Abstract
High-salinity, drought, and low temperature are three common environmental stress factors that seriously influence plant growth and development worldwide. Recently, microRNAs (miRNAs) have emerged as a class of gene expression regulators that have also been linked to stress responses. However, the relationship between miRNA expression and stress responses is just beginning to be explored. Here, we identified 14 stress-inducible miRNAs using microarray data in which the effects of three abiotic stresses were surveyed in Arabidopsis thaliana. Among them, 10 high-salinity-, four drought-, and 10 cold-regulated miRNAs were detected, respectively. miR168, miR171, and miR396 responded to all of the stresses. Expression profiling by RT-PCR analysis showed great cross-talk among the high-salinity, drought, and cold stress signaling pathways. The existence of stress-related elements in miRNA promoter regions provided further evidence supporting our results. These findings extend the current view about miRNA as ubiquitous regulators under stress conditions.
Keywords: microRNA, microarray, stress, gene expression regulators, Arabidopsis thaliana
INTRODUCTION
MicroRNAs (miRNAs) are a highly conserved class of small noncoding RNAs that regulate gene expression by post-transcriptional degradation or translational repression (Carrington and Ambros 2003; Bartel 2004). With the discovery of large numbers of miRNAs in both plants and animals, the important roles of these special small RNAs have been widely recognized (Llave et al. 2002; Park et al. 2002; Ambros 2004). Many processes, such as leaf development, auxin signaling, phase transition, flowering, and genome maintenance, are regulated in similar ways by different miRNAs (Aukerman and Sakai 2003; Palatnik et al. 2003; Vaucheret et al. 2004; Guo et al. 2005; Mallory et al. 2005).
Recently, there has been strong evidence leading to the proposal that miRNAs are hypersensitive to abiotic or biotic stress as well as to diverse physiological processes (Sunkar and Zhu 2004; Lu et al. 2005). The first report linking miRNA and stress tolerance was miR398, expression of which is transcriptionally down-regulated by oxidative stresses. In Arabidopsis, miR398 was found to target two closely related Cu/Zn superoxide dismutase coding genes, cytosolic CSD1 and chloroplastic CSD2, and a reduced level of miR398 led to improved tolerance of transgenic lines compared with the wild-type plants under oxidative stress conditions (Sunkar et al. 2006). Additionally, miR395 and miR399 were identified to be involved in sulfate and inorganic phosphate starvation responses, respectively (Jones-Rhoades and Bartel 2004; Fujii et al. 2005). In rice, miR169g was confirmed as the only member induced by drought among the miR169 family (Zhao et al. 2007). Furthermore, in line with the hypothesis of miRNA's response to environmental stimuli, 21 miRNAs belonging to 11 miRNA families in Arabidopsis were predicted to be up-regulated under UV-B stress condition (Zhou et al. 2007). However, systematic expression analysis for miRNAs under abiotic stress conditions in Arabidopsis has not been reported. Hence, efforts to identify novel stress-regulated miRNAs and determine their expression patterns could improve our understanding of their functions in stress adaptation.
Currently, a variety of biochemical, molecular, and bioinformatic approaches and technologies have been developed for miRNA analysis and detection (Schena et al. 1995; Eisen and Brown 1999). Using tiling path microarray analysis as a tool, it is now possible to perform high-throughput profiling of the expression of all the known miRNAs to examine their expression profiles under different environmental stresses (Garzon et al. 2006; Zhao et al. 2007). In this work, we analyzed the effects of 117 miRNAs under high-salinity, drought, and low-temperature stress conditions with miRNA chips representing nearly all known miRNAs cloned or identified in Arabidopsis. Fourteen stress-inducible miRNAs were detected, and our results were further confirmed by detecting their expression patterns and analyzing the cis-elements in their promoter sequences.
