Abstract
Bacterial pathogens have evolved sophisticated mechanisms to interact with their hosts. A specialized type III protein secretion system capable of translocating bacterial proteins into host cells has emerged as a central factor in the interaction between a variety of mammalian and plant pathogenic bacteria with their hosts. Here we describe AvrA, a novel target of the centisome 63 type III protein secretion system of Salmonella enterica. AvrA shares sequence similarity with YopJ of the animal pathogen Yersinia pseudotuberculosis and AvrRxv of the plant pathogen Xanthomonas campestris pv. vesicatoria. These proteins are the first examples of putative targets of type III secretion systems in animal and plant pathogenic bacteria that share sequence similarity. They may therefore constitute a novel family of effector proteins with related functions in the cross-talk of these pathogens with their hosts.
Keywords: bacterial pathogenesis, type III secretion, host response
Salmonella enterica causes diseases that range from self-limiting gastroenteritis (food poisoning) to systemic enteric infection (typhoid fever). The type of disease is largely determined by the species of the infected host and the serovar of the infecting bacteria. For example, S. enterica serovar typhimurium (S. typhimurium) causes a typhoid-like systemic disease in mice, although in humans it generally causes a self-limiting gastroenteritis.
During the process of adaptation to animal hosts, S. enterica has evolved a variety of mechanisms to colonize, invade, replicate, and survive within the host. These include mechanisms to respond to the low pH of the stomach and other compartments (1), various sensory and regulatory systems (2), as well as a complex protein secretion apparatus that can deliver effector molecules into host cells to modulate host cellular functions (3). In turn, the host has evolved ways to detect and control bacterial infection. This includes constitutive, nonspecific defense mechanisms that are most important during the initial stages of a primary bacterial infection, as well as specific humoral and cellular immune mechanisms that dominate the defense responses during the later stages of infection or during subsequent encounters with the pathogen. The constitutive defenses against primary infections are typically directed against a broad range of bacteria. Little is known, however, about mechanisms that might enable the naive mammalian host to recognize specifically certain common pathogens to respond immediately and in the most efficient way. A number of examples of such an adaptation of the host to the encounter with specific pathogens have been described for plants (reviewed in refs. 4 and 5). Recognition of pathogen-specific factors (termed “avirulence factors”) allows the plant to mount a so-called “hypersensitive response,” which is characterized by a localized necrosis that limits the infection to the area of initial encounter.
Recent studies suggest that most avirulence factors from plant pathogenic bacteria are presented to the host via type III protein secretion systems (reviewed in ref. 6). This specialized protein secretion system has been identified in several Gram-negative pathogenic bacteria including the plant pathogens Pseudomonas spp., Erwinia spp. and Xanthomonas spp. and the animal pathogens Salmonella spp., Pseudomonas aeruginosa, Shigella spp., Yersinia spp., and Escherichia coli spp. (reviewed in ref. 7). Although there is remarkable conservation among the components of this protein secretion apparatus across bacterial species, the effector proteins that travel through this secretory pathway are much less conserved, particularly among plant and animal bacterial pathogens. This has been interpreted as an adaptation to the biology of the different bacterial pathogens. Here we describe the identification and characterization of a S. typhimurium protein, termed AvrA, which is secreted and translocated into the host cell via the invasion-associated type III system. This protein shares sequence homology with YopJ, a target of a type III protein secretion system of Yersinia pseudotuberculosis and the avirulence factor AvrRxv from Xanthomonas campestris pv. vesicatoria. Our data suggest that AvrRxv, AvrA, and YopJ belong to a novel family of secreted effector proteins that might serve analogous functions in the cross-talk between pathogenic bacteria and their plant or animal hosts.
MATERIALS AND METHODS
Bacterial Strains, Recombinant DNA, Genetic Techniques, and Nucleotide Sequencing.
