue at the tRNA 3′ end. When peptidyl transfer is studied using native substrates, i.e., full-length pept-tRNA and aa-tRNA, the rate of peptide bond formation is determined by the preceding rate-limiting step of aa-tRNA binding to the A site (accommodation), which is relatively slow (in the range of 10 sec−1) (Pape et al. 1998). Given that the intrinsic rate of peptide bond formation is much faster than that, estimated to be > 300 sec−1 (Bieling et al. 2006), moderate changes in the rate of the chemistry step are likely to be obscured by the slow binding step. In model reactions in solution, the rate of aminolysis of different aa-tRNA varies significantly with the identity of the amino acid (Weber and Orgel 1979), and studies on short substrate fragments in the 50S-catalyzed reaction indicated that such differences may be maintained on the ribosome (Krayevsky and Kukhanova 1979). The rate of the reaction with the short aa-tRNA analog puromycin, which does not need to accommodate, varies within a factor of 50–100 depending on the length of the peptidyl moiety of the P-site pept-tRNA, the C-terminal amino acid of the peptide, or the identity of the tRNA, and ranges from about 1 sec−1 with fMet-tRNAfMet in the P site, 10 – 20 sec−1 with di- and tri-peptidyl-tRNAs fMetPhe-tRNAPhe and fMetPhePhe-tRNAPhe (Katunin et al. 2002; Brunelle et al. 2006), and 50 sec−1 with fMetAlaAsnMetPheAla-tRNAAla (Katunin et al. 2002). For the A-site substrate, a strong decrease in nucleophilicity due to replacement of the α-NH2 group with an OH group in either puromycin or aa-tRNA dramatically reduces the rate of peptidyl transfer (Katunin et al. 2002; Bieling et al. 2006).
Interestingly, Pro-tRNAPro seems to be a poor A-site substrate (Rychlik et al. 1970), probably because of its low nucleophilicity and because of the steric constraints a proline may exert on the geometry of the substrates and/or the conformation of active-site residues. Intriguingly, A-site-bound Pro-tRNAPro plays a crucial role in the inhibition of translation elongation by the SecM protein. SecM is a 170 amino acid protein that regulates the synthesis of protein SecA, which is part of the protein secretion machinery of the bacterial cell and is encoded by the same operon downstream of the SecM ORF (Oliver et al. 1998; Nakatogawa and Ito 2002). Translation of SecM mRNA stops at position Pro166 when the amount of SecA present in the cell is insufficient to promote the egression of the nascent SecM chain out of the ribosomal exit tunnel, preventing the translation of the downstream ORF coding for SecA. It was shown that ribosomal stalling in the case of SecM can be reconstituted in an in vitro translation system that contains ribosomes, tRNAs, translation factors, and aa-tRNA synthetases (Nakatogawa and Ito 2002). Several signals that are required for translation inhibition have been identified (Fig. 2A). First, the mRNA specifying for the internal peptide FxxxxWIxxxxGIRAGP, composed of residues 150–166 of the native SecM protein, autonomously leads to inhibition of elongation, also in the context of different reporter constructs (Nakatogawa and Ito 2002). The critical Trp155 residue in the nascent peptide is located 12 amino acids away from the PT center when ribosome stalling is induced, and it may be located in the immediate vicinity of the narrowest part of the exit tunnel, located ∼25 Å away from the active site. FRET results suggested that a functional stalling-competent SecM nascent peptide adopts a compact, rather than extended, conformation in the exit tunnel (Woolhead et al. 2006). The compaction of the nascent chain is necessary, though not sufficient, for ribosome stalling (Woolhead et al. 2006), and the distance between the conserved Trp155 residue and the C terminus of pept-tRNA appears to be critical for signaling from the tunnel toward the active site (Nakatogawa and Ito 2002). These observations suggest an important role of the nascent chain itself during signaling. The critical Pro166 residue is not incorporated into the nascent chain in stalled SecM-ribosome complexes, but unreacted Pro-tRNAPro is bound to the A site (Garza-Sanchez et al. 2006; Muto et al. 2006). The interpretation of these findings is that peptide bond formation between Pro-tRNAPro and FxxxxWIxxxxGIRAG-tRNAGly is inhibited in such complexes. The effect seems to be specific for Pro-tRNAPro, because puromycin can efficiently attack P-site pept-tRNA carrying that peptide, albeit with an unknown rate (Muto et al. 2006). It has been proposed that the presence of Pro-tRNAPro in the PT center predisposes the ribosome to stall upon synthesis of SecM (Woolhead et al. 2006). Such a model would imply that a slow reaction involving Pro-tRNAPro gives the signal coming from the tunnel sufficient time to trigger further conformational changes in the active site that lead to complete inhibition of the reaction.
