Abstract
Primary CD8+ T cells from HIV+ asymptomatics can suppress virus production from CD4+ T cells acutely infected with either non-syncytia-inducing (NSI) or syncytia-inducing (SI) HIV-1 isolates. NSI strains of HIV-1 predominantly use the CCR5 chemokine receptor as a fusion cofactor, whereas fusion of T cell line-adapted SI isolates is mediated by another chemokine receptor, CXCR4. The CCR5 ligands RANTES (regulated on activation, normal T cell expressed and secreted), macrophage inflammatory protein 1α (MIP-1α), and MIP-1β are HIV-1 suppressive factors secreted by CD8+ cells that inhibit NSI viruses. Recently, the CXC chemokine stromal cell-derived factor 1 (SDF-1) was identified as a ligand for CXCR4 and shown to inhibit SI strains. We speculated that SDF-1 might be an effector molecule for CD8+ suppression of SI isolates and assessed several SDF-1 preparations for inhibition of HIV-1LAI-mediated cell–cell fusion, and examined levels of SDF-1 transcripts in CD8+ T cells. SDF-1 fusion inhibitory activity correlated with the N terminus, and the α and β forms of SDF-1 exhibited equivalent fusion blocking activity. SDF-1 preparations having the N terminus described by Bleul et al. (Bleul, C.C., Fuhlbrigge, R.C., Casasnovas, J.M., Aiuti, A. & Springer, T.A. (1996) J. Exp. Med. 184, 1101–1109) readily blocked HIV-1LAI-mediated fusion, whereas forms containing two or three additional N-terminal amino acids lacked this activity despite their ability to bind and/or signal through CXCR4. Though SDF-1 is constitutively expressed in most tissues, CD8 T cells contained extremely low levels of SDF-1 mRNA transcripts (<1 transcript/5,000 cells), and these levels did not correlate with virus suppressive activity. We conclude that suppression of SI strains of HIV-1 by CD8+ T cells is unlikely to involve SDF-1.
Keywords: CD8 cells, virus suppression
HIV-1 infection causes a progressive disease typically marked by an acute phase with massive viremia, a subsequent asymptomatic phase where virus replication is confined mostly to lymphoid organs, and a final stage of immune deterioration, resurgence of viremia, and opportunistic infections usually followed by death (1, 2). While the mechanisms responsible for curtailing viremia and maintaining the asymptomatic phase have not been clearly defined, cellular immune responses appear to be involved. This perception is reinforced by studies demonstrating a temporal association between development of HIV-specific CD8+ cytotoxic T-lymphocytes (CTLs) and the resolution of viremia following acute HIV-1 infection (3, 4). In addition, these studies noted the absence of CTL reactivity associated with failure to resolve plasma viremia (4), and that declines in CTL reactivity may precede a resurgence of viremia (3). Strong HIV-specific CTL responses have also been detected in long-term nonprogressing HIV+ subjects (5).
Along with conventional major histocompatibility complex class I restricted CTL reactivities, cellular immune responses to HIV-1 include a noncytolytic CD8+ T cell-mediated suppression of virus not restricted by major histocompatibility complex class I antigens at the effector phase (6). It is well established that activated CD8+ T cells from HIV+ individuals cells can suppress HIV-1 in primary CD4+ lymphocytes (7), and that virus suppression is at least partly mediated by CD8+ cell-derived soluble factors (8). The CC chemokines RANTES (regulated on activation, normal T cell expressed and secreted), macrophage inflammatory protein 1α (MIP-1α), and MIP-1β, were recently identified as HIV-inhibitory substances secreted by CD8 cells (9). Chemokines are members of a family of related proinflammatory cytokines having a variety of biological properties including leukocyte chemotaxis and activation (10). They are secreted by a range of cell types that include activated CD4+ and CD8+ T cells. Certain chemokine receptors have recently been shown to serve as cofactors for HIV-1 fusion with CD4+ cells (11–16). Non-syncytia-inducing (NSI) strains predominantly use the CCR5 chemokine receptor whereas syncytia-inducing (SI) strains use another chemokine receptor, CXCR4 as a fusion cofactor (11–16). This accounts for the observation that the CCR5 ligands RANTES, MIP-1α, and MIP-1β can inhibit NSI but not SI strains such as NL4-3 or LAI (9, 12, 13, 16). The CXC chemokine stromal cell-derived factor 1 (SDF-1) was recently identified as a natural ligand for CXCR4 and shown to inhibit infection by SI but not NSI strains of HIV-1 (17, 18).
Whereas several studies indicate that the CC chemokines RANTES, MIP-1α, and MIP-1β may account for much of the CD8+ T cell-soluble activity against NSI strains of HIV-1 (9, 19), they cannot explain the full spectrum of suppressive activity exhibited by CD8+ cells (7, 19–21), particularly the inhibition of SI strains. These strains of HIV-1 are generally not associated with transmission or the initial phases of HIV-1 disease; however their development is often associated with more rapid declines in CD4+ T cells and progression to overt AIDS (22). The reasons why T cell tropic SI strains fail to emerge early during the course of HIV-1 disease is unclear, but the ability of CD8+ cells to suppress their replication may be a contributing factor.