The S. typhimurium wild-type strain SL1344 (8) and its isogenic derivatives SB147 (9), SB161 (10), SB169, SB220, SB241 (11), SB225 (12), SB302 (13), and SB303 (14) have been described. All recombinant DNA procedures were carried out following standard techniques. The centisome 63 pathogenicity island region located immediately downstream of orgA (15) was retrieved by chromosomal walking as follows. A 676 nt fragment of the 3′-terminal region of orgA was amplified by PCR using a pair of degenerate primers (5′-CATCCGCGGGCTAAGCGTATTTTG-3′ and 5′-CTATCTAGATTATTCAGCATAGCGGC-3′), digested with SacII and XbaI, and cloned into the suicide vector pSB377 (12). The resulting plasmid was integrated into the wild-type S. typhimurium strain SL1344 by homologous recombination. Genomic DNA from the resulting strain was digested with NheI and XbaI, ligated, and transformed into E. coli CC118λpir. A plasmid (pSB851) carrying an 11-kb fragment of S. typhimurium chromosome was recovered from one of the transformants. Southern blot analysis confirmed that the insert of pSB851 contains a contiguous region spanning from orgA through the fhlA boundary of the centisome 63 pathogenicity island (data not shown).
A strain carrying a nonpolar mutation in avrA was constructed by replacing a BstBI–AatII internal fragment of avrA with a terminatorless aphT cassette that confers resistance to kanamycin. The mutated allele was introduced into the Salmonella chromosome by homologous recombination using the suicide vector pGP704 (16) as described elsewhere (10), yielding the mutant strain SB733.
A set of primers (5′-TTTGGGGATGGACTCTTC-3′ and 5′-CGGGCGTTTATCTGTCTT-3′) complementary to 3′-terminal regions of the conserved neighboring genes orfX and orfY was used to amplify the chromosomal regions comprising avrA or a second alternative genetic element (see Results). The 1.5- or 0.6-kb PCR products were cloned into the SmaI site of pBluescript SKII+ (Stratagene) and sequenced using the dideoxy chain termination kit (United States Biochemical). To construct a fusion protein between glutathione S-transferase and AvrA, a DNA fragment containing the entire avrA sequence was amplified by PCR (primers: 5′-GGTGGAATTCAGATGATATTTTCGGTGCAGGAG-3′ and 5′-CCGATCCATGGATTTCCTCTGGCAGGCAACC-3′), cloned into pGEX-KG via EcoRI and NcoI, and purified as described (17).
An 18 aa epitope of adenovirus protein E4–6/7, which is recognized by the monoclonal antibody M45 (18), was fused to the C terminus of AvrA as follows. A 925 nt fragment containing avrA was amplified by PCR using two degenerate primers (5′-CGGAGAATTCAGATGATATTTTCGGTGCAGG-3′ and 5′-CCGATCCATGGGTTTAAGTAAAGACTTATATTCAGC-3′) and subsequently cloned into the tagging vector pSB504 (14). To bring the avrA epitope fusion under the control of its native promoter, the EcoRI–BstBI fragment of this vector was replaced by the 870 nt SnaBI–BstBI fragment of pSB851 that comprises the 5′ half of avrA and the entire upstream intergenic region, resulting in plasmid pSB885.
Immunofluorescence Staining and Detection of Translocated AvrA Protein in Cultured Henle-407 Cells.
Immunofluorescence staining and biochemical fractionation of S. typhimurium-infected Henle-407 cells was carried out as described (19).
Preparation of Culture Supernatant Proteins and Virulence Assays.
Preparation of culture supernatant proteins (11), bacterial invasion of cultured epithelial cells (20), macrophage cytotoxicity assays (21), and experimental animal infections (20) were carried out as described elsewhere.
RESULTS
Identification and Cloning of the avrA Gene.