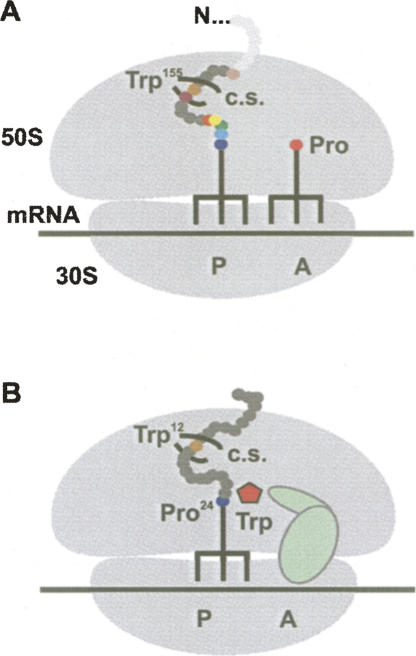
FIGURE 2.
Two examples of nascent peptides modulating the activity of the PT center. (A) Inhibition of peptide bond formation in the stalled SecM-ribosome complex requires the constriction site (c.s.) in the exit tunnel, Pro-tRNAPro bound to the A site, and several essential residues (colored) within the inhibiting peptide (dark gray). (B) Hydrolysis of TnaC-tRNA catalyzed by RF2 (pale green) is inhibited by the combined effects of the nascent TnaC peptide (dark gray) with two essential residues (Trp12 and Pro24) and the inhibition inducer free tryptophan (red pentagon), which binds presumably to the A site of the PT center.
Several residues connecting the tunnel constriction site and the PT center are candidate mediators of peptidyl transferase regulation. The tunnel constriction is composed of potentially flexible residues of ribosomal proteins L4 and L22, surrounded by a 23S rRNA region that is particularly rich in A residues. Mutations of Gly91 and Ala93 in L22, both pointing into the tunnel at the constriction site (Ban et al. 2000), relieve the stalling effect of the SecM nascent peptide (Nakatogawa and Ito 2002). Further suppressors of SecM signaling are A2058 and A749–A753 of 23S rRNA, which are located around the constriction site 20–25 Å away from the PT center at opposite sides of the exit tunnel (Nakatogawa and Ito 2002). Base–backbone interactions connect A2058 with G2505 and G2505 with U2506. The latter nucleotide belongs to the inner-shell layer of bases at the active site and appears to move upon binding of aa-tRNA substrate to the A site (Schmeing et al. 2005b). Furthermore, MD simulations indicate that U2506 can adopt a number of energetically equivalent orientations, suggesting its conformational flexibility (Trobro and Åqvist 2006). Thus, one may speculate that the signal leading to inhibition of peptidyl transfer upon synthesis of SecM at least in part may be transmitted through a 23S rRNA-mediated pathway that would target U2506. Based on cryo-EM data, a model for the transmission of the SecM signal toward the PT center via a cascade of rRNA rearrangements has been proposed (Mitra et al. 2006). Nucleotides in the PT center itself have not been identified as suppressors, most likely because mutations of most of these residues confer lethal phenotype. Experiments on ribosomes carrying lethal mutation at U2506 and/or G2505 using recently developed tagging systems (Leonov et al. 2003; Hesslein et al. 2004; Youngman and Green 2005) could clarify the role of these nucleotides in the communication of the inhibitory activity of the SecM nascent chain. The specificity of signaling may require that all three signals—compaction of the nascent chain, Pro-tRNAPro bound to the A-site substrate, and 23S rRNA rearrangements—are present simultaneously to trigger complete inhibition of peptidyl transfer activity.