RESULTS AND DISCUSSION
Arabidopsis miRNA microarray experiments
In order to test whether the expression of any of the currently known miRNAs was regulated by abiotic stresses, we prepared a miRNA microarray containing 117 probes that were complementary to known Arabidopsis miRNAs. In addition, eight short oligos possessing no homology with any existing miRNA sequences were designed as negative controls, and a transcriptional repressor Hex as positive control was also included. Total RNA was extracted from the whole plants, and low-molecular-weight miRNAs, which were used for preparation of labeled probes, were obtained. Microarrays were hybridized with Cy3 probe pairs of high-salinity-, drought-, and cold-treated plants and unstressed plants as described in Materials and Methods. The hybridized microarray was scanned by a separate laser channel for Cy3 emissions. The ratio of the treated and untreated plants' fluorescent signal intensities was then measured as a relative measure to determine changes in the differential expression of the oligomer sequence represented by miRNA spots on the microarrays. The quality of the microarray data was assessed, and standard quality-control measures indicated that the microarray used in this analysis showed a similar distribution of intensities. None of the negative control probes gave a detectable signal in any of the samples. The q value calculated from different samples was used as a measure of biological reproducibility, and for each gene, it was the lowest false discovery rate at which that gene was called significant.
Identification of stress-response miRNAs
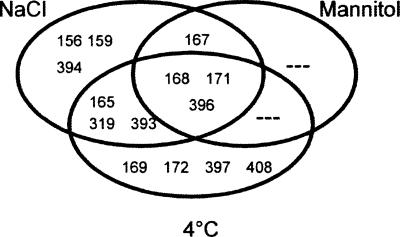
The miRNA populations from different stress treatment samples were compared to a mock sample. Significance analysis of microarrays (SAM) and a criterion of fold change >1.5 and q value <0.001 were used to examine the effects of various stress treatments. We found that 14 miRNAs (miR156, miR159, miR165, miR167, miR168, miR169, miR171, miR172, miR319, miR393, miR394, miR396, miR397, and miR408) on the microarray showed differential expression profiles in response to abiotic stresses (Table 1). No statistically significantly down-regulated miRNAs were found during this process. Among them, 10 high-salinity-, four drought-, and 10 cold-regulated miRNAs were detected (Fig. 1). miR165, miR319, and miR393 were up-regulated by both high salinity and cold, miR167 was induced by both high salinity and drought, and miR168, miR171, and miR396 were observed to respond remarkably to all three stresses.
TABLE 1.
Stress-response miRNAs identified by microarray analysis
FIGURE 1.
Venn diagram illustrates common and unique differential miRNAs expression under three treatment conditions.
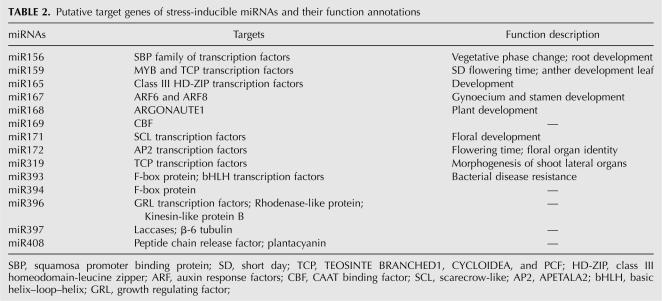
Based on the characteristic of their targets, the identified miRNAs could be classified into three major categories (Table 2). The first category includes eight miRNAs—miR156, miR159, miR165, miR169, miR171, miR172, miR319, and miR396—which target transcription factors involved in further regulation of gene expression and signal transduction that probably function in stress responses (Jones-Rhoades and Bartel 2004; Sunkar and Zhu 2004). Thus, it appears that the induction of these miRNAs would lead to the repression of some corresponding transcription factor genes which, in turn, target a set of specific protein coding genes and play defensive roles against stresses. The second category includes miR167, miR168, miR393, and miR394, which are related to direct response to stresses or external