We considered the possibility that in a manner analogous to CC chemokine-mediated suppression of NSI strains, CD8+ cells may produce one or more inhibitory factors that are ligands for the CXCR4 receptor used by SI strains. To investigate the possibility that the CXCR4 ligand SDF-1 may play a role in this suppression, we examined several SDF-1 preparations for their ability to block HIV-1-mediated cell–cell fusion. Our studies provide information on the structure-activity relationships of this CXC chemokine and demonstrate the sensitivity of SDF-1 antiviral activity to amino terminal alterations. We also investigated the levels of SDF-1 transcripts in primary CD8+ T cell effectors and a herpesvirus saimiri (HVS)-transformed CD8+ T cell line. Our findings lead us to conclude that SDF-1 is unlikely to account for the noncytolytic inhibitory activity of CD8+ T cells against SI strains of HIV-1.
MATERIALS AND METHODS
Chemokines and Antichemokine Antibodies.
Recombinant human RANTES, MIP-1α, and MIP-1β were from R & D Systems and Genzyme Diagnostics. Neutralizing antibodies to MIP-1α, MIP-1β, and RANTES and Quantikine immunoassay kits were from R & D Systems. Sources of SDF-1 preparations evaluated were as follows. Chemically synthesized SDF-1α (1–67) was a kind gift from Ian Clark-Lewis (17, 18). Chemically synthesized SDF-1α (−2 to 68) was prepared at Berlex Biosciences (Richmond, CA) (23). Recombinant SDF-1α (1–68) and SDF-1β (1–72) were obtained from Bob Goldman (Peprotek, Rocky Hill, NJ), and recombinant SDF-1β (−3 to 72) was obtained from R & D Systems. Numbering of SDF-1 amino acids is based on its ORF (24) after designating the N-terminal lysine of the mature protein described by Bleul et al. (25) as position number 1.
Preparation of Primary CD8+ T-Lymphocytes.
Venous blood was obtained from HIV− donors or asymptomatic HIV+ patients infected for >7 years. Peripheral blood mononuclear cells (PBMCs) were prepared by Ficoll-Hypaque density separation. CD8+ T cells were captured on anti-CD8 microCellector flasks (Applied Immune Sciences, Santa Clara, CA) according to the manufacturer’s recommendations, and activated for 3 days with a mixture of 50 ng/ml anti-CD3 and 100 ng/ml anti-CD28 antibodies in AIM-V medium supplemented with 10% fetal calf serum, 20 units/ml interleukin 2 (IL-2), and penicillin-streptomycin at 37°C in a humidified incubator.
Preparation of Cell Culture Supernatant Concentrates.
A HVS-transformed CD8+ cell line derived from a HIV+ asymptomatic patient (7) was cultured at 2 × 106/ml in serum-free AIM-V medium containing 20 units/ml IL-2 in a spinner culture vessel for 3–4 days. The cells were removed by centrifugation and the conditioned medium clarified with a 0.4-micron filter. Ammonium sulfate was added at 4°C to 65% saturation, and the precipitate was collected by centrifugation at 5,500 × g for 30 min, resuspended in a minimum volume of 0.1× PBS, and dialysed for 24 h against several changes of 0.1× PBS. The material was lyophilised before reconstitution in a minimal volume to yield ≈30-fold concentration over the starting supernatants. Quantikine immunoassay kits were used to quantify RANTES, MIP-1α, and MIP-1β levels in this material.
Virus Suppression Assays.
Purified CD8+ T cells from HIV+ and HIV− subjects were removed from the microCellector flasks after activation, washed, and expanded for 3 days before use. CD8+-depleted target cells were derived from cryopreserved PBMC pools from HIV− donors. PBMC were antibody-activated as above for 2–3 days, and depleted of CD8+ cells with anti-CD8 antibody coated magnetic beads (Dynal) according to the manufacturers’ procedures. CD8-depleted cells (1 × 105) were seeded per well in 96-well U-bottom plates. HIV-1 virus stocks and CD8+ effectors or their culture supernatants were added at varying ratios to a final volume of 100 μl. In antibody neutralization experiments, the CD8+ T cells or their concentrated supernatants were preincubated with antibodies for 1 h at room temperature before use. Cells were incubated in AIM-V medium supplemented as above in a humidified 37°C incubator. Every 3–4 days, 40 μl of supernatant was removed, adjusted to 1% Triton, and assayed for reverse transcriptase activity as described (26). Cultures were refed with fresh medium containing CD8 supernatants or antibodies to maintain the original conditions.
HIV-1-Mediated Cell–Cell Fusion Assay.
Fusion assays were performed as described (27) except that CEM cells were employed as the uninfected fusion partner. Briefly, uninfected CEM cells (7 × 104) were incubated with CEM cells (1 × 104) chronically infected with HIV-1LAI in 96-well half-area plates (Costar) in the presence or absence of various concentrations of the SDF-1 preparations in a final volume of 100 μl of culture medium. Concentrations of SDF-1 in stock solutions were determined with a BCA protein assay (Bio-Rad) using BSA as a standard. Dilutions of the SDF-1 preparations in 10 μl of culture medium were added to the cell mixtures at initial setup. After 24 h at 37°C, multinucleated syncytia were enumerated by microscopic examination, and the IC90 was estimated by interpolation.
Reverse Transcription–PCR (RT-PCR) Amplification.