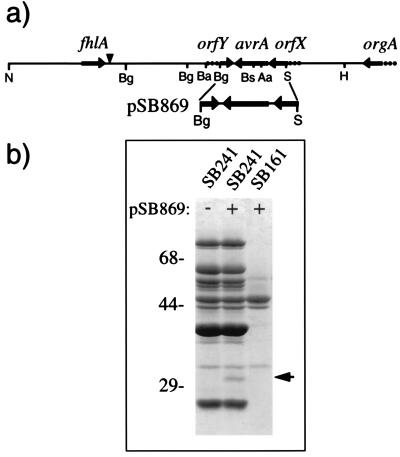
The centisome 63 type III secretion system of S. typhimurium directs the export and translocation into the host cell of several proteins, some of which remain unidentified (11, 19, 22, 23). Characterization of these target proteins is of great interest because it may lead to the identification of potential effectors of cellular responses. Genes encoding type III secreted proteins are frequently located in pathogenicity islands or plasmids in the vicinity of genes encoding the components of the secretion apparatus itself (reviewed in ref. 7). Therefore, we investigated the presence of genes encoding targets of this secretion apparatus in a so far uncharacterized region of the centisome 63 pathogenicity island of S. typhimurium located downstream of the previously identified gene orgA (15, 24) (Fig. 1a). An ≈8-kb contiguous DNA segment of this region was cloned by chromosome walking as indicated in Materials and Methods. Discrete segments of this region were subcloned into the expression vector pWKS30 (25) and the resulting plasmids examined for their potential to encode secreted proteins. One of these subclones (pSB869), carrying a 1.8 kb SnaBI–BglII fragment, was shown to encode an ≈32 kDa secreted protein when expressed in the S. typhimurium strain SB241 (Fig. 1b). This strain carries a mutation in sipD and therefore secretes target proteins via the centisome 63 type III secretion apparatus in the absence of stimulatory signals (12). The 32 kDa protein was absent from culture supernatants of the secretion-deficient isogenic invG mutant strain SB161 (10) carrying the same plasmid. These results indicated that pSB869 was likely to encode a novel target of the centisome 63 type III secretion apparatus or, alternatively, a regulator of its expression.
Figure 1.
(a) Genetic organization of the avrA locus. The genetic organization of the fhlA proximal region of the centisome 63 pathogenicity island of S. typhimurium and the location of avrA is shown. A vertical arrowhead indicates the estimated boundary of the centisome 63 pathogenicity island (24). N, NheI; Ba, BamHI; Bg, BglII; H, HindIII; S, SnaBI; Aa, AatII; Bs, BstBI. (b) Identification of AvrA in culture supernatants of S. typhimurium. Culture supernatant proteins from the hypersecreting S. typhimurium sipD mutant strain SB241 or the secretion-defective invG mutant strain SB161 carrying the plasmid pSB869 (which encodes avrA) were separated by SDS/PAGE and stained with Coomassie brilliant blue. The arrow indicates the position of AvrA. Numbers to the left indicate the position of the molecular weight standards.
The entire nucleotide sequence of this plasmid was determined and a single 906 nt ORF was identified capable of encoding a 302 aa polypeptide calculated from translated sequences of 32 kDa, which lacks a typical sec-dependent signal sequence. This ORF is flanked upstream by a 164 nt noncoding sequence and the 3′ end of what appears to be another ORF (orfX; unpublished results) (Fig. 1a). Downstream, this ORF is flanked by a 96 nt noncoding region and the 3′ end of another putative ORF (orfY; unpublished results) (Fig. 1a). Similar to the other genes from the centisome 63 pathogenicity island, the overall nucleotide composition of the 906 nt ORF (42% GC) significantly deviates from the average GC content of S. enterica.
Comparison of the predicted amino acid sequence of the protein encoded in the 906 nt ORF with that of other proteins present in the available databases revealed significant sequence similarity to YopJ (56% identity, 72% similarity) and AvrRxv (25% identity, 48% similarity) (Fig. 2). YopJ is a protein of unknown function that is secreted via a type III protein secretion system encoded in a large virulence-associated plasmid of the animal pathogen Yersinia pseudotuberculosis (27). AvrRxv is a protein found in some strains of the plant pathogen Xanthomonas campestris pathovar vesicatoria and induces a hypersensitive response in resistant lines of tomato plants (28, 29). The sequence similarity suggests that AvrRxv, YopJ, and the 906 nt ORF, which we have termed AvrA, belong to a family of secreted effector proteins that might serve related functions in the cross-talk between bacterial pathogens and their plant or animal hosts.
Figure 2.
Amino acid sequence alignment of S. typhimurium AvrA, Yersinia pseudotuberculosis YopJ, and Xanthomonas campestris pv. vesicatoria AvrRxv. Sequences were aligned using the bestfit program of the Genetics Computer Group software package from the University of Wisconsin (Version 7) (26). Black boxes indicate identical amino acids and shaded boxes indicate conservative substitutions.
AvrA Is a Target of the Centisome 63 Type III Protein Secretion and Translocation Apparatus.