stimuli. miR393 and miR394 target the messages of F-box proteins, which were recently reported to be differentially regulated by stress conditions and play significant roles in the abiotic stress-response pathway (Jones-Rhoades and Bartel 2004; Navarro et al. 2006; Jain et al. 2007). miR168 has been proven to target mRNA of ARGONAUTE 1 (AGO1), which might be the only member of the AGO family that is regulated by a miRNA (Vazquez et al. 2004). The direct role of AGO in plant abiotic stresses has not yet been demonstrated. However, one of its associated partners, Hsp90, is stress-sensitive (Tahbaz et al. 2001; Liu et al. 2005; Leung and Sharp 2007). miR167 targets two auxin response factors, ARF6 and ARF8 (Wu et al. 2006). Plant auxin regulates many agronomically important aspects of plant growth and development as well as responses to environmental stress conditions (Fedoroff 2002; Himmelbach et al. 2003; Achard et al. 2006). Hence, miR167 might play a potential role in stress-resistance progress by affecting auxin signaling pathways. The last category includes miR397 and miR408, whose target genes are hydrolase and oxidoreductase coding genes, which respond to many stresses (Kimura et al. 2003; Apel and Hirt 2004). However, a few of the miRNAs reported to be regulated by high salinity, drought, or cold stress were missed in our results, such as miR389 and miR400 (Sunkar and Zhu 2004). We speculate that this might have been caused by the differences between our treatment methods and theirs or by the technical differences between RNA gel blot and microarray analysis. Some other miRNAs failed to be detected, perhaps because they are required at low levels at this stage or their expression is limited to specific cell types or particular growth conditions, which make it difficult to identify the changes under stress conditions.
TABLE 2.
Putative target genes of stress-inducible miRNAs and their function annotations
In addition, most miRNAs were proven to be involved in the regulation of important developmental processes as shown in Table 2. Stressed plants often resemble abnormal developmental phenotypes, which can be viewed as entering a particular developmental phase (Cooper et al. 2003; Sunkar et al. 2007). Furthermore, there is evidence that many of the miRNA target genes are involved in both development and stress regulation, so we speculate that these miRNAs might be co-regulated by both environmental factors and developmental cues.
Expression profiles of stress-inducible miRNAs during stress treatments
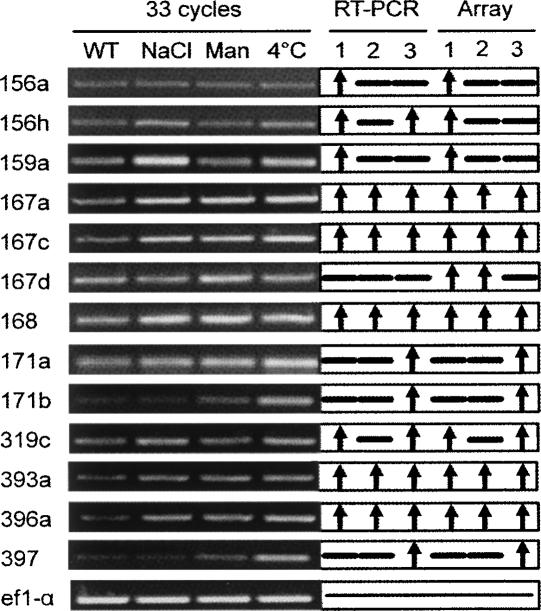
In microarray data, we found that miRNAs belonging to the same family have similar expression patterns, suggesting that they could not be differentiated in microarray analysis because of a potential cross-hybridization problem. To determine which locus is responsive to stress conditions, we performed semiquantitative RT-PCR analysis using specific primers designed to amplify fold-back precursor transcripts in Arabidopsis. Two-week-old wild-type Arabidopsis seedlings were subjected to stress treatments of 300 mM NaCl, 200 mM mannitol, or 4°C, respectively. To ensure the RT-PCR was accurate, elongation factor 1-α (ef1-α) gene was used as internal control, and varying PCR cycle numbers were determined to quantitatively amplify each transcript (Fig. 2). The RT-PCR results were ultimately consistent with the microarray data for all miRNAs tested except miR156h and miR167d. The induction of miR393 reported here was in agreement with previous studies using Northern blotting (Sunkar and Zhu 2004).
FIGURE 2.
