Abstract
Down-regulation of the initial burst of viremia during primary HIV infection is thought to be mediated predominantly by HIV-specific cytotoxic T lymphocytes, and the appearance of this response is associated with major perturbations of the T cell receptor repertoire. Changes in the T cell receptor repertoire of virus-specific cytotoxic T lymphocytes were analyzed in patients with primary infection to understand the failure of the cellular immune response to control viral spread and replication. This analysis demonstrated that a significant number of HIV-specific T cell clones involved in the primary immune response rapidly disappeared. The disappearance was not the result of mutations in the virus epitopes recognized by these clones. Evidence is provided that phenomena such as high-dose tolerance or clonal exhaustion might be involved in the disappearance of these monoclonally expanded HIV-specific cytotoxic T cell clones. These findings should provide insights into how HIV, and possibly other viruses, elude the host immune response during primary infection.
After infection of the host with a virus, a complex series of virologic and immunologic events characterize the phase of primary infection. The primary objective of the host is to eliminate the virus, and indeed a vigorous virus-specific immune response is rapidly generated to achieve this goal. In contrast, the primary objective of the virus is to persist and to survive in the infected host. In this regard, recent studies have demonstrated that viruses, including HIV, have evolved several strategies to evade the immune response and to establish chronic infection. These include: (i) viral latency (1, 2), (ii) inhibition of antigen presentation and processing (3), and (iii) mutations in the viral genome that may compromise recognition by neutralizing antibodies or by virus-specific cytotoxic T lymphocytes (CTL), and also may alter binding to the major histocompatibility complex, which, in turn, may lead to either anergy or generation of antagonists that may prevent virus-specific T cells from mediating their effector functions (4–9). Several of these mechanisms have been shown to be operative in HIV infection (6, 7, 9).
A substantial proportion (50–70%) of HIV-infected individuals experience a self-limited clinical syndrome of variable severity during primary infection (2–13). Despite the early appearance of robust humoral and cell-mediated immune response (10–18), HIV is virtually never completely eliminated and a state of chronic, persistent infection develops in most individuals (2, 10–13).
Major expansions of CD8+ T cells with a predominant Vβ usage have been observed during the primary immune response to HIV (17) and simian immunodeficiency virus (19). These expansions are of variable magnitude and may involve single or multiple Vβ families. The Vβ-specific expansions contain HIV-specific CTL and thus are thought to be part of the primary cell-mediated immune response to HIV (17). Furthermore, qualitative differences in the patterns of Vβ-specific expansions during acute HIV infection have been correlated with different rates of disease progression (20).
The present study has characterized the primary cell-mediated immune response to HIV infection, with particular focus on the changes in the T cell receptor (TCR) repertoire. Our results indicate that the HIV-specific T cell repertoire is severely compromised during primary infection due to the disappearance of the initially expanded virus-specific clonotypes expressed by CD8+ CTL.
MATERIALS AND METHODS
Patients with Primary HIV Infection.
Two patients with well documented historical, clinical, and laboratory parameters of primary HIV infection were studied serially at various times after infection. Both developed clinical symptoms within 4 to 6 weeks of the time of exposure to HIV and experienced a clinical syndrome of moderate severity with fever, diarrhea, fatigue, and lymphadenopathy. Resolution of symptoms and effective down-regulation of viremia (below 15,000 HIV RNA copies per ml of plasma) was observed in patient 15 within 10 weeks of the onset of symptoms. In contrast, patient 45 had high levels of viremia for a longer period of time (above 200,000 HIV RNA copies per ml of plasma until 16 weeks from the onset of symptoms). In patient 15, ddC (zalcitabine) monotherapy was initiated after day 42. In patient 45, antiviral therapy with AZT (zidovudine), 3TC (lamivudine), and ddI (didanosine) was initiated after day 150.
Isolation of Mononuclear Cells from Peripheral Blood and Cell Sorting.
Isolation of peripheral blood mononuclear cells (PBMC) and two-color cell sorting were performed as previously described (21, 22). For sorting of Vβ23+ T cells, PBMC were incubated with an anti-Vβ23 mAb (HUT-78, kindly provided by O. Kanagawa, Washington University, St. Louis). In all cases, purity of sorted populations was greater than 98%. All experiments were performed on an Elite cell sorter (Coulter).
Cell Culture.
CD8+ T cell clones isolated from PBMC of patients 15 and 45 were obtained by limiting dilution as described previously (23).
Analysis of HIV-Specific Cytotoxic Activity.