RT-PCR was performed using a Perkin–Elmer/Cetus GeneAmp RT-PCR kit and a Perkin–Elmer model 9600 thermocycler. SDF-1 mRNA was reverse transcribed with primer SDFα4 (5′-TTCTCCAGGTACTCCTGAATCC-3′), whereas reverse transcription for actin PCR was primed using oligo(dT). Reverse transcription occurred at 42°C for 15 min followed by 5 min at 99°C and cooling to 4°C. SDF-1 cDNAs were amplified using the SDFα4 3′ primer with the 5′ primer SDFα3 (5′-TGAGCTACAGATGCCCATGC-3′). PCR conditions were as follows: denaturation at 95°C for 2 min, followed by 35 cycles of 95°C for 15 s, 63°C for 45 s, and 72°C for 30 s, with a final 10 min incubation at 72°C. Actin PCR primers were as follows: forward, 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′; and reverse, 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′. Actin PCR conditions were the same as SDF-1 amplifications except the annealing temperature was 65°C and the number of cycles was 24. The actin PCR product had a predicted size from spliced mRNA of 661bp. PCR products were visualized with ethidium bromide in 2% agarose gels, and in some cases transferred to Hybond N+ membrane (Amersham) for hybridization. The SDF-1 PCR product was cloned into the vector pT7blue (Novagen) and sequenced using an Applied Biosystems Prism 377 automated DNA sequencer. SDF-1 RNA standards for quantitative RT-PCR were synthesized with a T7 RNA polymerase transcription kit (Novagen), using the restriction enzyme-linearized plasmid containing the cloned SDF-1 PCR product as a template. 32P-labeled riboprobes were made by incorporating [α-32P]CTP into the SDF-1 transcripts.
RESULTS
RANTES, MIP-1α, and MIP-1β Suppress NSI but Not SI Strains of HIV-1.
To facilitate studies of CD8+ T-lymphocyte suppression of HIV-1, we generated a HVS-transformed CD8+ cell line from the cells of a long-term asymptomatic HIV+ patient (7). This cell line, CD8(HVS), exhibits similar virus-suppressive reactivity as the primary CD8+ cells from which it was derived, although it lacks detectable CTL activity (7). Concentrated supernatants were prepared from CD8(HVS) cells to evaluate soluble factors as contributors to CD8-suppressive activity. Following the identification of RANTES, MIP-1α, and MIP-1β as HIV-1 suppressive factors secreted by CD8+ cells (9), we assayed the concentrated supernatants and found that HVS transformed-CD8+ cells secreted each of these CC chemokines as previously reported for HTLV-I-transformed and primary CD8+ cells (9).
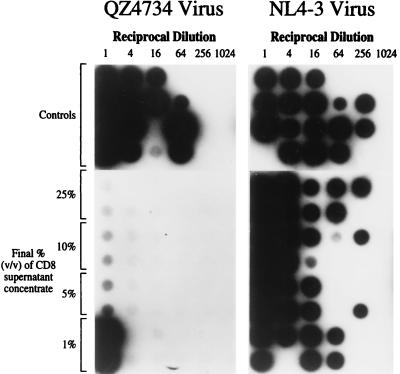
The concentrated supernatants were tested for inhibitory activity against NSI and SI viruses in assays that quantify virus titer on primary CD4+ lymphocyte targets. The concentrates had potent antiviral activity against NSI but not SI strains of HIV-1 (Fig. 1). A representative result from these experiments is shown in Fig. 1 for a concentrated supernatant that contained 1,300 ng/ml MIP-1α, 500 ng/ml MIP-1β, and 132 ng/ml RANTES. CD4+ lymphocytes were infected with serial dilutions of virus stocks in the presence of varying amounts of the concentrate, and virus titer (expressed as TCID50/ml) was estimated from end-point dilution (27). As little as 5% (vol/vol) of the concentrated supernatant reduced the titer of the QZ4734 NSI isolate by more than 100-fold. This isolate was derived from an individual during acute HIV infection and behaves similarly to several other low passage primary isolates in these assays. In contrast, the titer of NL4–3 was not significantly reduced by the concentrated supernatant even at high concentrations (Fig. 1).
Figure 1.
Sensitivity of the QZ4734 primary HIV-1 isolate and the NL4-3 strain to suppression by a concentrated supernatant from the CD8(HVS) cell line. The autoradiograph represents products of reverse transcriptase assays of culture supernatants from a 96-well plate. Each well contained 9 × 104 activated CD4+ cells from a pool of HIV− donors. Virus was titrated in the horizontal dimension, and the supernatant concentrate titrated in the vertical dimension. Cultures were sampled and refed at 3- to 4-day intervals. The figure is from day 7 when control virus titers were 3.2 × 103 and 9.3 × 103 TCID50/ml for the QZ4734 and NL4-3 isolates, respectively.
In agreement with reports from several groups (9, 12, 13, 16, 20), we found that the NL4-3 and LAI strains of HIV-1 were not inhibited by the CC chemokines. We determined that RANTES, MIP-1α, and MIP-1β were mostly responsible for the activity of the concentrated supernatant against NSI isolates based on the ability of a mixture of antibodies to all three CC chemokines to abrogate the suppressive activity. However, the same mixture of antibodies was unable to effect the virus-suppressive activity of the CD8(HVS) cells in coculture experiments at effector to target cell ratios of ≥1:1 (ref. 11; data not shown). This result suggested that CD8 cell-associated factors other than the three CC chemokines may also contribute to virus inhibition and is consistent with other recent studies (19–21).