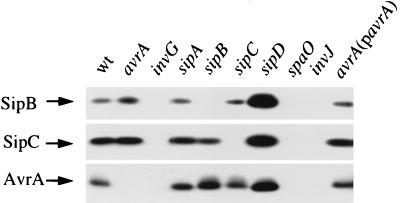
To confirm that AvrA is secreted by the centisome 63 type III system of S. enterica, we performed a Western blot analysis using a mouse polyclonal antiserum raised against a purified N-terminal glutathione S-transferase–AvrA fusion protein. AvrA was detected in supernatants of wild-type S. typhimurium strain SL1344 as well as in supernatants of isogenic derivative strains carrying mutations in sipA (12), sipB, or sipC (11), which encode other targets of the type III secretion system (Fig. 3). Higher levels of AvrA were found in culture supernatants of the hypersecreting sipD mutant strain SB241 (12). In contrast, no AvrA was detected in culture supernatants prepared from isogenic strains carrying mutations in invG (10), spaO (14), or invJ (13), which encode genes essential for secretion. Furthermore, an epitope-tagged AvrA protein was also secreted into the culture supernatant of S. typhimurium in an inv-dependent manner (data not shown). Cytoplasmic and secreted forms of the tagged protein migrated slightly slower than wild-type AvrA (data not shown), consistent with its larger size, due to the addition of the epitope tag. These results confirmed that avrA encodes a 32 kDa polypeptide that is secreted via the centisome 63 type III secretion system. A null mutation in avrA did not affect the secretion of other known targets of this system (Fig. 3; data not shown) nor the translocation of SipC into the host cell as determined by indirect immunofluorescence (data not shown). These results indicate that AvrA is not involved in either of these transport functions and therefore suggests that it is a potential effector molecule (14).
Figure 3.
Western blot analysis of AvrA secretion. Proteins from culture supernatants of wild-type S. typhimurium and the isogenic strains with nonpolar mutations in avrA (SB733), invG (SB161), sipA (SB225), sipB (SB169), sipC (SB220), sipD (SB241), spaO (SB302), and invJ (SB303), as well as from SB733 complemented with pSB869 were resolved by SDS/12% PAGE, transferred to nitrocellulose, and probed with antibodies directed to AvrA, SipB, and SipC as indicated in Materials and Methods. Reprobing of Western blots with an antibody directed to the cytoplasmic protein 6-phosphogluconate dehydrogenase indicated that the observed secretion patterns were not due to the different extent of bacterial cell lysis. Loading conditions were adjusted to obtain similar signal intensity. Thus, proteins from 100, 20, and 5,000 microliters of culture supernatants were loaded to visualize the Sip proteins, plasmid encoded (pSB869), and chromosomally encoded AvrA, respectively.
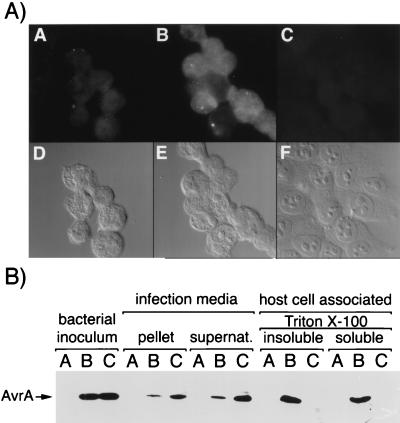
Effector proteins secreted via type III secretion systems are thought to get translocated inside the target host cell where they are expected to stimulate or modulate host cellular functions (23, 30–36). We therefore investigated the translocation of AvrA into cultured intestinal Henle-407 cells infected with wild-type S. typhimurium using two complementary approaches. Cultured intestinal Henle-407 cells were infected with S. typhimurium expressing an epitope-tagged AvrA protein. Two hours after infection, cells were fixed and processed for immunofluorescence staining with antibodies directed to the epitope tag (see Materials and Methods). The epitope-tagged AvrA protein was detected in cells infected with wild-type S. typhimurium SL1344 but not in cells infected with the hypersecreting, translocation-deficient sipD mutant strain SB241 (Fig. 4A). Confocal microscopy confirmed the intracellular location of AvrA (data not shown). To further demonstrate the translocation of AvrA into infected cells, cultured Henle-407 cells were infected with either SL1344 or SB241 expressing the epitope-tagged AvrA protein. Two hours after infection, a biochemical fractionation of the infected cells was carried out and the presence of AvrA was investigated in the infection medium and in the Triton X-100 soluble and insoluble fractions that contain translocated proteins and internalized bacteria, respectively. AvrA was present in the infection media of both strains, whereas it was detected only in Triton X-100 soluble and insoluble fractions of Henle-407 cells infected with wild-type bacteria (Fig. 4B). These results confirm that AvrA is translocated into cultured Henle-407 and that this translocation is dependent on the function of the type III protein secretion system encoded at centisome 63 on the S. typhimurium chromosome.