RT-PCR validation of the microarray. (RT-PCR) results from RT-PCR analysis; (Array) results from microarray; (Man) mannitol treatment; (lane 1) 300 mM NaCl treatment; (lane 2) 200 mM mannitol treatment; (lane 3) 4°C treatment; (↑) up-regulated compared to WT; and (—) no significant difference.
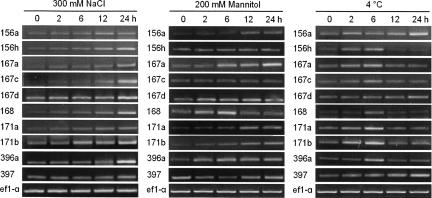
To further analyze the temporal expression patterns of these miRNAs, time course analysis by RT-PCR was performed to detect whether they can be regulated by stress after an extended treatment time (Fig. 3). Our data showed that the levels of miR156h, miR167a, miR167c, miR167d, miR168, and miR171b were gradually increased from 2 to 24 h after exposure to high-salinity treatment, while expression of miR396a peaked at 24 h. miR167a levels accumulated after 2 h of drought stress and were greatly increased after 24 h of treatment, while miR168 first increased then returned to a normal level after 6 h. During cold-stress treatment, most miRNA levels were higher after 6 h and declined with longer stress; the exception was miR156a, which had maximal expression at 24 h. The expression of additional members of miRNA families was also analyzed, but these transcripts were undetectable or did not change (data not shown). Members of the same miRNA family might have different functional roles in spite of their high similarity in the sequence. Taken together, the newly identified stress-response miRNAs were prone to being induced by all treatments, indicating the existence of great cross talk between high-salinity, drought, and cold stress signaling processes. Our results revealed that a single miRNA has the potential to regulate multiple functionally related mRNAs in response to stress.
FIGURE 3.
Time course of altered expression of selected miRNAs by RT-PCR analysis. PCR amplification cycles: ef1-α, 25 cycles; miRNA precursor, 33 cycles.
Stress-relevant cis-elements exist in the promoters of miRNA genes
Cis- and trans-acting elements involved in stress-induced gene expression have been analyzed extensively (Jaglo-Ottosen et al. 1998; Kasuga et al. 1999; Zhang et al. 2005). To further elucidate the inducibility of these products, we analyzed the 1000-base-pair (bp) upstream promoter sequence of 20 miRNAs by using the PlantCARE database (http://intra.psb.ugent.be:8080/PlantCARE) (Lescot et al. 2002). Among the elements listed in Table 3, we identified several known stress-responsive elements, such as the ABA-response elements (ABREs), anaerobic induction elements (AREs), MYB binding site involved in drought-inducibility (MBS), heat-stress-responsive elements (HSEs), low-temperature-responsive elements (LTRs), and defense- and stress-responsive elements (TC-rich repeats). Some other regulatory elements were also identified, such as those possibly involved in regulation in response to gibberellic acid (GA), ethylene, salicylic acid (SA), or methyl jasmonate (MeJA). In plants, most ABA-responsive genes have the conserved ABREs in their promoters, which are significant cis-elements for genes responsive to abiotic stress in Arabidopsis (Mundy et al. 1990; Xu et al. 1996). Of the 20 miRNAs analyzed, six had ABREs, suggesting these miRNAs might be involved in ABA-mediated stress-response processes. Sixteen miRNAs had AREs, which respond to hypoxic, low-temperature, and dehydration stress (Dolferus et al. 1994). The presence of ABREs and AREs suggests that these miRNA might be regulated by stress conditions as protein coding genes. For example, miR393 was reported to be strongly up-regulated by cold, dehydration, and NaCl treatments. We found seven AREs and two ABREs in the promoters of three miR393 family members (miR393a, -b, and -c), which might provide the evidence of the induction of this miRNA. Likewise, the responses of miR167, miR168, and miR396 to high-salinity, drought, and cold treatments are correlated with the enrichment of AREs and ABREs in their promoters. Stress-related elements also existed in miR408 promoters and the role of this miRNA needs to be determined experimentally. Taken together, analysis of stress-response enrichment cis-elements provides additional evidence that these 14 miRNA genes are very likely to be involved in the responses to abiotic stresses.
