Abstract
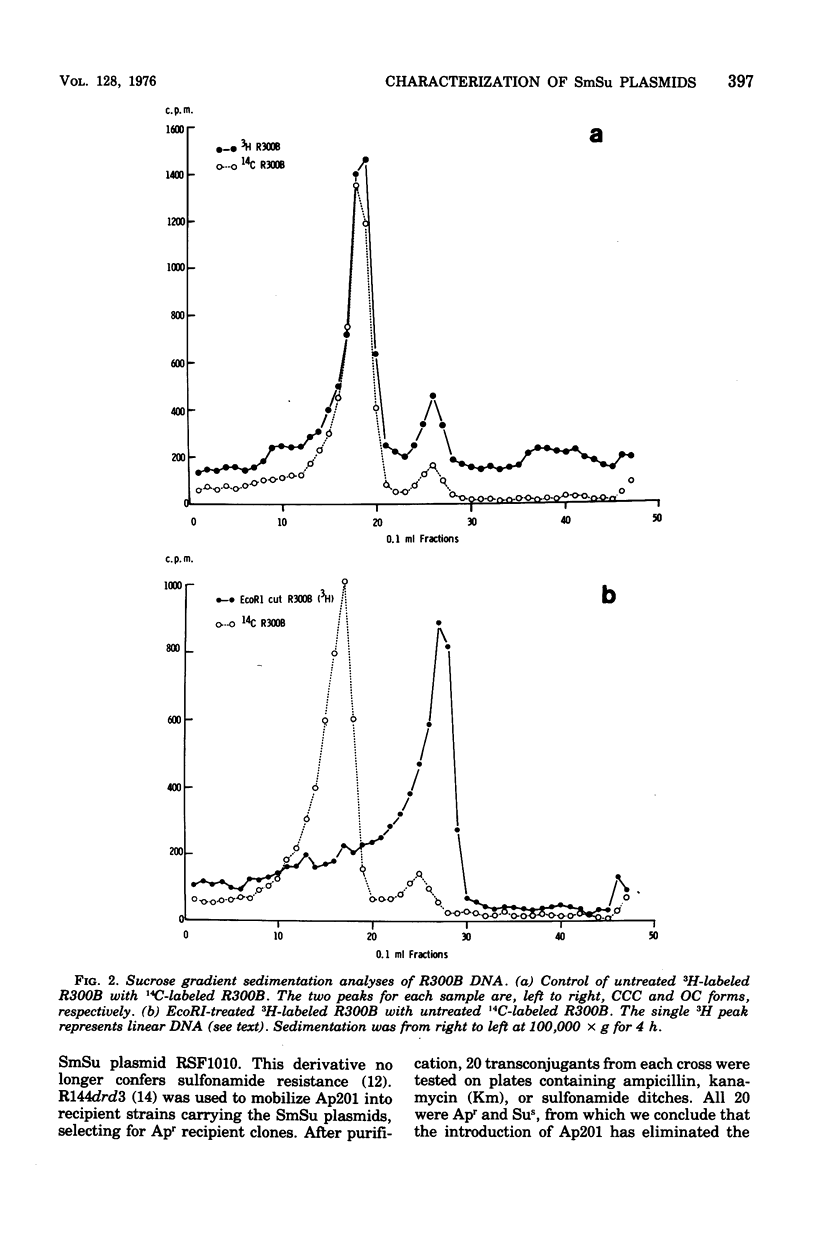
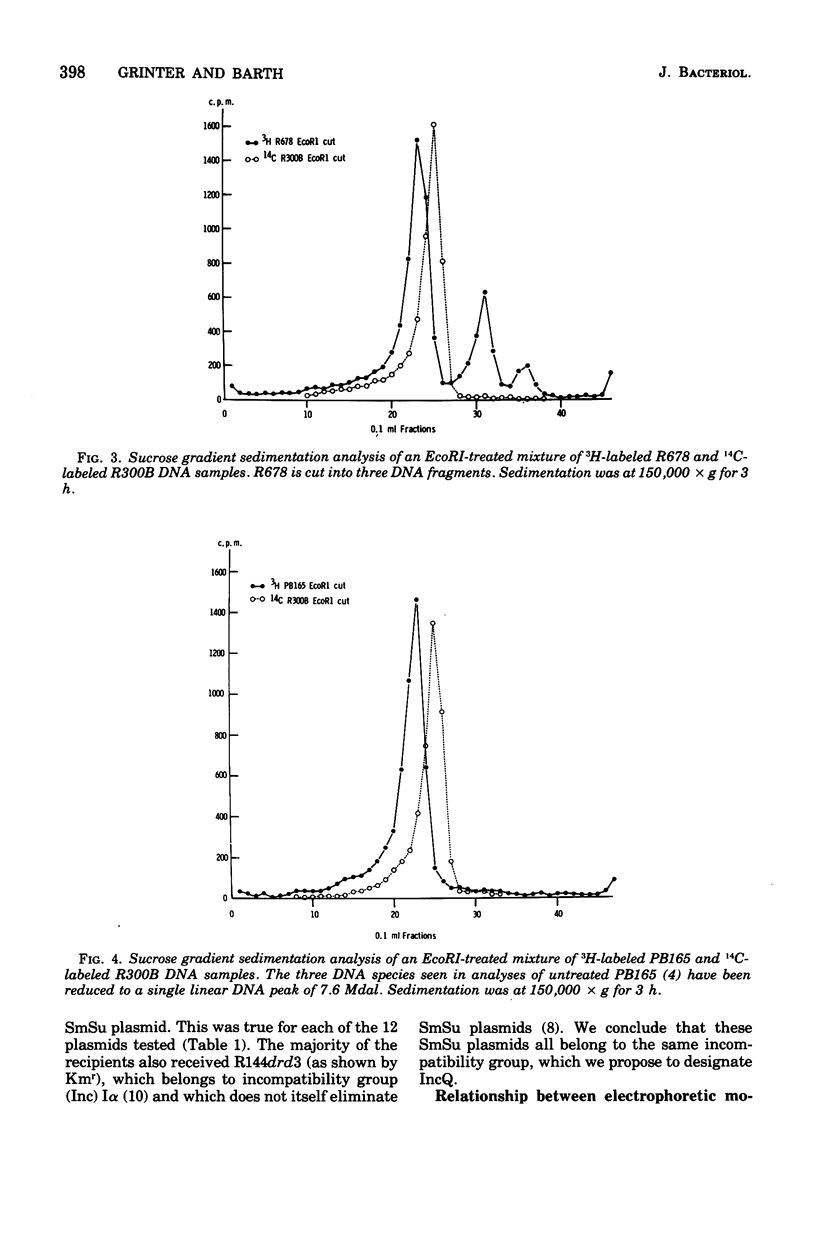
Twelve plasmids carrying genes for streptomycin and sulfonamide resistance were studied for the number and distribution of sites on the plasmid moleucles susceptible to cleavage by the restriction endonuclease EcoRI. Ten of the twelve were found to have a single cut site, one plasmid (R678) had three such sites, and plasmid PB165, which was isolated as three supercoiled deoxyribonucleic acid species with molecular weights 7.4 x 10(6), 14.7 x 10(6), and 21.4 x 10(6) was reduced to a single (linear) species of molecular weight 7.6 x 10(6) after cutting with EcoRI. We conclude that PB165 forms oligomers in Escherichia coli and that the number of copies of these per chromosome is more consistant and that the number of copies of these per chromosome is more consistent with a negative than a positive control mechanism for plasmid replication. Compatibility testing of a positive control mechanism for plasmid replication. Compatibility testing of these plasmids showed they all belong to the same incompatibility group, which we designate IncQ, suggesting that they may have come from a common ancestor.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J., Healing D., Hutchison J. G. Characteristics of some co-trimoxazole-resistant Enterobacteriaceae from infected patients. J Clin Pathol. 1972 Dec;25(12):1086–1088. doi: 10.1136/jcp.25.12.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Datta N., Hedges R. W., Grinter N. J. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J Bacteriol. 1976 Mar;125(3):800–810. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello F., Timmis K., Cohen S. N. Replication control in a composite plasmid constructed by in vitro linkage of two distinct replicons. Nature. 1976 Jan 29;259(5541):285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- Coetzee J. N., Datta N., Hedges R. W. R factors from Proteus rettgeri. J Gen Microbiol. 1972 Oct;72(3):543–552. doi: 10.1099/00221287-72-3-543. [DOI] [PubMed] [Google Scholar]
- Falkow S., Guerry P., Hedges R. W., Datta N. Polynucleotide sequence relationships among plasmids of the I compatibility complex. J Gen Microbiol. 1974 Nov;85(1):65–76. doi: 10.1099/00221287-85-1-65. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Datta N. Plasmids determining I pili constitute a compatibility complex. J Gen Microbiol. 1973 Jul;77(1):19–25. doi: 10.1099/00221287-77-1-19. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynell E., Datta N. Mutant drug resistant factors of high transmissibility. Nature. 1967 May 27;214(5091):885–887. doi: 10.1038/214885a0. [DOI] [PubMed] [Google Scholar]
- Smith H. R., Humphreys G. O., Anderson E. S. Genetic and molecular characterisation of some non-transferring plasmids. Mol Gen Genet. 1974 Mar 27;129(3):229–242. doi: 10.1007/BF00267915. [DOI] [PubMed] [Google Scholar]
- Thompson R., Hughes S. G., Broda P. Plasmid identification using specific endonucleases. Mol Gen Genet. 1974;133(2):141–149. doi: 10.1007/BF00264835. [DOI] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. Plasmid incompatibility and control of replication: copy mutants of the R-factor R1 in Escherichia coli K-12. J Bacteriol. 1975 Nov;124(2):641–649. doi: 10.1128/jb.124.2.641-649.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]