Figure 4.
Translocation of AvrA into host cells. (A) Henle-407 cells were infected with wild-type S. typhimurium without (A) or with (B) a plasmid encoding an epitope-tagged AvrA or the isogenic translocation defective, hypersecreting sipD mutant strain carrying the same plasmid (C). Infected cells were subsequently stained with an antibody directed to the epitope tag. Micrographs of the AvrA stained cells (A–C) and the corresponding DIC images (D-F) are shown. (B) Detection of AvrA in different fractions of Henle-407 cells infected with wild-type and sipD S. typhimurium strains. The different fractions obtained as indicated in Materials and Methods were probed with an antibody directed to the epitope tag of AvrA. A, wild-type S. typhimurium strain SL1344; B, SL1344 carrying a plasmid encoding an epitope-tagged AvrA (pSB885); C, S. typhimurium sipD strain SB241 carrying pSB885. Arrow to the left indicates the position of AvrA. Only the relevant part of the blot is shown.
Genetic Organization and Distribution of avrA Among S. enterica Serovars.
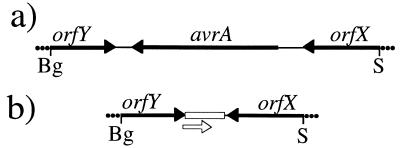
The distribution of avrA among different S. enterica serovars was determined by dot blot analysis of chromosomal DNA with a probe corresponding to the avrA coding sequence. The probe strongly hybridized to only a subset of the strains tested (23 of 30) (Table 1). Among the serovars that failed to hybridize were the host-adapted S. enterica serovar typhi (Salmonella typhi) and S. enterica serovar choleraesuis (Salmonella choleraesuis). In contrast, sequences upstream and downstream of avrA (orfX and orfY) were detected in all strains tested (Fig. 5). Southern hybridization analysis of the avrA region of several avrA− serovars revealed that these strains lack an ≈900 nt fragment corresponding to avrA (data not shown). Nucleotide sequence analysis of PCR products amplified from the avrA− S. typhi and S. choleraesuis strains with primers complementary to the 3′ termini of orfX and orfY revealed that avrA and its noncoding flanking regions are replaced by a 261 nt element that is highly conserved (97% identical) between these two strains. This element harbors a 207 nt ORF that encodes a polypeptide with limited homology to the orfG gene product encoded in several plasmids of Borrelia burgdorferi (37). The flanking fragments of orfX and orfY are nearly identical in all strains examined (the alignment of these sequences can be viewed at our laboratory’s home page at the following URL http://www.informatics.sunysb.edu/microbiology/galanpage.html).
Table 1.
Dot blot hybridization to assess distribution of avrA
| Hybridization signal | Serotype |
|---|---|
| Positive | |
| S. enterica sv. agona | B |
| S. enterica sv. brandenburg | B |
| S. enterica sv. bredeney | B |
| S. enterica sv. duisburg | B |
| S. enterica sv. heidelberg | B |
| S. enterica sv. java | B |
| S. enterica sv. schwarzengrund | B |
| S. enterica sv. typhimurium (four isolates) | B |
| S. enterica sv. braenderup | C1 |
| S. enterica sv. infantis | C1 |
| S. enterica sv. tennessee | C1 |
| S. enterica sv. thompson | C1 |
| S. enterica sv. virchov | C1 |
| S. enterica sv. bovis morbidificans | C2 |
| S. enterica sv. hadar | C2 |
| S. enterica sv. manhattan | C2 |
| S. enterica sv. newport | C2 |
| S. enterica sv. dublin | D1 |
| S. enterica sv. enteritidis (two isolates) | D1 |
| S. enterica sv. gallinarum | D1 |
| S. enterica sv. panama | D1 |
| S. enterica sv. pullorum (two isolates) | D1 |
| S. enterica sv. anatum | E1 |
| Negative | |
| S. enterica sv. choleraesuis | C1 |
| S. enterica sv. montevideo | C1 |
| S. enterica sv. ohio | C1 |
| S. enterica sv. othmarschen | C1 |
| S. enterica sv. nienstaedten | C4 |
| S. enterica sv. typhi (three isolates) | D1 |
| S. enterica sv. arizona | — |
Unless otherwise indicated, one isolate for each serovar was tested.