CD8+ Cells Suppress SI HIV-1 Strains.
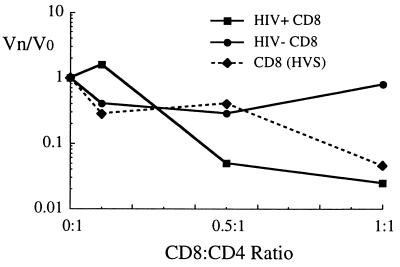
To focus on CD8+ cell virus-suppressive activities that were not mediated by the CC chemokines, we investigated a variety of CD8+ effector cells for their ability to inhibit CC chemokine-resistant SI strains of HIV-1. Using a 1 log reduction in virus titer as a criteria for suppression, we found that CD8+ cells from a majority of HIV-1+ asymptomatic individuals as well as the transformed CD8(HVS) cells inhibit the SI viruses HIV-1LAI and NL4-3 when tested at effector to target cell ratios of ≥1:1. In contrast, CD8+ cells from HIV-1− normal donors rarely inhibit these viruses. An example of suppression of NL4-3 is shown in Fig. 2. CD8+ cells from an HIV+ donor as well as the CD8(HVS) cells potently inhibited NL4-3 infection, whereas CD8+ cells from the HIV− normal donor did not significantly alter NL4-3 titer (Fig. 2).
Figure 2.
Suppression of NL4-3 replication in primary CD4+ cells by primary CD8+ lymphocytes from an HIV+ asymptomatic individual, from an HIV− donor, and the CD8(HVS) cell line. The Vn/Vo ratio is defined as the apparent virus TCID50/ml in the presence of the effector (Vn), divided by the TCID50/ml in the control wells (Vo). Vo was 8,063 TCID50/ml for this experiment.
SDF-1 Suppresses SI Strains of HIV-1.
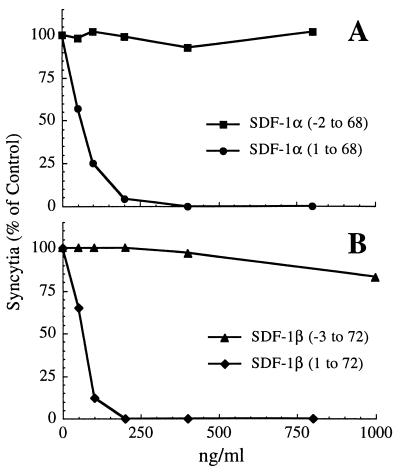
The recent identification of SDF-1 as a natural CXCR4 ligand, and its reported activity against SI strains of HIV-1, led us to speculate that in a manner analogous to CC chemokine inhibition of NSI strains, SDF-1 might be an effector molecule for CD8-mediated inhibition of SI strains. However, initial experiments with a recombinant SDF-1β (−3 to 72) from R & D Systems failed to demonstrate antiviral activity against the HIV-1LAI isolate in either PBMC or CEM T cell targets. The reason for the discrepancy between our results with recombinant SDF-1β (−3 to 72) and those with a chemically synthesized SDF-1α (1–67) (17, 18) was not immediately apparent, but we noted that the SDF-1 preparations differed in sequence at both the amino and carboxyl termini. To investigate this further, we assembled a panel of SDF-1α and SDF-1β preparations, including SDF-1α (1–67), to test their ability to inhibit HIV-1LAI-mediated fusion. The sources of these preparations and their N- and C-terminal sequences are described in Materials and Methods and Table 1, respectively. Infectivity experiments with HIV-1LAI in PBMC confirmed that SDF-1α (1–67) was active while SDF-1β (−3 to 72) was not (data not shown). We compared activities for the full panel of SDF-1 preparations in cell–cell fusion assays employing CEM cells chronically infected with HIV-1LAI and uninfected CEM cells as fusion partners (Fig. 3 and Table 1). In stark contrast to the inactive SDF-1β (−3 to 72), a recombinant SDF-1β (1–72) having the same N terminus as SDF-1α (1–67) (25) exhibited potent fusion blocking activity (Fig. 3B). The activity of SDF-1β (1–72) was nearly identical to that of two SDF-1α preparations differing only by 1 amino acid at the C terminus [SDF-1α (1–67) and SDF-1α (1–68), Table 1]. These results demonstrate that fusion blocking activity is not influenced by amino acids differentiating the C-terminus of SDF-1α and SDF-1β. Furthermore, they suggested that the lack of antiviral activity of SDF-1β (−3 to 72) (Fig. 3B and Table 1) was likely due to the N-terminal amino acid extension. The N-terminal region was previously shown to be crucial for the biological activity of another CXC chemokine, IL-8 (28), and a 5-amino acid truncation of the N terminus of SDF-1 resulted in loss of Ca2+ signaling, chemotaxis, and anti-HIV-1 activity (18). To examine the influence of extending the N terminus of SDF-1, we compared the fusion blocking activity of SDF-1α (1–68) and SDF-1α (−2 to 68) differing only by two amino acids at their N terminus, and found that SDF-1α (−2 to 68) did not block fusion of HIV-1LAI-infected cells in our assays (Fig. 3A). Table 1 summarizes the fusion blocking activity (expressed as IC90) of the SDF-1 preparations and compares their N- and C-terminal sequences with biological activity. No toxicity was observed with any of the SDF-1 preparations when examined at concentrations up to 1.6 μM in the cell-fusion assay. We found that fusion blocking activity was associated with forms of SDF-1α or SDF-1β that had the same N terminus described by Bleul et al. (25) for the mature form of SDF-1α. An extension of two or three amino acids at the N terminus abolished this activity, whereas C-terminal extensions of up to 5 amino acids did not influence the antiviral properties of SDF-1. Several SDF-1 preparations that did not block fusion were nevertheless able to induce a signal or stimulate chemotaxis, presumably through their interaction with CXCR4, suggesting that the requirements for antiviral activity and receptor mediated signaling may not be identical.