TABLE 3.
Kinds and numbers of known stress-related elements in the upstream regions of miRNA genes
In conclusion, we analyzed 117 miRNAs expression profiles under three abiotic stress conditions in Arabidopsis and found that 14 of them were differentially regulated by one or more stress conditions. Information about miRNA expression patterns and the biochemical function of their targets is essential and provides a valuable contribution toward our understanding of the function of these tiny noncoding genes and their cooperation in complex biological networks. Based on the RT-PCR and promoter analysis, we speculate that miR167, miR168, and miR396 might play important roles in plant abiotic stress responses. However, due to the potential uncertainty in this experiment, it is not feasible to just estimate the exact effects of these miRNAs in plants. These data provide a starting point for future studies, and continued efforts are needed to confirm the function of miRNAs in stress response and stress adaptation.
MATERIALS AND METHODS
Plant materials
A. thaliana ecotype Columbia seeds were surface-sterilized and sown on MS-agar plates. Seeds were stratified at 4°C for 2 d and then transferred to 22°C for 2 wk. For different stresses, seedlings were transferred to blotting paper without stress treatment or saturated with 300 mM NaCl, or 200 mM mannitol, or treated with cold (4°C). For RNA extraction and microarray experiment, the whole plants were frozen and stored in liquid nitrogen immediately after harvest.
MicroRNA microarray construction
Mature miRNA sequences were downloaded in the miRBase (http://microrna.sanger.ac.uk/, as of June 2005). There were 117 miRNAs from Arabidopsis after redundant sequences were discarded. In addition, eight short oligos were designed to possess no homology with any existing RNA sequence, and their corresponding synthetic miRNAs were produced by in vitro transcription using a miRNA Probe Construction Kit (Ambion). Various amounts of synthetic miRNAs were spiked into the RNA samples. All miRNA probe sequences were designed to be complementary to the full-length mature miRNA. To facilitate probe immobilization on aldehyde-modified glass slides, the probe sequences were concatenated up to 40 nt and modified with 5′-amino-modifier C6. Oligonucleotide probes were synthesized at MWG Biotech and dissolved in EasyArray spotting solution at 40 μM concentration. Each probe was printed in triplicate using a SmartArray microarrayer (CapitalBio Corporation).
RNA extraction and miRNA microarray experiments
Total RNA was isolated from different Arabidopsis seedlings with TRIZOL reagent (Invitrogen) and the low-molecular-weight RNA was isolated using the miRNA Isolation Kit (Ambion). Target RNA labeling was performed as described (Thomson et al. 2004). In brief, 4 μg of low-molecular-weight RNA was labeled with 500 ng of 5′-phosphate-cytidyl-uridyl-cy3-3′ (Dharmacon) with 2 units of T4 RNA ligase (NEB). The labeling reaction was performed at 4°C for 2 h. Labeled RNA was precipitated with 0.3 M sodium acetate and 2.5 volumes of ethanol and was then resuspended in 15 μL of hybridization buffer containing 3× SSC, 0.2% SDS, and 15% formamide. Hybridization was performed using LifterSlip (Erie), which allowed for even dispersal of hybridization solutions between the microarray and coverslip. The hybridization chamber was laid on a three-phase tiling agitator (BioMixer II) to prompt the microfluidic circulation under the coverslip. Hybridization was performed in a water bath at 42°C overnight. After the array had been hybridized at 42°C overnight, it was washed with two consecutive washing solutions (0.2% SDS, 2× SSC at 42°C for 5 min, and 0.2% SSC for 5 min at room temperature).