Figure 5.
Genetic organization of the two genetic elements found between orfX and orfY in different S. enterica serovars. (a) S. enterica serovar typhimurium and serovar enteritidis. (b) S. enterica serovar choleraesuis and serovar typhi. Open box indicates the location of a 264 nt region with no homology to avrA. Open arrow indicates the position of a potential ORF.
Virulence of S. typhimurium avrA Mutants.
To examine the role of AvrA in Salmonella virulence we constructed a S. typhimurium strain carrying a nonpolar mutation in avrA as indicated. The resulting strain, termed SB733, was tested in a variety of in vitro and in vivo virulence assays. SB733 induced membrane ruffling, entered into cultured intestinal Henle-407 cells, and induced macrophage cytotoxicity in a manner indistinguishable from that of the wild-type strain (Table 2). When inoculated at various doses into the highly susceptible BALB/c mice or the less susceptible Swiss mice, both strains displayed the same level of virulence (Table 2 and data not shown).
Table 2.
Effect of avrA on S. typhimurium virulence phenotypes
| Strain | Relevant genotype | Henle-407 invasion* | J774 toxicity† | Mouse virulence‡ (mean time to death) |
|---|---|---|---|---|
| SL1344 | Wild type | 69 ± 10 | 30 ± 5 | 6 |
| SB147 | invA::aphT | 0.15 ± 0.03 | 2 ± 1 | ND |
| SB733 | avrA::aphT | 56 ± 10 | 25 ± 5 | 6 |
| SB733(pSB869) | avrA::aphT(avrA+) | 74 ± 8 | 24 ± 2 | 6 |
ND, not determined
Entry into Henle-407 cells was determined by the gentamicin resistance assay. Values are the mean ± the SD of six independent determinations and represent the percentage of the bacterial inoculum that survived 2 hr of gentamicin treatment.
Macrophage J774 cytotoxicity was determined by a dye exclusion assay (21). Values are the percent of cells exhibiting cytotoxicity after 30 min of infection. At least 200 cells were evaluated in each assay.
BALB/c mice (six per group) were infected orally with 106 bacteria. There were no survivors in any group. The mean time to death is expressed in days.
To further investigate a potential role for AvrA in avirulence, we amplified by PCR the avrA gene from the S. enterica serovars agona (Salmonella agona) and hadar (Salmonella hadar), which are not virulent for mice. The amplified DNA fragments were cloned into an expression vector and the resulting plasmids were introduced into the S. typhimurium avrA mutant strain SB733. Western blot analysis confirmed that S. typhimurium strain SB733 efficiently expressed and secreted the heterologous AvrA proteins (data not shown). The virulence of the SB733 derivative strains expressing the heterologous avrA alleles was compared with that of SB733 with the expression vector alone and the wild-type parent S. typhimurium strain SL1344. All strains exhibited indistinguishable virulence phenotypes in orally inoculated Swiss mice as indicated by the number of survivors and the mean time to death (data not shown). However, these results do not rule out a role for AvrA in avirulence (or virulence) as detection of this putative function will require the identification of a host in which this protein may be able to exert such an effect.