In preliminary experiments, fresh unfractionated PBMCs from patient 15 were tested for cytotoxic activity against a panel of HLA-matched and mismatched Epstein–Barr virus-transformed lymphoblastoid cell lines (B-LCL) infected with various strains of recombinant vaccinia virus (kindly provided by B. Moss, Laboratory of Molecular Virology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda). Vaccinia strains used included vPE16, which expresses gp160 from the BH8 clone of HIV-1 IIIB (25); vPE8, which expresses gp120 from the BH8 clone of HIV-1 IIIB (26); vVKI, which expresses a gag-pol fusion protein (27); vVTFnef, which contains the complete nef ORF of HIV-1 (28); and vSC8, which expresses only Escherichia coli β-galactosidase (28). Labeling of target cells with Na51CrO4 (Amersham), cytotoxic assays and calculation of 51Cr release were carried out as described (24). All CTL assays were performed in triplicate. Target cells, including autologous and allogeneic B-LCL, HMY-B7 (29) (gift of J. Lopez de Castro, Consejo Superior de Investigaciones Cientificas, Madrid) and HMY-B52 cell lines were pulsed with 1 μM peptide (30 min at room temperature) before the addition of effector cells (sorted Vβ23+ T cell populations and CD8+ T cell clones). Peptides of gp41 were designed from HIV-1 env consensus B sequences (30). Peptide gp41 842–850 (IPRRIRQGL) was selected based on the published HLA-B7 peptide-binding motif of P2 = P, P3 = R, and P9 = hydrophobic (31). HLA-A32 and HLA-B8-restricted epitopes have been described in the 837–856 gp41 region (32).
Vβ Repertoire Typing and Diversity Analysis.
The Vβ repertoire in fresh, unfractionated PBMC collected at different time points from the onset of symptoms was analyzed by semiquantitative PCR assay as previously described (17, 33, 34). Sequences of Vβ-specific oligonucleotides and the control Cβ and Cα oligonucleotides have been reported elsewhere (33). To monitor the migration of PCR products, 3′ Cβ and 3′ Cα primers were labeled with [γ-32P]ATP (Amersham). PCR products were resolved on 10% polyacrylamide gels containing 7 M urea. Gels were exposed overnight on Kodak storage phosphor screens, and the radioactive signals were quantitated using a PhosphorImager (Molecular Dynamics).
To analyze the clonality of T cell populations expressing specific Vβ families, total cDNA was amplified for 27 cycles in a GenAmp PCR system 9600 (Perkin–Elmer). DNA sequence of the recombinant clones was determined using an Applied Biosystems 377 automated sequencer (Perkin–Elmer), following manufacturer’s instructions. Sequences were aligned to the published sequences of Vβ17.1 (35), Vβ22.1, and Vβ23.1 (35, 36).
Prevalence and persistence of specific T cell clonotypes in PBMC and sorted Vβ23+ T cells was determined by using diversity-specific semiquantitative PCR (DS-PCR). cDNA samples then were amplified for 25 cycles by using a constant 5′ primer, and a 3′ clonotype-specific primer complementary to the TCR β chain CDR3 hypervariable region. For the Vβ23 clonotype SVGNYL (see below), primers used were Vβ23 SalI (5′-GGCCAGGTCGACGACATCTGATCAAAGAAAAGA-3′) and 8Rdiv (5′-CTGCTCGTAGAGATAGTTACCCACTGA-3′). For clonotype RETSL, primers used were 5′Fex (5′-GGCCAGGTCGACCTGCCATGGGCACCAGGCTCCTCTGC-3′) and 44Udiv (5′-CTGGGTCTCCAAACTAGTCTCTC-3′). A 200-bp coamplified fragment of the TCR α gene served as internal control. Separation and quantitation of PCR products were performed as above.
Analysis of HIV-1 Epitope Variation.
HIV was directly isolated from 500 μl of precleared plasma by ultracentrifugation. RNA was extracted from the viral pellet by the guanidium isothiocyanate method, as described (37), and was reverse-transcribed using 0.8 μM HIV-specific antisense primers and 200 units of SuperScript II reverse transcriptase (Life Technologies, Gaithersburg, MD) (unpublished work). cDNA then was subjected to 50 cycles of PCR amplification. PCR products were ligated into PCKII (Invitrogen), and transformed into STBL2 competent cells (Life Technologies). DNA sequence of recombinant clones was determined using an Applied Biosystems 377 automated sequencer, as above.
RESULTS
Perturbations in the TCR Repertoire During Primary HIV Infection and After Transition to the Chronic Phase of Infection.