MODULATION OF THE HYDROLYTIC ACTIVITY OF THE PT CENTER
The other activity of the PT center, hydrolysis of pept-tRNA, can also be modulated by nascent peptide. In Escherichia coli, the 24-residue-long leader peptide TnaC, together with the inducer free tryptophan, regulates the attenuated expression of the tna operon that encodes tryptophan-metabolizing enzymes. The mRNA region coding for the leader peptide is followed by a UGA stop codon that is read by RF2. At high concentration of free tryptophan, hydrolysis of the nascent TnaC-tRNAPro is inhibited, leading to a stalled ribosomal complex, which, in turn, blocks the access of the transcription terminator Rho to its binding sites on the mRNA so that the leader peptide and, subsequently, the Trp-metabolizing enzymes are synthesized (Gong and Yanofsky 2001, 2002). When the concentration of tryptophan is low, hydrolysis of TnaC-tRNAPro and ribosome recycling take place, thus allowing Rho-mediated transcription termination, resulting in expression of only the leader peptide, and not the downstream enzymes (Gong and Yanofsky 2002; Gong et al. 2007). The main features of TnaC and of the translating ribosome required for inhibition of the hydrolytic activity have been identified (Fig. 2B). As in the case of the SecM-induced ribosome stalling, interactions between the TnaC nascent peptide and the constriction site in the ribosomal tunnel are important for inhibition of termination. Crucial residues in the nascent peptide are Trp12 and, intriguingly, the C-terminal Pro24, esterified to the P-site tRNA (Stewart and Yanofsky 1986; Gollnick and Yanofsky 1990). Trp12 in TnaC-tRNA is responsible for changes in the accessibility to chemical modification of residue A788 in vicinity of the constriction site, and the neighboring Lys11 residue in the TnaC nascent chain can be cross-linked to A750 (Cruz-Vera et al. 2005), i.e., the same region in 23S rRNA as involved in SecM signaling. Trp12 may thus contribute to recognition of the nascent peptide by elements of the tunnel, similarly to Trp155 of SecM. Because the spacing between Trp12 and the active site is important, an involvement of the nascent peptide itself in the signaling process appears likely (Gong and Yanofsky 2002). On the ribosome, Lys90 of L22 and U2609 of 23S rRNA are essential (Cruz-Vera et al. 2005). Lys90 may interact with Trp12, while U2609 is located more closely to the PT center (∼15 Å), and thus may be involved in communication rather than recognition (Cruz-Vera et al. 2005). Although both TnaC- and SecM-mediated signaling are likely to involve ribosomal RNA or protein residues at the tunnel, the sets of mutants that affect the respective process only partially overlap. Generally, the region of the tunnel involved in TnaC recognition seems to be located somewhat nearer to the PT center than in the SecM case. This may reflect a difference in the conformation of the nascent chain or in the location of the parts of the nascent chain that interact with the tunnel.
A fundamental difference between SecM and TnaC signaling is that the stalled TnaC–ribosome complex creates a binding site for the inducer, free tryptophan, in the active site (Gong and Yanofsky 2002). The changes in ribosome structure that result from the interaction of the nascent peptide with ribosomal residues forming the tunnel, especially the interactions involving Trp12 of TnaC, seem to be important for the formation of the tryptophan binding site and for inhibition of hydrolysis (Cruz-Vera et al. 2005, 2006, 2007). The exact location of the binding site of free tryptophan is not known, but its binding changes the accessibility for chemical modification of A2572 (Cruz-Vera et al. 2006), the residue which may act as a sensor of conformational changes of the active site (Beringer et al. 2005). Trp-tRNATrp in the A site can substitute for tryptophan as an inducer and inhibit peptidyl transfer, suggesting that the tryptophanyl moiety of Trp-tRNATrp and free tryptophan may bind to the same site (Gong and Yanofsky 2002). Tryptophan binding also interferes with peptidyl transfer from TnaC-tRNAPro to A site-bound puromycin, possibly because tryptophan and puromycin may compete for the same binding site (Cruz-Vera et al. 2006). Peptidyl transfer of TnaC to Ile-tRNAIle is inhibited when an mRNA construct is used with an AUA codon corresponding to a rare Ile-tRNAIle introduced instead of the stop codon (Cruz-Vera et al. 2006). The inhibition of peptidyl transfer to Ile-tRNAIle (AUA) depends on the presence of Trp12 in the TnaC nascent chain (Cruz-Vera et al. 2006). The decoding of an AUA codon by rare Ile-tRNAIle is expected to be slow, and the adjustment of the 3′ end of the tRNA in the PT center may be impaired once the interaction of TnaC with the exit tunnel creates the tryptophan binding site. However, peptidyl transferase activity per se appears not to be inhibited, because TnaC is efficiently transferred to Ile-tRNAIle on a more common—and presumably rapidly decoded—AUU codon (Cruz-Vera et al. 2006).