DISCUSSION
We have identified a protein of S. typhimurium, termed AvrA, that is secreted via the centisome 63 type III secretion system. AvrA exhibited significant sequence similarity to YopJ of Yersinia pseudotuberculosis and AvrRxv of Xanthomonas campestris, two previously identified proteins that are likely to depend on type III secretion systems to exert their function. Although the components of the type III secretion machinery are remarkably conserved between animal and plant pathogenic bacteria, the target proteins that had been identified thus far share little homology (reviewed in ref. 7). Indeed, AvrRxv, YopJ, and AvrA constitute the first example of putative targets of type III secretion systems that are conserved between plant and animal pathogenic bacteria. It is possible that this family of proteins may exert related functions in the cross-talk between these pathogens and their hosts. Consistent with this hypothesis is the observation that AvrA is translocated into host cells, a property expected for effector proteins that are presented to the host via a type III secretion system. Although the translocation of YopJ and AvrRxv into host cells has not been investigated, it is likely that intracellular delivery by a type III protein secretion system is an essential requirement for the function of these proteins. YopJ has been shown to be secreted via a type III secretion system in Yersinia pseudotuberculosis (27) and the function of most avirulence proteins in plant pathogenic bacteria has also been shown to depend on such systems (reviewed in ref. 6). The finding of similarities between virulence factors of animal and plant pathogenic bacteria is not unprecedented. Rahme et al. (38) have previously shown that common genes are required for Pseudomonas aeruginosa to cause disease in both plant and animal hosts.
What is the function(s) of this family of effector proteins? Xanthomonas campestris pv. vesicatoria is the causal agent of leaf spot disease in pepper and tomato and the AvrRxv protein has been shown to be required for the induction of the “hypersensitive response” in resistant lines of host plants (29). This response is characterized by the activation of a variety of plant defense mechanisms that lead to the localized formation of necrotic lesions due to programmed cell death at the point of initial bacterial encounter, which halts any further bacterial growth (4, 5, 39). Bacterial proteins capable of stimulating such a response are known as “avirulence factors” and are thought to exert their function by interacting with specific resistance gene products in the nonpermissive host plant. Interestingly, unlike most avirulence proteins, AvrRxv is rather broadly recognized, eliciting a hypersensitive response not only in “resistant” lines of X. campestris pv. vesicatoria natural host plants but also in a variety of other plant species that are normally not infected by this bacterial pathogen (28, 29). Besides their ability to induce a hypersensitive response, avirulence genes of plant pathogens share the following characteristics: (i) they are present in only a subset of bacterial strains; (ii) they are usually not associated with virulence phenotypes (although not all avr genes have been examined for a role in virulence); and (iii) they are most often presented to the host via type III protein secretion systems. Is AvrA an example of an avirulence gene in S. enterica? Although the existence of “avirulence” genes with similar properties to those of plant pathogenic bacteria has not been demonstrated in mammalian pathogens, this may simply reflect the fact that studies have been most often aimed at the identification of genes involved in virulence. We have shown here that besides the sequence homology, AvrA has features that resemble avirulence determinants from phytopathogenic bacteria: (i) AvrA is a target of a type III protein secretion system; (ii) in contrast to other identified targets of this system, AvrA is not associated with virulence in the assay systems examined; and (iii) AvrA exhibits a scattered distribution among S. enterica serovars, being notably absent from the host-adapted serovars typhi and choleraesuis. However, we were unable to demonstrate a functional equivalent of the plant hypersensitive response in two different strains of mice infected with a mouse virulent S. typhimurium strain encoding different avrA alleles from mouse nonpathogenic serovars of S. enterica. This is indeed not surprising considering that it required testing of several hundred tomato lines to identify one that was able to mount an AvrRxv- specific hypersensitive response upon infection with Xanthomonas campestris pv. vesicatoria (29). Demonstration of the ability of AvrA to induce a hypersensitive response will require the identification of a specific host in which AvrA may be able to exert its putative avirulence function. Nevertheless, at this point, we cannot completely rule out a role for AvrA in Salmonella virulence. It is possible that a protein with redundant functions to AvrA may be encoded elsewhere in the Salmonella chromosome or that our assays have not been able to detect a virulence phenotype. For example, AvrA may help to ensure that infection of certain Salmonella serovars remains localized in some compartment (e.g., intestinal tract) of selected hosts. Indeed, different Salmonella serovars are capable of causing localized, systemic, acute, or cronic infections depending on the infected host. A localized or persistent infection may be advantageous for spread, because it may allow shedding of the organism for longer periods of time. It has been proposed that the original function of avirulence gene products may not have included the induction of defense responses in nonpermissive hosts but rather it may have been to improve the fitness of the pathogens or their ability to cause disease in certain permissive hosts. In this context, then, the hypersensitive response may well be considered to be an evolutionary adaptation of certain hosts to frequent contact with certain bacterial “virulence” factors.