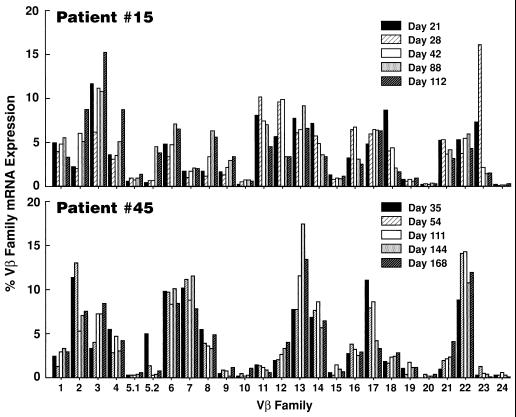
In preliminary experiments, determination of the relative usage of Vβ families was assessed by a semiquantitative PCR assay in two patients with primary infection. Perturbations, i.e., increments or declines, of a Vβ family was considered to be significant only if there was at least a 2-fold increase or decrease of the Vβ family in question over sequential time points (10, 20). However, 2-fold changes were not considered to be significant if they involved Vβ families that were expressed at very low levels (i.e., 1–2%). On the basis of these criteria, the Vβs that were found to be perturbed in patient 15 include Vβ5.2 (from 0.5% at day 21 to 4.8% at day 88), Vβ8 (from 1.7% at day 21 to 6.3% at day 88), Vβ18 (from 8.7% at day 21 to 1.7% at day 112), and Vβ23 (from 16.1% at day 28 to 1.6% at day 112) (Fig. 1). In patient 45, Vβ17 varies from 11.2% at day 35 to 3.3% at day 168 (Fig. 1). Because the percentage of Vβ22 is generally below 5% (unpublished observation), this Vβ was considered to be expanded in patient 45 even if a doubling in the relative expression of Vβ22 among sequential time points was not observed (Fig. 1). Indeed, the analysis of the recombinant molecular clones of Vβ22 at day 35 (see below) showed that all the clones analyzed had identical clonotypes, thus indicating a monoclonal expansion of this Vβ.
Figure 1.
Analysis of TCR repertoire during primary HIV infection in patients 15 and 45. The Vβ repertoire in freshly unfractionated PBMC collected at different time points from the onset of symptoms was analyzed by semiquantitative polymerase PCR as described in Materials and Methods. A minimum of a doubling in the relative expression of Vβ families among sequential time points by PCR was considered to be significant.
These results indicate that in HIV infection Vβ perturbations occur not only during primary infection but also after the transition to the chronic phase (Vβ22 in patient 45), and these perturbations are the result of antigenic stimulations.
Dynamics of Amplification and Deletion of Specific TCR Clonotypic Determinants During Primary Infection and After the Transition to the Chronic Phase of Infection.
Molecular clones of the above-mentioned expanded Vβ families were derived from PBMC collected at different time points during the acute and chronic phases of infection and sequenced.
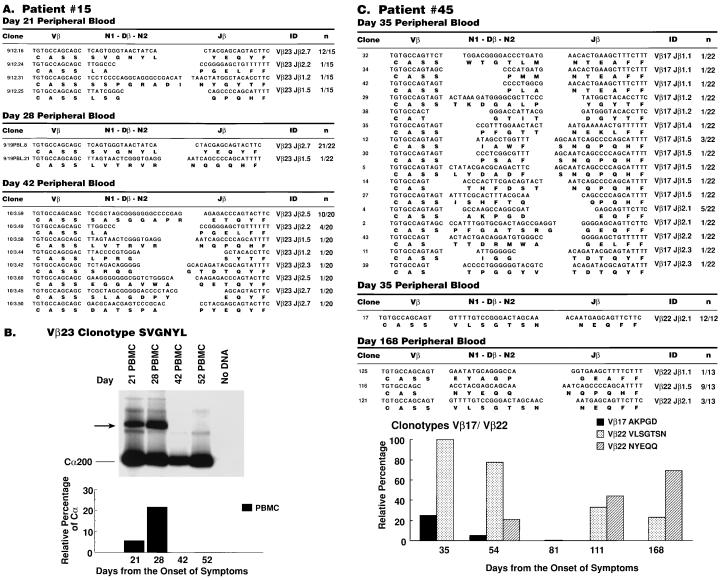
A dramatic monoclonal Vβ expansion was identified in patient 15. Vβ23 was monoclonally expanded with 80% and 95% of the recombinant clones exhibiting identical rearrangement (SVGNYL) at day 21 and 28, respectively (Fig. 2A). However, this clonotype was no longer represented in more than 50 molecular clones sequenced at day 42 (Fig. 2A) and day 52 (data not shown), whereas two other clonotypes (LA and LVTRVR), which were minimally represented at day 21 and 28, were still detected (Fig. 2A), thus indicating that Vβ-specific expansions and deletions can occur simultaneously with an otherwise stable clonotype background.
Figure 2.
Diversity analysis of expanded Vβ families in PBMC of patients 15 and 45 by DNA sequencing. (A) Patient 15. Clone 9/12.16 is representative of 12 Vβ23 clones with identical clonotypes (SVGNYL), derived from PBMC collected at day 21. Clone 9/19.PBL.8 is representative of 21 of 22 Vβ23 clones with clonotype SVGNYL derived at day 28, and clone 10/3.59 is representative of 10 of 20 identical Vβ23 clones (clonotype SASGGAPR) derived at day 42. (B) Dynamics of amplification and deletion of HIV-specific clonotype SVGNYL in patient 15. DS-PCR was performed as in Materials and Methods. (C) Patient 45. Clone 4 is representative of 5 of 22 clones of Vβ17 with identical clonotype (AKPGD) derived from PBMC at day 35, whereas this clonotype was present in 1 of 20 clones at day 54, and disappeared by day 81 (0 of 19 clones). Clone 17 is representative of 12 Vβ22 clones with identical clonotypes (VLSGTSN) derived from PBMC collected at day 35, and by day 168 this clonotype was significantly reduced (3 of 13 clones). Clone 116 is representative of nine clones with identical clonotypes (NYEQQ) derived at day 168. Temporal changes in Vβ17 and Vβ22 clonotypic representation expressed as relative percentages (Bottom). Analysis of clonotypes NYEQQ and VLSGTSN was not performed on day 81.