In the cell, hydrolysis of TnaC-tRNAPro depends on the presence of RF2. The inhibition of hydrolysis at high concentration of tryptophan could occur at any of the steps during RF2-dependent termination. Conformational changes induced by the concerted action of the TnaC nascent chain and tryptophan may interfere with the correct binding of the GGQ motif of RF2 in the active site, with conformational changes of RF2 and the ribosome essential for efficient catalysis of hydrolysis (Youngman et al. 2007) or with the correct positioning of the hydrolytic water molecule.
The involvement of nascent protein chains in modulation of ribosome function may represent a fairly widespread mechanism of regulated gene expression. In several bacterial antibiotic resistance operons, the synthesis of a nascent leader peptide leads to stalled ribosomal complex in presence of the antibiotic (Lovett and Rogers 1996). Similarly, in fungi, certain nascent peptides involved in amino acid metabolism—together with their respective amino acids—seem to target termination or elongation of translation (Tenson and Ehrenberg 2002). In mammalian cells, the protein gp48 of the cytomegalovirus inhibits its own termination (Cao and Geballe 1996). These examples indicate that modulation of the activity of the PT center may allow for a flexible and rapid regulation of gene expression.
ACKNOWLEDGMENTS
I thank M.V. Rodnina and W. Wintermeyer for discussions and critical reading of the manuscript and H.-J. Lipps for financial support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.980308.
REFERENCES
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bashan, A., Agmon, I., Zarivach, R., Schluenzen, F., Harms, J., Berisio, R., Bartels, H., Franceschi, F., Auerbach, T., Hansen, H.A., et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- Bayfield, M.A., Dahlberg, A.E., Schulmeister, U., Dorner, S., Barta, A. A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc. Natl. Acad. Sci. 2001;98:10096–10101. doi: 10.1073/pnas.171319598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayfield, M.A., Thompson, J., Dahlberg, A.E. The A2453–C2499 wobble base pair in Escherichia coli 23S ribosomal RNA is responsible for pH sensitivity of the peptidyltransferase active site conformation. Nucleic Acids Res. 2004;32:5512–5518. doi: 10.1093/nar/gkh888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer, M., Rodnina, M.V. Importance of tRNA interactions with 23S rRNA for peptide bond formation on the ribosome: Studies with substrate analogs. Biol. Chem. 2007a;388:687–691. doi: 10.1515/BC.2007.077. [DOI] [PubMed] [Google Scholar]
- Beringer, M., Rodnina, M.V. The ribosomal peptidyl transferase. Mol. Cell. 2007b;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Beringer, M., Adio, S., Wintermeyer, W., Rodnina, M.V. The G2447A mutation does not affect ionization of a ribosomal group taking part in peptide bond formation. RNA. 2003;9:919–922. doi: 10.1261/rna.5600503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer, M., Bruell, C., Xiong, L., Pfister, P., Bieling, P., Katunin, V.I., Mankin, A.S., Bottger, E.C., Rodnina, M.V. Essential mechanisms in the catalysis of peptide bond formation on the ribosome. J. Biol. Chem. 2005;280:36065–36072. doi: 10.1074/jbc.M507961200. [DOI] [PubMed] [Google Scholar]
- Bieling, P., Beringer, M., Adio, S., Rodnina, M.V. Peptide bond formation does not involve acid–base catalysis by ribosomal residues. Nat. Struct. Mol. Biol. 2006;13:423–428. doi: 10.1038/nsmb1091. [DOI] [PubMed] [Google Scholar]
- Brunelle, J.L., Youngman, E.M., Sharma, D., Green, R. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA. 2006;12:33–39. doi: 10.1261/rna.2256706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J., Geballe, A.P. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol. 1996;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changalov, M.M., Ivanova, G.D., Rangelov, M.A., Acharya, P., Acharya, S., Minakawa, N., Foldesi, A., Stoineva, I.B., Yomtova, V.M., Roussev, C.D., et al. 2′/3′-O-peptidyl adenosine as a general base catalyst of its own external peptidyl transfer: Implications for the ribosome catalytic mechanism. ChemBioChem. 2005;6:992–996. doi: 10.1002/cbic.200400349. [DOI] [PubMed] [Google Scholar]