Image acquisition and data processing
Clustering arrays were scanned with a confocal LuxScan scanner. The scanning setting was adjusted to obtain a visualized equal intensity of U6 spots across arrays. Data was extracted from the TIFF images using LuxScan 3.0 software (CapitalBio). Low-intensity spots were removed for which fewer than 30% of the signal pixels exceeded the median background plus two times its standard deviation. Then normalization was performed based on the mean array intensity for inter-array comparison. For each sample, consideration of two hybridizations was carried out and each miRNA probe had three replicate spots on a microarray. The mean intensity value of each probe was used for cluster analysis. The raw data were Log 2 transformed and median centered by arrays and genes using the Adjust Data function of CLUSTER 3.0 software and then further analyzed with hierarchical clustering with average linkage (Eisen et al. 1998). Original microarray data are deposited at the Gene Expression Omnibus. To determine the significant differentially expressed miRNAs, Significance Analysis of Microarrays (SAM, version 2.1) was performed using two-class unpaired comparison in the SAM procedure (Tusher et al. 2001).
RT-PCR analysis
Total RNA isolated with RNase Plant Mini Kit (Qiagen) was processed. Reverse transcription reactions were performed in 20 μL using 5 μg of RNA by M-MLV (NEB). RT-PCR conditions for elongation factor 1-α gene ef1-α amplification were as follows: 94°C for 5 min, 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min, and 72°C for 5 min, 25 cycles. Primers for ef1-α were as follows: forward, 5′-GTATGGTTGTTACCTTTGCTCCCACAG and reverse, 3′-CATCATTTGGCACCCTTCTTCACTGC (500 bp). For miRNA precursor amplification, essentially the same conditions were used except the number of PCR cycles was increased to 33. The primer pairs used for RT-PCR and predicted amplicon sizes were as follows:
156a, 5′-AGAATTTGGTATGCAGAGACAGAT; 3′-CGGTTTCTGGACTAATTGGAA (298 bp);
156h, 5′-TGACACGATCACACAACATGG; 3′-CCACCGTCACTGCTTACTTA (209 bp);
159a, 5′-CACGCTAAACATTGCTTCGGAAT; 3′-ATCCCATAAGCCCTAATCCTTGT (290 bp);
159c, 5′-ACCAAGTTTTGAAGAACAGAGACT; 3′-CATAGAGAGTGCGCGGTGTT (169 bp);
167a, 5′-GTGTAGTCAACTGTGTGCGTT; 3′-GCACAACTTGTTGCTCAGGT (234 bp);
167c, 5′-TTCATGCTACAATCATTAGCAGGT; 3′-AGTCGTCTTCATGTCTGTATGT (205 bp);
167d, 5′-GAGTTGTGGCCATTAAGAGCT; 3′-CTTCTTGTTAATGTTTGCTCTCTCCT (216 bp);
168, 5′-TGATAGTAGAGTCTCACCATCG; 3′-GAAGGAGAAGCGTAGAAATCTTC (205 bp);
171a, 5′-TCCAAAATAGAGACGAGAGAGT; 3′-CTCCTCCTCACACTTCACAT (215 bp);
171b, 5′-CGAGTGCCTGTAGAGTAAAAAC; 3′-TTCTGGAGCTAAGTGGAGATT (278 bp);
319c, 5′-AAACACTCGTGGTAGAGAAACGAT; 3′-AGAGGTTGAAAATGCAAATCCAGT (238 bp);
393, 5′-CAAAGAGATAGCATGATCCAA; 3′-AAGAGGAACACGATCCATTGAC (214 bp);
396a, 5′-AGGGTTTCGTCTGCTCTACAT; 3′-TCTGATTATGGAATCAATCACGCT (242 bp);
397, 5′-CCCCTGGGTTTGAATGAACAT; 3′-AGAACTCTCAAGGTCTTTTAAGTG (180 bp).
ACKNOWLEDGMENTS
We thank Dr. Jianqing Zhao (CapitalBio, Beijing, China) for his technical help. This work was supported by the National Basic Research Program (Grant 2006CB1001006), Program for Changjiang Scholars and Innovative Research Team in University (Grant IRT0635), and the National Natural Science Foundation (Grants 30570144 and 30500042) in China.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.895308.