The distribution of clonotype SVGNYL was determined at various time points using DS-PCR. This method uses the uniqueness of the CDR3 region at the DNA level to derive clonotype-specific antisense primers that can be used in semiquantitative PCR assays to determine the representation of specific T cell clones in a given T cell population. Kinetic analysis using these techniques confirmed the disappearance of clonotype SVGNYL by day 42 (Fig. 2B). Of note, DS-PCR analysis showed that on day 28, the frequency of SVGNYL clonotype in lymph nodes, mononuclear cells were 10-fold lower as compared with peripheral blood (unpublished results).
In patient 45, both Vβ17 and Vβ22 were expanded at day 35 (Fig. 2C), and each comprised a dominant clonotype. The Vβ17 clonotype AKGPD, which was predominant at day 35 (23% of clones sequenced), was significantly reduced at day 54 (5% of clones), and was no longer detected at day 81 (Fig. 2C, Bottom). Interestingly, Vβ22 also showed a dominant clonotype (VLSGTSN) at day 35 (Fig. 2C). However, while this clonotype progressively declined over time (100% of the clones at day 35, 71% at day 54, 33% at day 111, and 23% at day 168 (Fig. 2C, Bottom), it never disappeared as was shown for Vβ17 AKGPD and Vβ23 SVGNYL clonotypes (Fig. 2 B and C, Bottom). The progressive decline of the VLSGTSN clonotype coincided with the rise of a new dominant clonotype (NYEQQ), which represented 21% of the clones at day 54, 56% at day 111, and peaked at day 168 (69% of clones) (Fig. 2C, Bottom) when transition to the chronic phase already had occurred.
These data indicate that the expanded CD8+ TCR clonotypes that were dominant during primary HIV infection rapidly decline (clonotype VLSGTSN in patient 45) or totally disappear (clonotype SVGNYL in patient 15, and clonotype AKPDG in patient 45) during primary infection or after transition to the chronic phase of infection.
Differential Fate of Distinct HIV-Specific Cytotoxic T Cell Clones with Identical Viral Epitope Specificity.
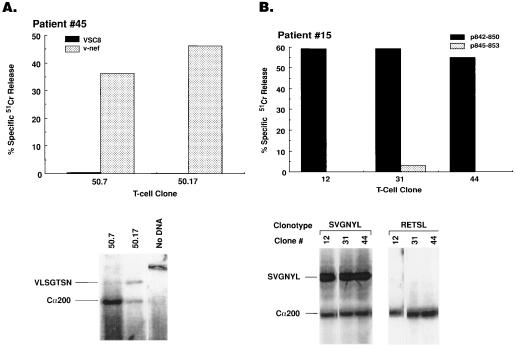
It has been previously demonstrated that the CD8+ Vβ families that were expanded during the primary immune response to HIV contain HIV-specific CTL (17). Similarly, a number of nef-specific CTL clones derived from patient 45 at day 54, expressed the expanded Vβ22 clonotype VLSGTSN (Fig. 3). With regard to the characterization of HIV-specific cytotoxicity in patient 15, analysis of the cytotoxic activity mediated by CD8+ T cells was performed using a panel of 25 peptides (twentymers with 10 overlapping amino acids) spanning more than two-thirds of gp41. The epitope specificity of the CD8+ CTL was mapped to the p842–850 of gp41 (data not shown). In fact, sorted Vβ23+ T cells derived from PBMC collected at day 28 when Vβ23 was monoclonally expanded, lysed autologous B-LCL pulsed with gp41 p842–850 (data not shown). Further analysis revealed that this epitope was HLA-B7 restricted (the HLA type of patient 15 is A1, A3, B7, B60, C3, C7) (data not shown). More importantly, the Vβ23 clonotype SVGNYL was expressed on CTL clones specific for gp41 p842–850 (Fig. 3).
Figure 3.
HIV-specific cytotoxicity mediated by expanded Vβ23+ and Vβ22+ T cell clones in patients 45 and 15, respectively. (A) HIV-specific cytotoxicity mediated by CD8+ T cell clones derived from PBMC of patient 45 at day 35 against autologous B-LCL infected with nef-expressing vaccinia constructs. Clonotype VLSGTSN is expressed by nef-specific CTL clones from patient 45, determined by DS-PCR (Bottom). (B) HIV-specific cytotoxicity mediated by CD8+ T cell clones derived from PBMC of patient 15 on day 28, using autologous B-LCL targets pulsed with gp41 p842–850. Clonotype SVGNYL, but not RETSL, is expressed by p842–850-specific CTL clones derived from patient 15 on day 28, as determined by DS-PCR (Bottom). CTL activity was expressed as % specific 51Cr release.