- Cruz-Vera, L.R., Rajagopal, S., Squires, C., Yanofsky, C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Cruz-Vera, L.R., Gong, M., Yanofsky, C. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. 2006;103:3598–3603. doi: 10.1073/pnas.0600082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Vera, L.R., New, A., Squires, C., Yanofsky, C. Ribosomal features essential for tna operon induction: Tryptophan binding at the peptidyl transferase center. J. Bacteriol. 2007;189:3140–3146. doi: 10.1128/JB.01869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, G.K., Bhattacharyya, D., Burma, D.P. A possible mechanism of peptide bond formation on ribosome without mediation of peptidyl transferase. J. Theor. Biol. 1999;200:193–205. doi: 10.1006/jtbi.1999.0987. [DOI] [PubMed] [Google Scholar]
- Dorner, S., Polacek, N., Schulmeister, U., Panuschka, C., Barta, A. Molecular aspects of the ribosomal peptidyl transferase. Biochem. Soc. Trans. 2002;30:1131–1136. doi: 10.1042/bst0301131. [DOI] [PubMed] [Google Scholar]
- Frolova, L.Y., Tsivkovskii, R.Y., Sivolobova, G.F., Oparina, N.Y., Serpinsky, O.I., Blinov, V.M., Tatkov, S.I., Kisselev, L.L. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sanchez, F., Janssen, B.D., Hayes, C.S. Prolyl-tRNAPro in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J. Biol. Chem. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick, P., Yanofsky, C. tRNATrp translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J. Bacteriol. 1990;172:3100–3107. doi: 10.1128/jb.172.6.3100-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, F., Yanofsky, C. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 2001;276:1974–1983. doi: 10.1074/jbc.M008892200. [DOI] [PubMed] [Google Scholar]
- Gong, F., Yanofsky, C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- Gong, M., Cruz-Vera, L.R., Yanofsky, C. Ribosome recycling factor and release factor 3 action promotes TnaC-peptidyl-tRNA dropoff and relieves ribosome stalling during tryptophan induction of tna operon expression in Escherichia coli . J. Bacteriol. 2007;189:3147–3155. doi: 10.1128/JB.01868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J.L., Schmeing, T.M., Moore, P.B., Steitz, T.A. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. 2002;99:11670–11675. doi: 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesslein, A.E., Katunin, V.I., Beringer, M., Kosek, A.B., Rodnina, M.V., Strobel, S.A. Exploration of the conserved A+C wobble pair within the ribosomal peptidyl transferase center using affinity purified mutant ribosomes. Nucleic Acids Res. 2004;32:3760–3770. doi: 10.1093/nar/gkh672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunin, V.I., Muth, G.W., Strobel, S.A., Wintermeyer, W., Rodnina, M.V. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol. Cell. 2002;10:339–346. doi: 10.1016/s1097-2765(02)00566-x. [DOI] [PubMed] [Google Scholar]
- Klaholz, B.P., Pape, T., Zavialov, A.V., Myasnikov, A.G., Orlova, E.V., Vestergaard, B., Ehrenberg, M., van Heel, M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- Krayevsky, A.A., Kukhanova, M.K. The peptidyltransferase center of ribosomes. Prog. Nucleic Acid Res. Mol. Biol. 1979;23:1–51. doi: 10.1016/s0079-6603(08)60130-0. [DOI] [PubMed] [Google Scholar]
- Leonov, A.A., Sergiev, P.V., Bogdanov, A.A., Brimacombe, R., Dontsova, O.A. Affinity purification of ribosomes with a lethal G2655C mutation in 23 S rRNA that affects the translocation. J. Biol. Chem. 2003;278:25664–25670. doi: 10.1074/jbc.M302873200. [DOI] [PubMed] [Google Scholar]
- Lovett, P.S., Rogers, E.J. Ribosome regulation by the nascent peptide. Microbiol. Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, K., Schaffitzel, C., Fabiola, F., Chapman, M.S., Ban, N., Frank, J. Elongation arrest by SecM via a cascade of ribosomal RNA rearrangements. Mol. Cell. 2006;22:533–543. doi: 10.1016/j.molcel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Muto, H., Nakatogawa, H., Ito, K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol. Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Nakatogawa, H., Ito, K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Noller, H.F. Ribosomal RNA and translation. Annu. Rev. Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Noller, H.F., Hoffarth, V., Zimniak, L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Oliver, D., Norman, J., Sarker, S. Regulation of Escherichia coli secA by cellular protein secretion proficiency requires an intact gene X signal sequence and an active translocon. J. Bacteriol. 1998;180:5240–5242. doi: 10.1128/jb.180.19.5240-5242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., Rodnina, M.V. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry, S., Brodersen, D.E., Murphy, F.V., Dunham, C.M., Selmer, M., Tarry, M.J., Kelley, A.C., Ramakrishnan, V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Polacek, N., Gaynor, M., Yassin, A., Mankin, A.S. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature. 2001;411:498–501. doi: 10.1038/35078113. [DOI] [PubMed] [Google Scholar]
- Polacek, N., Gomez, M.J., Ito, K., Xiong, L., Nakamura, Y., Mankin, A. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol. Cell. 2003;11:103–112. doi: 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- Rawat, U.B., Zavialov, A.V., Sengupta, J., Valle, M., Grassucci, R.A., Linde, J., Vestergaard, B., Ehrenberg, M., Frank, J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- Rychlik, I., Cerna, J., Chladek, S., Pulkrabek, P., Zemlicka, J. Substrate specificity of ribosomal peptidyl transferase. The effect of the nature of the amino acid side chain on the acceptor activity of 2′(3′)-O-aminoacyladenosines. Eur. J. Biochem. 1970;16:136–142. doi: 10.1111/j.1432-1033.1970.tb01064.x. [DOI] [PubMed] [Google Scholar]
- Schmeing, T.M., Huang, K.S., Kitchen, D.E., Strobel, S.A., Steitz, T.A. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell. 2005a;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Schmeing, T.M., Huang, K.S., Strobel, S.A., Steitz, T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005b;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Shaw, J.J., Green, R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol. Cell. 2007;28:458–467. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, A., Beringer, M., Rodnina, M.V., Wolfenden, R. The ribosome as an entropy trap. Proc. Natl. Acad. Sci. 2004;101:7897–7901. doi: 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, H., Mugnier, P., Das, A.K., Webb, H.M., Evans, D.R., Tuite, M.F., Hemmings, B.A., Barford, D. The crystal structure of human eukaryotic release factor eRF1—Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Stewart, V., Yanofsky, C. Role of leader peptide synthesis in tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 1986;167:383–386. doi: 10.1128/jb.167.1.383-386.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson, T., Ehrenberg, M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- Thompson, J., Kim, D.F., O'Connor, M., Lieberman, K.R., Bayfield, M.A., Gregory, S.T., Green, R., Noller, H.F., Dahlberg, A.E. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl. Acad. Sci. 2001;98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro, S., Åqvist, J. Mechanism of peptide bond synthesis on the ribosome. Proc. Natl. Acad. Sci. 2005;102:12395–12400. doi: 10.1073/pnas.0504043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro, S., Åqvist, J. Analysis of predictions for the catalytic mechanism of ribosomal peptidyl transfer. Biochemistry. 2006;45:7049–7056. doi: 10.1021/bi0605383. [DOI] [PubMed] [Google Scholar]
- Trobro, S., Åqvist, J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol. Cell. 2007;27:758–766. doi: 10.1016/j.molcel.2007.06.032. [DOI] [PubMed] [Google Scholar]
- Weber, A.L., Orgel, L.E. The formation of dipeptides from amino acids and the 2′(3′)-glycyl ester of an adenylate. J. Mol. Evol. 1979;13:185–192. doi: 10.1007/BF01739478. [DOI] [PubMed] [Google Scholar]
- Weinger, J.S., Parnell, K.M., Dorner, S., Green, R., Strobel, S.A. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- Woolhead, C.A., Johnson, A.E., Bernstein, H.D. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Youngman, E.M., Green, R. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods. 2005;36:305–312. doi: 10.1016/j.ymeth.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Youngman, E.M., Brunelle, J.L., Kochaniak, A.B., Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Youngman, E.M., He, S.L., Nikstad, L.J., Green, R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol. Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]