Table 1.
Sequences and activities of the SDF-1α and SDF-1 β preparations
| Preparation | Derivation | Syncytia blocking | IC90 μM | Biologic activity | N-terminal sequence | C-terminal sequence |
|---|---|---|---|---|---|---|
| SDF1-α 1–67 | Synthetic | + | ≤0.12 | +* | KPVSLSYR | ALN |
| SDF1-α 6–67 | Synthetic | ND | ND | −† | SYR | ALN |
| SDF1-α 1–68 | Recombinant | + | ≤0.13 | +‡ | KPVSLSYR | ALNK |
| SDF1-α −2 to 68 | Synthetic | − | ≥1.6 | +§ | DGKPVSLSYR | ALNK |
| SDF1-β 1–72 | Recombinant | + | ≤0.11 | +‡ | KPVSLSYR | ALNKKRFM |
| SDF1-β −3 to 72 | Recombinant | − | ≥1.0 | +¶ | SDGKPVSLSYR | ALNKKRFM |
The sources of the preparations are given in Materials and Methods.
Measured by chemotaxis of human lymphocytes; maximal at ≈125 nm (26).
Measured by chemotaxis of human lymphocytes; inactive up to ≈1 μM (26).
Measured by microphysiometer in Jurkat T cells EC50 ≈100 nM (R.H and M. Liang unpublished data).
Measured by chemotaxis in human hNT neurons EC50 ≈50 nM (33).
Measured by chemotaxis of human T-lymphocytes in IL-2 for 8-10 days, ED50 ≈ 60 nM (R & D Systems).
Figure 3.
Comparison of (A) two SDF-1α and (B) two SDF-1β preparations for their ability to inhibit HIV-1LAI-induced syncytia formation in an assay employing CEM cells. The results are expressed as percent of the numbers of syncytia formed in control wells containing no SDF-1. Assay details are described in Materials and Methods.
CD8+ Cells Contain Extremely Low Levels of SDF-1 Transcripts.
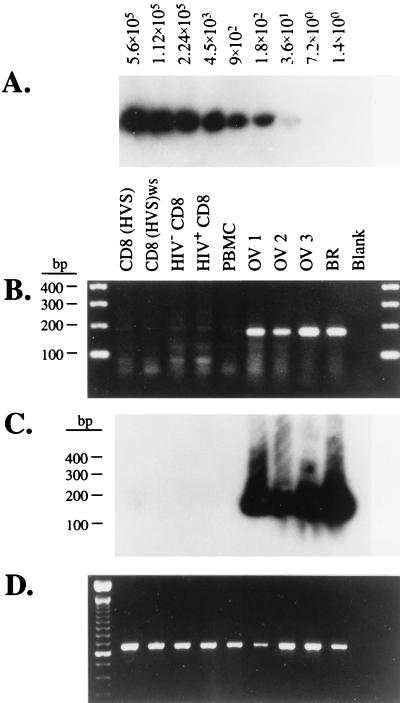
The results described above demonstrate that both SDF-1α and SDF-1β can inhibit SI viruses. To determine whether SDF-1 was indeed an effector molecule for CD8+ cell inhibition of SI viruses, we examined CD8+ effector cells for SDF-1 mRNA transcripts, because an antibody to SDF-1 was unavailable. We prepared total RNA from aliquots of each effector cell population in Fig. 2 and probed these RNAs for transcripts to either SDF-1α or -1β using RT-PCR with primers designed to span an mRNA splice junction. The predicted RT-PCR processed product size from SDF-1α or SDF-1β mRNA was 178 bp. We failed to amplify an obvious SDF-1 product from any CD8+ cell or PBMC RNA preparation (Fig. 4B), although RT-PCR for actin transcripts indicated the samples contained comparable amounts of amplifiable actin mRNA to control RNA samples from ovarian and breast tissue (Fig. 4D). By contrast, we readily amplified the predicted 178-bp SDF-1 RT-PCR product from the control RNA preparations (Fig. 4B). We cloned and sequenced this 178-bp fragment, verifying its identity with the published SDF-1 cDNA sequence (24). To ascertain the sensitivity limits of our RT-PCR procedures, transcripts were made from the cloned SDF-1 sequence and used as an external quantitation standard. We were able to detect ≥150 copies of the in vitro RNA transcript by ethidium bromide fluorescence, and further increased detection sensitivity with Southern blots of RT-PCR amplifications using radiolabeled riboprobes made from the cloned SDF-1 sequence. Hybridization with radiolabeled probes permitted detection of as few as 35 in vitro SDF-1 transcripts in a non-nested amplification (Fig. 4A), as well as the observation of a very faint band at a position corresponding to the SDF-1 PCR product in some amplifications. This faint product was seen with RNA from both CD8+ and CD4+ cells and there was no correlation of its presence or intensity with cell suppressive activity. Assuming a detection limit of 35 transcripts (Fig. 4A), we estimated an upper limit of one SDF-1 transcript per 8,500 cells from the assay depicted in Fig. 4 C and A because the amplifications contained 300,000 cell equivalents. Analysis of additional RNA samples using twice the number of cell equivalents provided a more conservative upper limit of one SDF-1 transcript per 5,000 CD8+ cells. Examination of CD8+ lymphocytes from four other HIV+ individuals for SDF-1 transcripts yielded similar results (data not shown). Although it remains a formal possibility that SDF-1 protein levels may not correlate with SDF-1 transcript levels, the extremely low levels of SDF-1 mRNAs detected in virus-suppressive CD8+ cells leads us to conclude that neither antibody-activated primary CD8+ lymphocytes nor the CD8(HVS) cells are likely to produce sufficient amounts of SDF-1 to account for their inhibition of SI viruses (Fig. 2).