Despite the fact that the expanded Vβ23 clonotype, which recognized gp41 p842–850 in an HLA-B7 restricted manner, had disappeared by day 42 (Fig. 2 A and B), HIV-specific cytotoxicity against autologous target cells infected with vaccinia constructs expressing gp160 but not gp120, was consistently observed in PBMC collected at different time points until day 245 (the last time point analyzed) (data not shown). There are three possibilities that could explain this observation: (i) the disappearance of the expanded Vβ23 clonotype was the result of mutations in the p842–850 epitope of the HIV gp41, and so the stimulus for the persistence of this clonotype was no longer present; (ii) the persistence of gp41-specific cytotoxicity after the disappearance of the expanded Vβ23 clonotype was due to selection for new T cell clones specific for gp41 epitopes different from p842–850; and (iii) the persistence of gp41-specific cytotoxicity after the disappearance of the Vβ23 clonotype was due to the presence or appearance of T cell clones with different TCR specific for p842–850. To test the first possibility, HIV RNA was extracted from plasma collected at day 21 and day 42, when the Vβ23 clonotype SVGNYL had disappeared; the extracted RNA was reverse-transcribed with a primer pair specific for a gp41 fragment containing the p842–850 CTL epitope, and several clones (10 at day 21 and 9 at day 42) of this fragment were obtained and sequenced. This analysis showed that 44% (4 of 9) of the clones derived from day 42 had no mutations in the CTL epitope sequence (Fig. 4A). Therefore, disappearance of the originally expanded HIV-specific clonotype could not be explained solely by mutations in the viral epitope recognized by cells expressing the specific clonotype.
Figure 4.
(A) DNA sequencing of the HIV gp41 fragment containing the p842–850 epitope recognized by Vβ23+ T cells expressing clonotype SVGNYL. DNA sequencing was performed on cloned plasma virus isolated on day 21 when clonotype SVGNYL was expanded, and on day 42 when clonotype SVGNYL had disappeared. Nucleotide sequences of clone HM04 (day 21) and of nine clones from day 42 are shown. Changes in the sequence of CTL epitope p842–850 are in bold. (B) HIV-specific cytotoxicity mediated by CD8+ T cell clones derived from PBMC of patient 15 on day 245, using autologous B-LCL targets pulsed with gp41 overlapping peptides spanning residues 841–855. (C) DNA sequence of five Vβ6.5 recombinant clones (clone 44 is representative of five clones with clonotype RETSL) derived from CD8+ T cell microculture 100–7, and of four Vβ6.5 recombinants (clone 67 is representative of four clones with clonotype RETSL) derived from CD8+ T cell microculture 100–17. Both clones were obtained by limiting dilution at day 245. (D) Dynamics of amplification and deletion of HIV-specific clonotype RETSL in patient 15. DS-PCR was performed as in Materials and Methods. (Bottom) Normalization of the Vβ-specific signal to the coamplified Cα fragment.
With regard to the second possibility, sorted CD8+ T cells were isolated from patient 15 at day 245, and a number of clones were obtained by limiting dilution analysis. CD8+ T cell proliferating microcultures efficiently lysed target cells sensitized with p842–850 (Fig. 4B), indicating that they recognized the same epitope as that recognized by the expanded Vβ23 clonotype. Nucleotide sequencing of the Vβ chains from these clones was performed in cells from representative microcultures (nos. 100–7 and 100–17), and showed that these cells expressed Vβ6.5 rearranged to Jβ2.5. The CDR3 junction of several molecular clones derived from these microcultures was identical at DNA level (RETSL sequence), hence indicating that these microcultures comprised clonal populations (Fig. 4B). These results indicate that after the disappearance of the expanded Vβ23 clonotype, the HIV-specific CTL clones that were identified later in the course of infection recognized the same HIV epitope even though they expressed a different Vβ family than the originally expanded clonotype (Vβ6.5 versus Vβ23).
It was then important to determine whether the Vβ6.5 RETSL clonotype was a component of the initial primary immune response. The kinetics of the RETSL Vβ6.5 clonotype in PBMC and its distribution in blood were analyzed by using a clonotype-specific primer and DS-PCR. This analysis demonstrated that the RETSL Vβ6.5 clonotype was expressed at the time of primary infection; however, in contrast to the expanded Vβ23 clonotype SVGNYL, it persisted after the transition to the chronic phase of infection (Fig. 4D).
DISCUSSION
In the present study, we have characterized the primary cell-mediated immune response to HIV infection to identify the potential strategies that allow HIV to escape the immune response and persist in the host despite the fact that a vigorous HIV-specific immune response is readily detected during primary infection. A potential mechanism of viral escape from the immune response during primary HIV infection has been identified. This involves disappearance/deletion of HIV-specific CTL clones.
