Abstract
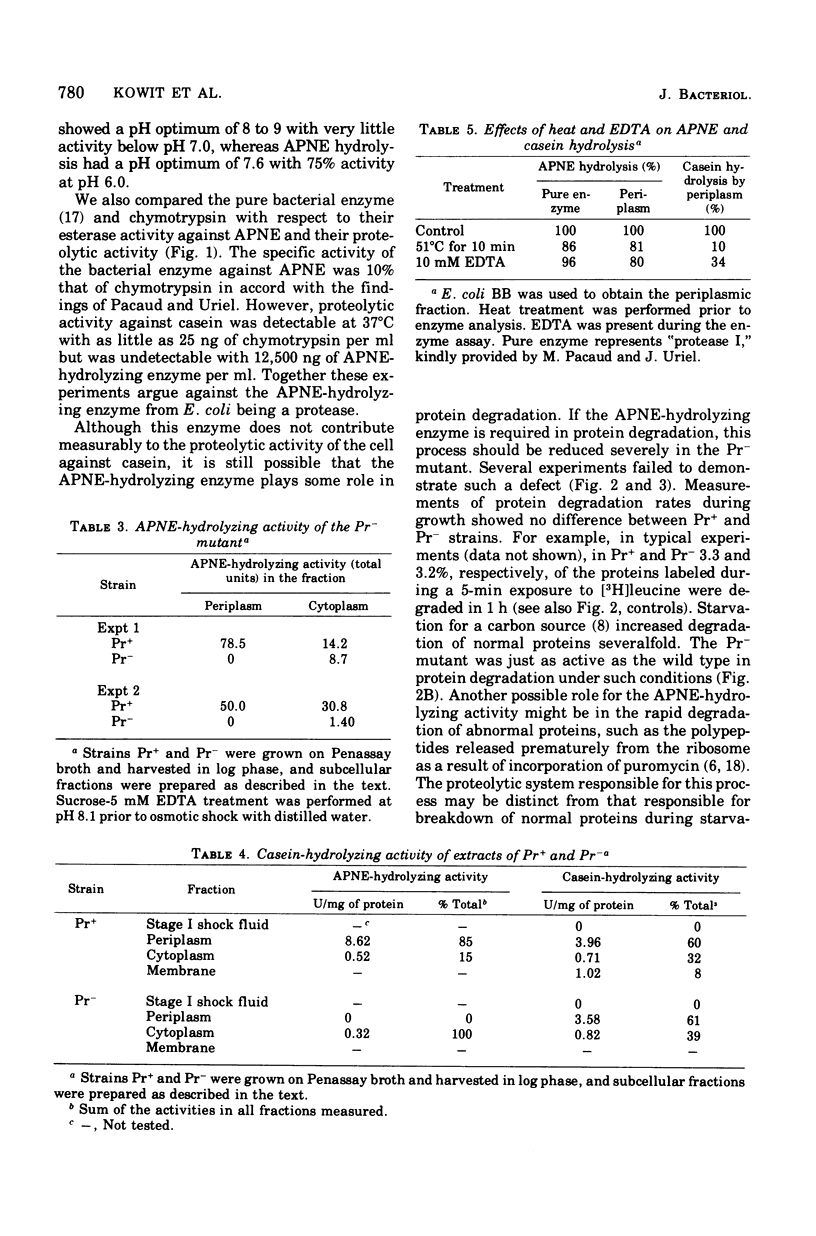
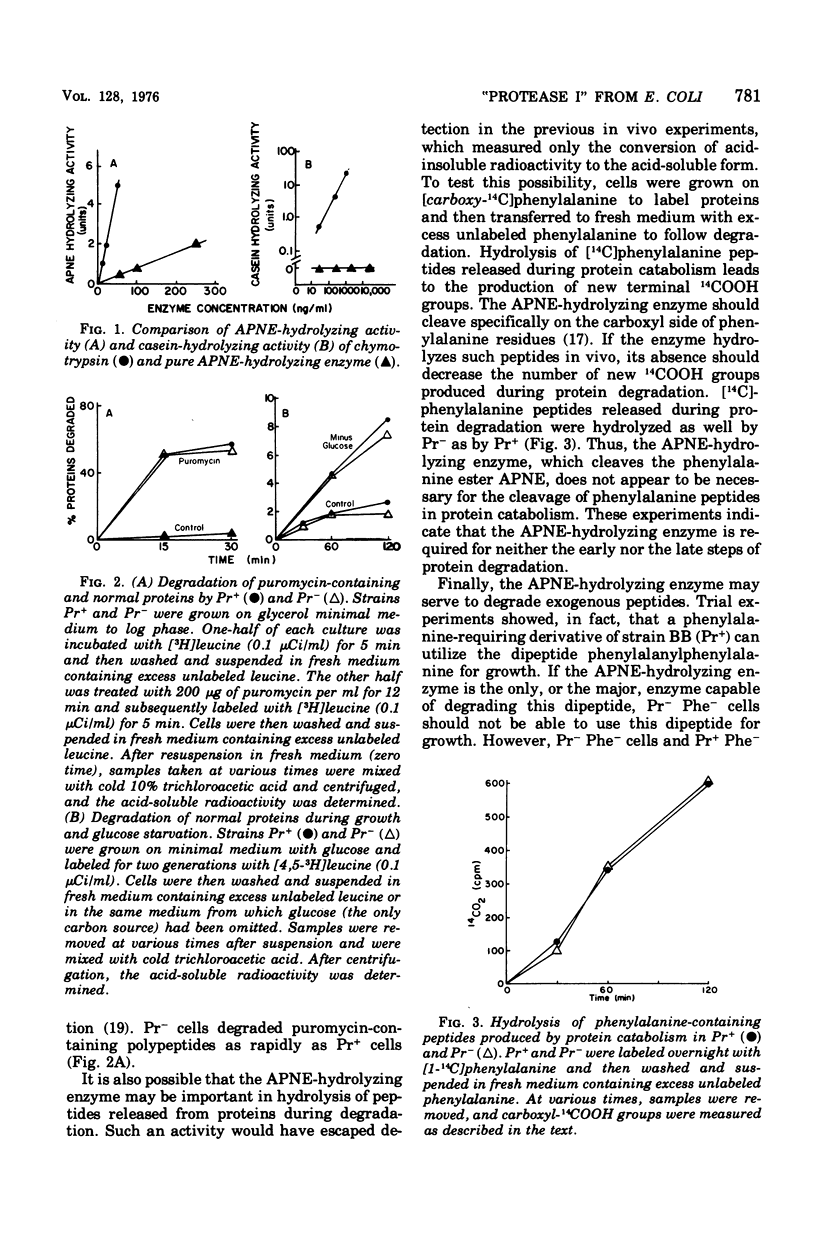
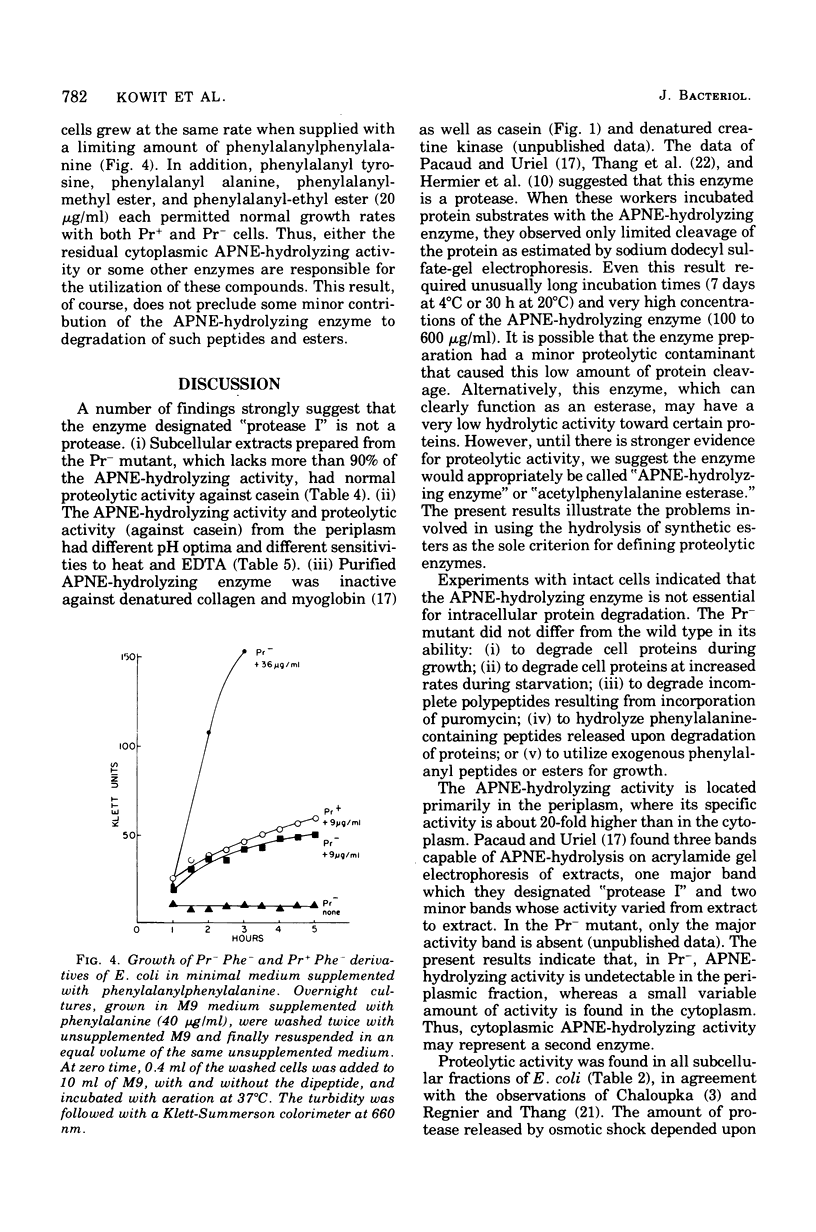
Pacaud and Uriel described an enzyme from Escherichia coli ("protease I") that hydrolyzes acetyl phenylalanine naphthyl ester (APNE). We examined the possible involvement of this enzyme in intracellular protein degradation, its subcellular distribution, and its proteolytic activity. Although the APNE-hydrolyzing activity is localized primarily in the periplasm, proteolytic activity against casein was found in the periplasm, membrane, and cytoplasm with similar specific activities. The APNE-hydrolyzing enzyme did not appear to contribute to the proteolytic activity of the periplasm. A mutant deficient in APNE-hydrolyzing activity lacked all activity in the periplasm but showed a slight percentage of residual activity in the cytoplasm. Extracts of such cells were normal in their ability to hydrolyze casein. The mutant was indistinguishable from wild-type cells in its rate of protein degradation during growth or glucose starvation and in the ability to rapidly degrade puromycin-containing polypeptides. Nitrogen starvation, which increased protein breakdown severalfold, affected neither the total amount nor the distribution of APNE-hydrolyzing activity. The mutant showed no defect in its ability to cleave small phenylalanine-containing peptides released during protein degradation. The mutant and wild-type cells are equally able to hydrolyze exogenously supplied phenylalanyl peptides. These experiments suggest that the APNE-hydrolyzing enzyme is not required for protein degradation and that "protease I" is probably not a protease.