Figure 4.
RT-PCR analysis of SDF-1 mRNA content of primary and transformed CD8+ effector cells. (A) Autoradiograph of a Southern blot of RT-PCR products from a dilution series of an in vitro SDF-1 transcript. The blot was probed with a 32P-labeled SDF-1 riboprobe. (B) Ethidium bromide-stained agarose gel of RT-PCR products of total RNA extracted from either HVS-transformed CD8+ cells, primary CD8+ cells from HIV+ and HIV− subjects, unfractionated PBMC from a pool of HIV− donors, and control tissues as noted. OV, ovarian tumor cells; BR, breast tumor cells. (C) Autoradiograph of a Southern blot of the gel in B probed with a 32P-labeled SDF-1 riboprobe. (D) Ethidium bromide-stained agarose gel bearing actin RT-PCR products amplified from the same RNA preparations used for the amplifications in B.
DISCUSSION
In this study we have addressed the question of whether the CXC chemokine SDF-1 plays a part in the noncytolytic suppression of SI strains of HIV-1 by CD8+ cells. The CC chemokines RANTES, MIP-1α, and MIP-1β have been a focus of attention since their identification as HIV-1-suppressive factors produced by CD8 cells (10) and the finding that their receptors are cofactors for virus–cell fusion (11–16). Despite their obvious importance, the CC chemokines are specific inhibitors only for NSI strains that use the CCR5 receptor. Such viruses are associated with transmission of HIV-1 and predominate early in the course of disease (29). Nevertheless, progression to AIDS is often associated with emergence of SI viruses (22, 29) that utilize the CXCR4 receptor as a cofactor for entry (11, 29). There may also exist transitional viruses between the SI and NSI phenotypes that can use a wider range of receptors and infect both macrophages and transformed CD4+ cell lines (14, 15, 29). Viruses evolving to utilize the CXCR4 receptor would presumably find an expanded range of host cells as this receptor is found on a wide range of cell types (30).
The reasons why SI variants are not associated with transmission of HIV-1 or not found during the early phases of infection are unclear, but may reflect the microenvironments where transmission and replication of HIV-1 takes place. SI variants have the capacity to use CXCR4, and as demonstrated above, SDF-1 effectively inhibits these strains (Fig. 3 and Table 1). This CXC chemokine is produced by a wide variety of cell types (ref. 24; Fig. 4), which may be present at relevant sites of viral replication. The inhibition of SI strains by SDF-1 suggests the possible design of therapeutic strategies targeted at CXCR4. In this regard our structure-function studies of SDF-1 are informative.
The SDF-1 proteins, in common with all other chemokines, are synthesized with an N-terminal leader that is enzymatically removed to yield the mature form of the molecule (31). Correct processing at the N terminus is essential for the generation of biologically active forms of the protein (32). Studies have shown that the biological activity of the CXC chemokine IL-8 (28), and the CC chemokines RANTES (33) and monocyte chemotactic protein 1 (MCP-1) (34), are critically dependent on the N-terminal region. The importance of the N-terminal region of chemokines for receptor activity has been further underscored by mutagenesis studies. Specifically, the addition of an extra amino acid to the N terminus of RANTES converts this chemokine into a potent receptor antagonist (33). Cleavage of one amino acid from the N terminus of MCP-1 generates a truncated molecule, MCP-12–76, that is able to activate eosinophils but that has lost its ability to activate basophils (35). Removal of 4 amino acids from the N terminus of IL-8 generates a mutant that has low receptor binding affinity and is totally unable to stimulate elastase release from neutrophils (36). These data suggest that N-terminal modification of chemokines can modify their bioactivity and also unmask a potential to switch their effector cell type. Here we have shown that the correct processing of SDF-1 proteins is important for their ability to suppress viral infectivity via competition for binding to the chemokine receptor CXCR4. A 2-amino acid extension at the N terminus of SDF-1 renders the molecule almost totally inactive in its ability to block viral infectivity of the HIV-1LAI strain, yet it is still capable of high affinity binding and activation of target cells. These data suggest that amino terminal modification of SDF-1 can discriminate between the ability of SDF-1 to signal and to inhibit HIV-1 infection. More detailed studies with SDF-1 will undoubtedly yield valuable information regarding the ability of the chemokine to act as an HIV-1 inhibitor, and could point the way toward designing novel therapeutics.