Mutations in the viral epitope recognized by CTL are considered the most common strategy used by viruses including HIV to elude virus-specific CTL (3, 4, 6–9). We have demonstrated that during primary infection disappearance of individual HIV-specific CTL clones occurs despite the fact that approximately 50% of the cognate epitope remains unmutated. A trivial explanation for the disappearance of these virus-specific CTL clones is that the frequency of these clones is very low (1 in 104–105 cells) after effective down-regulation of virus replication and, therefore, it may be very difficult to detect them in the circulation. The latter possibility is unlikely for two reasons: (i) we show that disappearance of these clones occurs when the levels of viremia are still very high; and (ii) virus-specific CTL clones of a different Vβ family that recognize the same epitope can, in fact, be detected in the circulation even several months after primary infection. The likely scenario during primary HIV infection is that the demonstrated disappearance of HIV-specific CTL clones reflects a deletion process caused by high levels of antigen, in this case virus. Clonal exhaustion/deletion is a well documented phenomenon in lymphocytic choriomenigitis virus infection and other murine experimental systems (39–43). The occurrence of clonal exhaustion is strictly dependent on the presence of high levels of viral antigens throughout the lymphoid system (38), which is the anatomic site where antigen presentation, stimulation, and generation of the immune response occur (44). High levels of antigen stimulate a rapid and vigorous immune response that is associated with the mobilization of all the T cell clones that are able to recognize a specific epitope. Because the fate of activated cells involved in the immune response is to die (45), the ultimate consequence of this massive mobilization is the deletion of those clones that have received this exhaustive stimulation (38). Of interest, a recent study has demonstrated that relatively few clones (approximately 15–20) may be specific for a certain antigen and yet may still generate massive T cell responses to that antigen, suggesting that stimulation of only a limited number of clones may be sufficient for the induction of clonal exhaustion (46). Clonal exhaustion virtually never occurs during a classical immune response (i.e., in the presence of an optimal concentration of antigen), where only a minor percentage of clones specific for a certain antigen are mobilized and the antigen is rapidly cleared (38).
Primary HIV infection is associated with overwhelming virus replication throughout the lymphoid system (10, 47); as mentioned above, this is a fundamental condition for the occurrence of clonal deletion. We conclude that the disappearance of HIV-specific CTL clones during primary infection is due to an exhaustive stimulation based on two observations in the present study: (i) the rapidity of disappearance/deletion of the monoclonally expanded cytotoxic clones (i.e., within 2 weeks for the Vβ23 HIV-specific clonotype in patient 15, and within 10 weeks for the Vβ17 clonotype in patient 45); and (ii) the magnitude of these expansions.
It is important to underscore that the deletion/exhaustion of clones involves individual virus-specific CTL clones, and thus does not necessarily result in a total loss of HIV-specific cytotoxic function. In fact, we have shown that HIV-specific CTL clones are still present several months after primary infection despite the fact that they originally had appeared during primary infection. It is conceivable that differences in the affinity for the specific antigen may play an important role in influencing the deletion process, and clones with higher binding affinity for the antigen will be more rapidly deleted compared with those with lower affinity.
An obvious question relates to the relevance of the selective deletion/exhaustion of initially expanded HIV-specific CTL clonotypes to virus escape from the immune response during primary infection. This is of particular interest in light of the fact that measurable HIV-specific CTL function persists, despite the disappearance of these clonotypes. However, it is highly likely that deletion/exhaustion of individual HIV-specific CTL clones will have a major impact. For example, the quantitative potential of the immune response will be affected because the overall precursor frequency of virus-specific CTL will have been greatly reduced, and the HIV-specific TCR repertoire already will be compromised in magnitude and competence during the early stage of HIV infection. It is also important to point out that genetic variability of the virus is an important component of HIV disease (48). This variability is already detected during primary infection and together with the inexorable process of clonal deletion contributes substantially to viral escape from the immune response.
With regard to the possibility that deletion/exhaustion of virus-specific T cell clones may occur in primary viral infections other than HIV, it is important to mention that this phenomenon has not been observed in subjects with acute mononucleosis, despite the fact that the primary immune response in these subjects also is associated with oligo-monoclonal expansions of certain Vβ T cell subsets (unpublished work).
In conclusion, we provide evidence that during the early stage of infection HIV tilts the delicate balance between spreading of virus and curtailment of virus by the immune system toward spreading of virus by inducing exhaustive stimulation and deletion of virus-specific CTL cell clones. These results provide insights for understanding the events involved in the generation of specific antiviral immune responses and in the strategies that certain viruses, in this case HIV, may use to escape otherwise vigorous immune responses, allowing for persistence in the host.
Acknowledgments
We thank Drs. J.-C. Cerottini, H. R. McDonald, and G. P. Corradin for helpful discussion. The part of the work performed by H.S. was in partial fulfillment of the requirements of the Ph.D degree, Department of Microbiology and Immunology, McGill University.