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang G. W., Fenton K. A simple method for measuring protein degradation in bacteria. Anal Biochem. 1974 May;59(1):185–189. doi: 10.1016/0003-2697(74)90024-4. [DOI] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L. Degradation of abnormal proteins in Escherichia coli (protein breakdown-protein structure-mistranslation-amino acid analogs-puromycin). Proc Natl Acad Sci U S A. 1972 Feb;69(2):422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L., Dice J. F. Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem. 1974;43(0):835–869. doi: 10.1146/annurev.bi.43.070174.004155. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., Howell E. M., Li J. B., Martel S. B., Prouty W. F. Physiological significance of protein degradation in animal and bacterial cells. Fed Proc. 1974 Apr;33(4):1112–1120. [PubMed] [Google Scholar]
- Hermier B., Pacaud M., Dubert J. M. Action of protease I of Escherichia coli on RNA polymerase of same origin. Eur J Biochem. 1973 Oct 5;38(2):307–310. doi: 10.1111/j.1432-1033.1973.tb03063.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- MANDELSTAM J. The intracellular turnover of protein and nucleic acids and its role in biochemical differentiation. Bacteriol Rev. 1960 Sep;24(3):289–308. doi: 10.1128/br.24.3.289-308.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossal N. G., Heppel L. A. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J Biol Chem. 1966 Jul 10;241(13):3055–3062. [PubMed] [Google Scholar]