The viral envelope sequence appears to be the major determinant of receptor usage, in particular the sequences of the V3 loop (15, 37) which largely determine the SI/NSI phenotype. It will be important to understand the immunological constraints and pressures influencing the evolution and receptor usage of HIV-1 within the host. One factor may be CD8+-mediated noncytolytic inhibition of replication of NSI viruses by secretion of the CC chemokines RANTES, MIP-1α, and MIP-1β. The possibility exists that the switch from an NSI to an SI phenotype is partly an evasion of this suppression. Nonetheless, CD8+ cells from infected individuals also have the ability to suppress production of SI strains such as NL4-3 and LAI (Fig. 2; ref. 20). Although we did not recover soluble materials from the CD8(HVS) cells that suppressed SI viruses, other investigators have reported such activities for CD8+ cell derived supernatants (20, 21), suggesting that this activity may not have been captured in the transformed population. The mechanisms of this suppression are as yet unknown, but by analogy with the CC chemokines, some part of this effect may be mediated by production of CXCR4 ligands. The recent identification of SDF-1α as a CXCR4 ligand and inhibitor of SI strains of HIV-1 (17, 18) led us to investigate whether it might be a component of CD8+ suppression. We find that while SDF-1α is active (Fig. 3 and Table 1), the SDF-1 gene is not significantly transcribed in activated CD8+ cells from a range of sources (Fig. 4). Prior work employing less sensitive Northern analysis indicated that SDF-1 mRNA was expressed in a wide range of tissues, but not in PBMC (24). Our findings do not exclude the possibility that SDF-1 may be important for inhibiting HIV replication in some compartments of the body where cells that produce this chemokine may be present, or in part account for the lack of transmission of SI strains.
Because SDF-1 is not the effector molecule in CD8+ suppression of SI isolates, it is unclear how CD8+ cells exert their effects, and which points of the virus life cycle are targeted. One possibility is that suppression is indeed at the level of virus entry, perhaps mediated by an as yet to be identified CXCR4 ligand. In preliminary experiments using PCR to assess virus entry, we observed that CD8 cells can decrease entry of HIV-1LAI (S. A. Stanfield-Oakley and M.L.G., unpublished observations). However, the magnitude of this effect did not appear sufficient to account for the extent of virus suppression (Fig. 2), suggesting that CD8 suppression of SI viruses, like that of NSI viruses, may be multifactoral (29). We and others have previously provided evidence that CD8 cells may also inhibit SI viruses at a later step in the infection process, and Moriuchi et al. (20) recently reported that CD8+ cell supernatants suppressed HIV expression from chronically infected promonocytic U1 cells. Our group found that primary CD8+ cells, and to some extent their soluble factors, inhibit the HIV-1 LTR promoter from the LAI strain (26). These findings have largely been confirmed by others (38, 39), although the contribution of transcriptional inhibition to CD8+ cell-mediated suppressive activity remains to be established. IL-16, a lymphocyte chemoattractant expressed as both a membrane and soluble form, exhibits anti-HIV activity (40) and may repress HIV-1 promoter activity directly or indirectly through effects on T cell activation (41). We have yet to investigate levels of IL-16 or whether it is involved in the suppression of SI isolates.
Despite rapid advances in the fields of HIV-1 coreceptors and chemokines, identification of the molecule(s) and mechanisms involved in CD8+-mediated virus suppression remains incomplete. As reported here, the CXCR4 ligand SDF-1 is unlikely to account for the CD8+ suppression of SI viruses. CD8 cells exhibit CC chemokine-independent suppressive activities toward NSI viruses (7, 19–21). Whether or not the mechanisms by which CD8 cells inhibit SI viruses will also account for their chemokine-independent activities against NSI viruses remains to be determined.
Acknowledgments
We thank Drs. Dani P. Bolognesi and Kent J. Weinhold for many insightful discussions, and Dr. Coreen Oei for the kind gift of RNA from ovarian and breast tissues. This work was supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health: RO1-AI32393-5 and RO1-AI40017-01A1 (M.L.G.), and P30-AI28662-08 (Center For AIDS Research).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: SDF-1, stromal cell-derived factor 1; MIP-1, macrophage inflammatory protein 1; RANTES, regulated on activation, normal T cell expressed and secreted; PBMC, peripheral blood mononuclear cells; HVS, herpesvirus saimiri; CTL, cytotoxic T-lymphocytes; SI, syncytia-inducing; NSI, nonsyncytia-inducing; RT-PCR, reverse transcription–PCR; IL, interleukin.