The genetic organization of the avrA locus in different serovars as well as its base composition, which is significantly deviated from the rest of the Salmonella chromosome, suggest that this gene was acquired by horizontal transmission. It is intriguing that the avrA locus has been replaced by a highly conserved 287 bp genetic element in the host adapted S. enterica serovars typhi and choleraesuis. However, it is not known whether avrA was acquired in conjunction with the rest of the centisome 63 pathogenicity island and later replaced by the 287 bp genetic element in these serovars or whether this gene may have been acquired by an independent genetic event after the divergence of S. enterica. It is also unknown whether the acquisition of this element (or loss of avrA) in these strains is in any way related to their host adaptation. Further studies on AvrA function may contribute to the understanding of pathogen-specific defense responses of the naive animal host and may lead to novel concepts for the development of therapeutic strategies to combat infectious agents.
Acknowledgments
We would like to thank C. Roy, D. Zhou, and K. Eichelberg for critical review of this manuscript. This work was supported by Public Health Service Grants AI30492 and GM52543 from the National Institutes of Health to J.E.G., who is an investigator from the American Heart Association. W.-D.H. was supported in part by a fellowship from the German Bundesministerium für Forschung und Technologie.
Footnotes
References
- 1.Foster J W, Spector M P. Annu Rev Microbiol. 1995;49:145–174. doi: 10.1146/annurev.mi.49.100195.001045. [DOI] [PubMed] [Google Scholar]
- 2.Mahan M J, Slauch J M, Mekalanos J J. Escherichia coli Salmonella: Cell Mol Biol. 1996;2:2803–2815. [Google Scholar]
- 3.Galán J E. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 4.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leach J E, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 6.Alfano J R, Collmer A. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galán J E, Bliska J B. Annu Rev Cell Dev Biol. 1996;12:219–253. doi: 10.1146/annurev.cellbio.12.1.221. [DOI] [PubMed] [Google Scholar]
- 8.Hoiseth S K, Stocker B A. Nature (London) 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 9.Ginocchio C, Galán J E. Infect Immun. 1995;63:729–732. doi: 10.1128/iai.63.2.729-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaniga K, Bossio J C, Galán J E. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 11.Kaniga K, Tucker S C, Trollinger D, Galán J E. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaniga K, Trollinger D, Galán J E. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collazo C M, Zierler M K, Galán J E. Mol Microbiol. 1995;15:25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 14.Collazo C, Galán J E. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones B D, Falkow S. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller V L, Mekalanos J J. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan K-L, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 18.Obert S, O’Connor R J, Schmid S, Hearing P. Mol Cell Biol. 1994;14:1333–1346. doi: 10.1128/mcb.14.2.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collazo C, Galán J E. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 20.Galán J E, Curtiss R., III Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L M, Kaniga K, Galán J E. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 22.Hueck C J, Hantman M J, Bajaj V, Johnston C, Lee C A, Miller S I. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 23.Wood M H, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 24.Mills D B, Bajaj V, Lee C A. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang R F, Kushner S R. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 26.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galyov E E, Hakansson S, Wolf-Watz H. J Bacteriol. 1994;176:4543–4548. doi: 10.1128/jb.176.15.4543-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whalen M C, Stall R E, Staskawicz B J. Proc Natl Acad Sci USA. 1988;85:6743–6747. doi: 10.1073/pnas.85.18.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whalen M C, Wang J F, Carland F M, Heiskell M E, Dahlbeck D, Minsavage G V, Jones J B, Scott J W, Stall R E, Staskawicz B J. Mol Plant Microbe Interact. 1993;6:616–627. doi: 10.1094/mpmi-6-616. [DOI] [PubMed] [Google Scholar]
- 30.Rosqvist R, Magnusson K E, Wolf-Watz H. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Persson C, Nordfelth R, Holmström A, Hakansson S, Rosqvist R, Wolf-Watz H. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 32.Sory M-P, Cornelis G R. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 33.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 34.Leister R T, Ausubel F M, Katagiri F. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandenackerveken G, Marois E, Bonas U. Cell. 1996;87:1307–1316. doi: 10.1016/s0092-8674(00)81825-5. [DOI] [PubMed] [Google Scholar]
- 36.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 37.Zuckert W R, Meyer J. J Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 39.Greenberg J T, Guo A, Klessig D F, Ausubel F M. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]