- Odessey R., Goldberg A. L. Oxidation of leucine by rat skeletal muscle. Am J Physiol. 1972 Dec;223(6):1376–1383. doi: 10.1152/ajplegacy.1972.223.6.1376. [DOI] [PubMed] [Google Scholar]
- Pacaud M., Richaud C. Protease II from Escherichia coli. Purification and characterization. J Biol Chem. 1975 Oct 10;250(19):7771–7779. [PubMed] [Google Scholar]
- Pacaud M., Uriel J. Isolation and some propeties of a proteolytic enzyme from Escherichia coli (protease I). Eur J Biochem. 1971 Dec 10;23(3):435–442. doi: 10.1111/j.1432-1033.1971.tb01638.x. [DOI] [PubMed] [Google Scholar]
- Prouty W. F., Goldberg A. L. Effects of protease inhibitors on protein breakdown in Escherichia coli. J Biol Chem. 1972 May 25;247(10):3341–3352. [PubMed] [Google Scholar]
- RAVIN H. A., BERNSTEIN P., SELIGMAN A. M. A colorimetric micromethod for the estimation of chymotrypsin activity. J Biol Chem. 1954 May;208(1):1–15. [PubMed] [Google Scholar]
- Regnier P., Thang M. N. Subcellular distribution and characterization of endo and exo-cellular proteases in E. coli. Biochimie. 1972;54(10):1227–1236. doi: 10.1016/s0300-9084(72)80063-4. [DOI] [PubMed] [Google Scholar]
- ZIPSER D. A STUDY OF THE UREA-PRODUCED SUBUNITS OF BETA-GALACTOSIDASE. J Mol Biol. 1963 Aug;7:113–121. doi: 10.1016/s0022-2836(63)80040-6. [DOI] [PubMed] [Google Scholar]