References
- 1.Schnittman S M, Fauci A S. Adv Intern Med. 1994;39:305–355. [PubMed] [Google Scholar]
- 2.Pantaleo G, Fauci A S. Annu Rev Microbiol. 1996;50:825–854. doi: 10.1146/annurev.micro.50.1.825. [DOI] [PubMed] [Google Scholar]
- 3.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B A. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrer T, Harrer E, Kalams S A, Elbeik T, Staprans S I, Feinberg M B, Cao Y, Ho D D, Yilma T, Caliendo A M, Johnson R P, Buchbinder S P, Walker B D. AIDS Res Hum Retroviruses. 1996;12:585–592. doi: 10.1089/aid.1996.12.585. [DOI] [PubMed] [Google Scholar]
- 6.Walker C M, Moody D J, Stites D P, Levy J A. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg M L, Lacey S F, Chen C H, Bolognesi D P, Weinhold K J. Semin Immunopathol. 1997;18:355–369. doi: 10.1007/BF00813503. [DOI] [PubMed] [Google Scholar]
- 8.Walker C M, Levy J A. Immunology. 1989;66:628–630. [PMC free article] [PubMed] [Google Scholar]
- 9.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;279:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 10.Schall T J, Bacon K B. Curr Opin Immunol. 1994;6:865–873. doi: 10.1016/0952-7915(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 12.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C, Peiper S C, Schall T J, Littman D R, Landau N R. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 13.Dragic T, Litwin V, Allaway G P, Martin S, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 14.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 15.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, Larosa G, Newman W, Gerard N, Gerard C, Sodroski J. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 16.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 17.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwarz O, Heard J-M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. Nature (London) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 18.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. Nature (London) 1996;382:829–832. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 19.Kinter A L, Ostrowski M, Goletti D, Oliva A, Weissman D, Gantt K, Hardy E, Jackson R, Ehler L, Fauci A S. Proc Natl Acad Sci USA. 1996;93:14076–14081. doi: 10.1073/pnas.93.24.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriuchi H, Moriuchi M, Combadiere C, Murphy P M, Fauci A S. Proc Natl Acad Sci USA. 1996;93:15341–15345. doi: 10.1073/pnas.93.26.15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy J A, Mackewicz C M, Barker E. Immunol Today. 1996;17:217–224. doi: 10.1016/0167-5699(96)10011-6. [DOI] [PubMed] [Google Scholar]
- 22.Tersmette M, Lange J M, de Goede R E, de Wolf F, Eeftink-Schattenkerk J K, Schellekens P T, Coutinho R A, Huisman J G, Goudsmit J, Miedema F. Lancet. 1989;i(8645):983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 23.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. Curr Biol. 1997;7:1121–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 24.Shirozu M, Nakano T, Inazawa J, Tashiro K, Tada H, Shinohara T, Honjo T. Genomics. 1995;28:495–500. doi: 10.1006/geno.1995.1180. [DOI] [PubMed] [Google Scholar]
- 25.Bleul C C, Fuhlbrigge R C, Casasnovas J M, Aiuti A, Springer T A. J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C-H, Weinhold K J, Bartlett J A, Bolognesi D, Greenberg M L. AIDS Res Hum Retroviruses. 1993;9:1079–1086. doi: 10.1089/aid.1993.9.1079. [DOI] [PubMed] [Google Scholar]
- 27.Chen C-H, Matthews T J, McDanal C B, Bolognesi D P, Greenberg M L. J Virol. 1995;69:3771–3777. doi: 10.1128/jvi.69.6.3771-3777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moser B, Dewald B, Barella L, Schumacher C, Baggiolini M, Clark-Lewis I. J Biol Chem. 1993;268:7125–7128. [PubMed] [Google Scholar]
- 29.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loetscher M, Geiser T, O’Reilly T, Zwahlen R, Baggiolini M, Moser B. J Biol Chem. 1994;269:232–237. [PubMed] [Google Scholar]
- 31.Schall T. In: The Cytokine Handbook. 2nd Ed. Thompson A, editor. San Diego: Academic; 1994. pp. 419–460. [Google Scholar]
- 32.Baggiolini M, Dewald B, Moser B. Adv Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- 33.Proudfoot A E, Power C A, Hoogewerf A J, Montjovent M O, Borlat F, Offord R E, Wells T N. J Biol Chem. 1996;271:2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- 34.Gong J H, Clark-Lewis I. J Exp Med. 1995;181:631–640. doi: 10.1084/jem.181.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber M, Uguccioni M, Baggiolini M, Clark-Lewis I, Dahinden C A. J Exp Med. 1996;183:681–685. doi: 10.1084/jem.183.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. J Biol Chem. 1991;266:23128–23134. [PubMed] [Google Scholar]
- 37.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 38.Mackewicz C E, Blackbourn D J, Levy J A. Proc Natl Acad Sci USA. 1995;92:2308–2312. doi: 10.1073/pnas.92.6.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Copeland K F T, McKay P J, Rosenthal K L. AIDS Res Hum Retroviruses. 1995;11:1321–1326. doi: 10.1089/aid.1995.11.1321. [DOI] [PubMed] [Google Scholar]
- 40.Baier M, Werner A, Bannert N, Metzner K, Kurth R. Nature (London) 1995;378:563. doi: 10.1038/378563a0. [DOI] [PubMed] [Google Scholar]
- 41.Theodore A C, Center D M, Nicoll J, Fine G, Kornfeld H, Cruikshank W W. J Immunol. 1996;59:1958–1964. [PubMed] [Google Scholar]