ABBREVIATIONS
- B-LCL
Epstein–Barr virus-transformed B-lymphoblastoid cell line
- CTL
cytotoxic T lymphocyte
- PBMC
peripheral blood mononuclear cell
- TCR
T cell receptor
- DS-PCR
diversity-specific semiquantitative PCR
References
- 1.Butera S T, Roberts B D, Lam L, Hodge T, Folks T M. J Virol. 1994;68:2726–2730. doi: 10.1128/jvi.68.4.2726-2730.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graziosi C, Pantaleo G, Butini L, Demarest J F, Saag M S, Shaw G M, Fauci A S. Proc Natl Acad Sci USA. 1993;90:6405–6409. doi: 10.1073/pnas.90.14.6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.del Val M, Engel H, Hacker H, Hartlaub U, Ruppert T, Licin P, Koszinowski U H. J Exp Med. 1992;176:729–738. doi: 10.1084/jem.176.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Phillips R E, Rowland-Jones S, Nixon D F, Gotch F M, Edwards J P, Ogunlesi A O, Elvin J G, Rothbard J A, Bangham C R M, Rizza C R, McMichael A J. Nature (London) 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 4.Niewiesk S, Daenke S, Parker C E, Taylor G, Weber J, Nightingale S, Bangham C R. J Virol. 1995;69:2649–2653. doi: 10.1128/jvi.69.4.2649-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Campos Lima P O, Gavioli R, Zhang Q J, Wallace L E, Dolcetti R, Rowe M, Rickinson A B, Masucci M G. Science. 1993;260:98–100. doi: 10.1126/science.7682013. [DOI] [PubMed] [Google Scholar]
- 6.Klenerman P, Rowland-Jones S, McAdam S, Edwards J, Daenke S, Lalloo D, Köppe B, Rosenberg W, Boyd D, Edwards A, Giangrande P, Phillips R E, McMichael A J. Nature (London) 1994;369:403–407. doi: 10.1038/369403a0. [DOI] [PubMed] [Google Scholar]
- 7.Bertoletti A, Sette A, Chisari F V, Penna A, Levrero M, De Carli M, Fiaccadori M, Ferrari C. Nature (London) 1994;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 8.Allen P M, Zinkernagel R M. Nature (London) 1994;369:355–356. doi: 10.1038/369355a0. [DOI] [PubMed] [Google Scholar]
- 9.Meier U-C, Klenerman P, Griffin P, James W, Koppe B, Larder B, McMichael A, Phillips R. Science. 1995;270:1360–1362. doi: 10.1126/science.270.5240.1360. [DOI] [PubMed] [Google Scholar]
- 10.Pantaleo G, Fauci A S. Annu Rev Immunol. 1995;13:487–512. doi: 10.1146/annurev.iy.13.040195.002415. [DOI] [PubMed] [Google Scholar]
- 11.Tindall B, Cooper D A. AIDS. 1991;5:1–14. [PubMed] [Google Scholar]
- 12.Daar E S, Moudgil T, Meyer R D, Ho D D. N Engl J Med. 1991;324:961–964. doi: 10.1056/NEJM199104043241405. [DOI] [PubMed] [Google Scholar]
- 13.Clark S J, Saag M S, Decker W D, Campbell-Hill S, Roberson J L, Veldkamp P J, Cappes J C, Hahn B H, Shaw G M. N Engl J Med. 1991;324:954–960. doi: 10.1056/NEJM199104043241404. [DOI] [PubMed] [Google Scholar]
- 14.Pantaleo G, Graziosi C, Fauci A S. N Engl J Med. 1993;328:327–335. doi: 10.1056/NEJM199302043280508. [DOI] [PubMed] [Google Scholar]
- 15.Koup R A, Safrit J T, Cao Y, Andres C A, McLeod G, Borkowsky W, Farthing C, Ho D D. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantaleo G, Demarest J F, Soudeyns H, Graziosi C, Denis F, Adelsberger J W, Borrow P, Saag M S, Shaw G M, Sékaly R P, Fauci A S. Nature (London) 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 18.Moore J P, Cao Y, Ho D D, Koup R A. J Virol. 1994;68:5142–5155. doi: 10.1128/jvi.68.8.5142-5155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z W, Kou Z C, Lekutis C, Shen L, Zhou D, Halloran M, Li J, Sodroski J, Lee-Parritz D, Letvin N L. J Exp Med. 1995;182:21–31. doi: 10.1084/jem.182.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantaleo G, Demarest J F, Schacker T, Vaccarezza M, Cohen O J, Daucher M, Graziosi C, Schnittman S S, Quinn T C, Shaw G M, Perrin L, Tambussi G, Lazzarin A, Sekaly R P, Soudeyns H, Corey L, Fauci A S. Proc Natl Acad Sci USA. 1997;94:254–258. doi: 10.1073/pnas.94.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantaleo G, Graziosi C, Butini L, Pizzo P A, Schnittman S M, Kotler D P, Fauci A S. Proc Natl Acad Sci USA. 1991;88:9838–9842. doi: 10.1073/pnas.88.21.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pantaleo G, Koenig S, Baseler M, Lane H C, Fauci A S. J Immunol. 1990;144:1696–1704. [PubMed] [Google Scholar]