REFERENCES
- Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., Van Der Straeten, D., Peng, J., Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Apel, K., Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Ambros, V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Cooper, B., Clarke, J.D., Budworth, P., Kreps, J., Hutchison, D., Park, S., Guimil, S., Dunn, M., Luginbuhl, P., Ellero, C., et al. A network of rice genes associated with stress response and seed development. Proc. Natl. Acad. Sci. 2003;100:4945–4950. doi: 10.1073/pnas.0737574100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus, R., Jacobs, M., Peacock, W.J., Dennis, E.S. Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol. 1994;105:1075–1087. doi: 10.1104/pp.105.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., Brown, P.O. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff, N.V. Cross-talk in abscisic acid signaling. Sci. STKE. 2002;2002:RE10. doi: 10.1126/stke.2002.140.re10. [DOI] [PubMed] [Google Scholar]
- Fujii, H., Chiou, T.J., Lin, S.I., Aung, K., Zhu, J.K. A miRNA involved in phosphate-starvation response in Arabidopsis . Curr. Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Garzon, R., Pichiorri, F., Palumbo, T., Iuliano, R., Cimmino, A., Aqeilan, R., Volinia, S., Bhatt, D., Alder, H., Marcucci, G., et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc. Natl. Acad. Sci. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H.S., Xie, Q., Fei, J.F., Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach, A., Yang, Y., Grill, E. Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 2003;6:470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jain, M., Nijhawan, A., Arora, R., Agarwal, P., Ray, S., Sharma, P., Kapoor, S., Tyagi, A.K., Khurana, J.P. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007;143:1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kimura, M., Manabe, K., Abe, T., Yoshida, S., Matsui, M., Yamamoto, Y.Y. Analysis of hydrogen peroxide-independent expression of the high-light-inducible ELIP2 gene with the aid of the ELIP2 promoter–luciferase fusions. Photochem. Photobiol. 2003;77:668–674. doi: 10.1562/0031-8655(2003)077<0668:aohpeo>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., Rouze, P., Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, A.K., Sharp, P.A. MicroRNAs: A safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Liu, J., Rivas, F.V., Wohlschlegel, J., Yates J.R., 3rd, Parker, R., Hannon, G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., Carrington, J.C. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S., Sun, Y.H., Shi, R., Clark, C., Li, L., Chiang, V.L. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis . Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Bartel, D.P., Bartel, B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy, J., Yamaguchi-Shinozaki, K., Chua, N.H. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc. Natl. Acad. Sci. 1990;87:1406–1410. doi: 10.1073/pnas.87.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., Weigel, D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., Chen, X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana . Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., Chinnusamy, V., Zhu, J., Zhu, J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., Kapoor, A., Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by down-regulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis . Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahbaz, N., Carmichael, J.B., Hobman, T.C. GERp95 belongs to a family of signal-transducing proteins and requires Hsp90 activity for stability and Golgi localization. J. Biol. Chem. 2001;276:43294–43299. doi: 10.1074/jbc.M107808200. [DOI] [PubMed] [Google Scholar]
- Thomson, J.M., Parker, J., Perou, C.M., Hammond, S.M. A custom microarray platform for analysis of microRNA gene expression. Nat. Med. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Gasciolli, V., Crete, P., Vaucheret, H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Wu, M.F., Tian, Q., Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- Xu, D., Duan, X., Wang, B., Hong, B., Ho, T., Wu, R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Ruan, J., Ho, T.H., You, Y., Yu, T., Quatrano, R.S. Cis-regulatory element based targeted gene finding: Genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana . Bioinformatics. 2005;21:3074–3081. doi: 10.1093/bioinformatics/bti490. [DOI] [PubMed] [Google Scholar]
- Zhao, B., Liang, R., Ge, L., Li, W., Xiao, H., Lin, H., Ruan, K., Jin, Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007;354:585–590. doi: 10.1016/j.bbrc.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Zhou, X., Wang, G., Zhang, W. UV-B responsive microRNA genes in Arabidopsis thaliana . Mol. Syst. Biol. 2007;3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]