- 23.Moretta A, Pantaleo G, Moretta L, Cerottini J C, Mingari M C. J Exp Med. 1983;157:743–754. doi: 10.1084/jem.157.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantaleo G, DeMaria A, Koenig S, Butini L, Moss B, Baseler M, Lane H C, Fauci A S. Proc Natl Acad Sci USA. 1990;87:4818–4822. doi: 10.1073/pnas.87.12.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Earl P L, Hugin A, Moss B. J Virol. 1990;64:2448–2451. doi: 10.1128/jvi.64.5.2448-2451.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karacostas V, Nagashima K, Gonola M A, Moss B. Proc Natl Acad Sci USA. 1989;86:8964–8967. doi: 10.1073/pnas.86.22.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabarti S, Brechling K, Moss B. Mol Cell Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storkus W J, Alexander J, Payne J A, Dawson J R, Cresswell P. Proc Natl Acad Sci USA. 1989;86:2361–2364. doi: 10.1073/pnas.86.7.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiBrino M, Parker K C, Margulies D H, Shiloach J, Turner R V, Biddison W E, Coligan J E. Biochemistry. 1995;34:10130–10138. doi: 10.1021/bi00032a005. [DOI] [PubMed] [Google Scholar]
- 30.Myers G, Hahn B H, Mellors J W, Henderson L E, Korber B, Jeang K-T, McCutchan F E, Pavlakis G N, editors. Human Retroviruses and AIDS: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. Los Alamos, NM: Los Alamos National Laboratory; 1995. [Google Scholar]
- 31.Huczko E L, Bodnar W M, Benjamin D, Sakaguchi K, Zhu N Z, Shabanowitz J, Henderson R A, Appella E, Hunt D F, Engelhard V H. J Immunol. 1993;151:2572–2587. [PubMed] [Google Scholar]
- 32.Lieberman J, Fabry J A, Kuo M C, Earl P, Moss B, Scolnick P R. J Immunol. 1992;148:2738–2747. [PubMed] [Google Scholar]
- 33.Labrecque N, McGrath H, Subramanyam M, Huber B T, Sekaly R-P. J Exp Med. 1993;177:1735–1743. doi: 10.1084/jem.177.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebai N, Pantaleo G, Demarest J F, Ciurli C, Soudeyns H, Adelsberger J W, Vaccarezza M, Walker R E, Sekaly R P, Fauci A S. Proc Natl Acad Sci USA. 1994;91:1529–1533. doi: 10.1073/pnas.91.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura N, Toyonaga B, Yoshikai Y, Du R-P, Mak T W. Eur J Immunol. 1987;17:375–383. doi: 10.1002/eji.1830170312. [DOI] [PubMed] [Google Scholar]
- 36.Ferradini L, Roman-Roman S, Azocar J, Michalaki H, Triebel F, Hercend T. Eur J Immunol. 1991;21:935–942. doi: 10.1002/eji.1830210412. [DOI] [PubMed] [Google Scholar]
- 37.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 38.Zinkernagel R M. Science. 1996;271:173–178. doi: 10.1126/science.271.5246.173. [DOI] [PubMed] [Google Scholar]
- 39.Mitchison N A. Proc R Soc London. 1964;161:275–293. doi: 10.1098/rspb.1964.0093. [DOI] [PubMed] [Google Scholar]
- 40.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel R M. Nature (London) 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 41.Rammensee H-G, Kroschewski R, Frangoulis B. Nature (London) 1989;339:541–544. doi: 10.1038/339541a0. [DOI] [PubMed] [Google Scholar]
- 42.Moskophidis D, Lechner F, Pircher H, Zinkernagel R M. Nature (London) 1993;362:758–761. doi: 10.1038/362758a0. [DOI] [PubMed] [Google Scholar]
- 43.von Boehmer H. Nature (London) 1993;362:696. doi: 10.1038/362696a0. [DOI] [PubMed] [Google Scholar]
- 44.Parrot D M V, Wilkinson P C. Prog Allergy. 1981;28:193–284. [PubMed] [Google Scholar]
- 45.Cohen J J, Duke R C, Fadok V A, Sellins K S. Annu Rev Immunol. 1992;10:267–293. doi: 10.1146/annurev.iy.10.040192.001411. [DOI] [PubMed] [Google Scholar]
- 46.Maryanski J L, Jongeneel C V, Bucher P, Casanova J-L, Walker P R. Immunity. 1996;4:47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- 47.Pantaleo G, Graziosi C, Demarest J F, Butini L, Montroni M, Fox C H, Orenstein J M, Kotler D P, Fauci A S. Nature (London) 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 48.Coffin J M. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]