Abstract
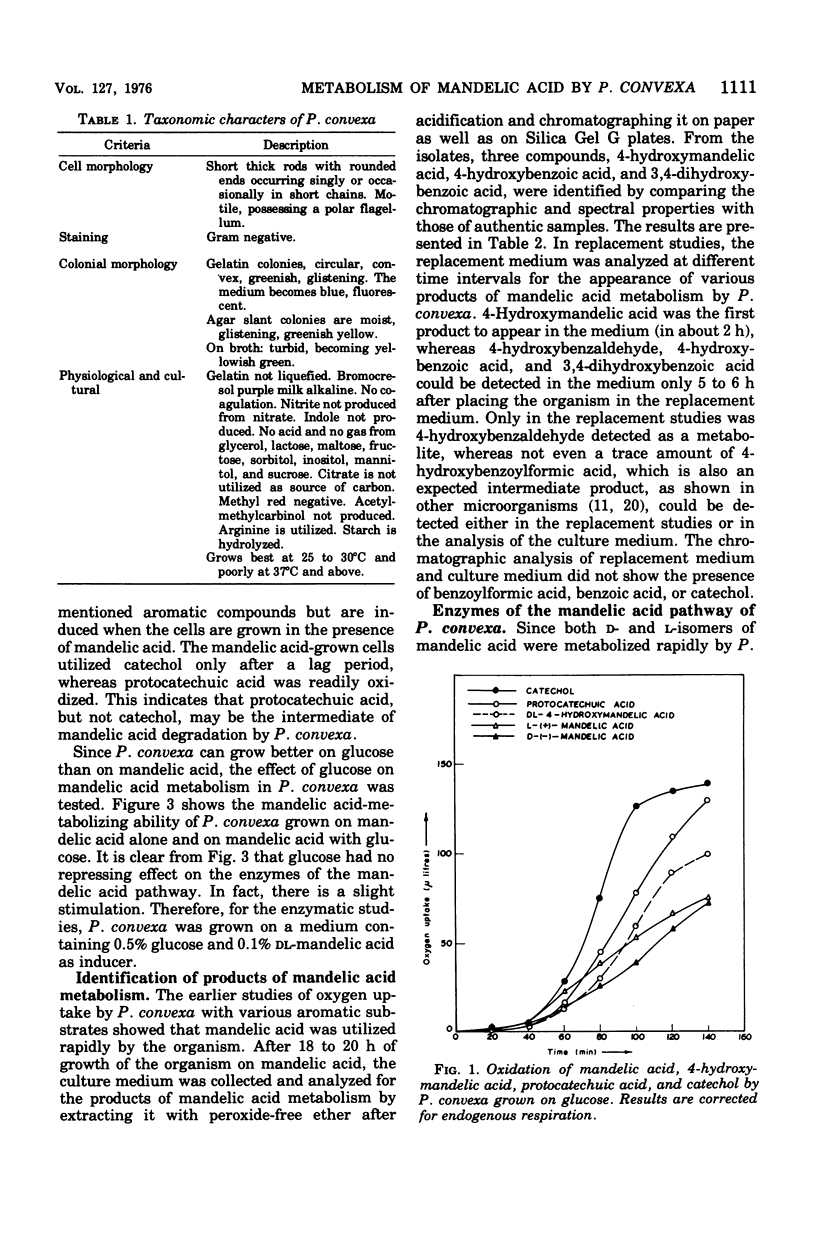
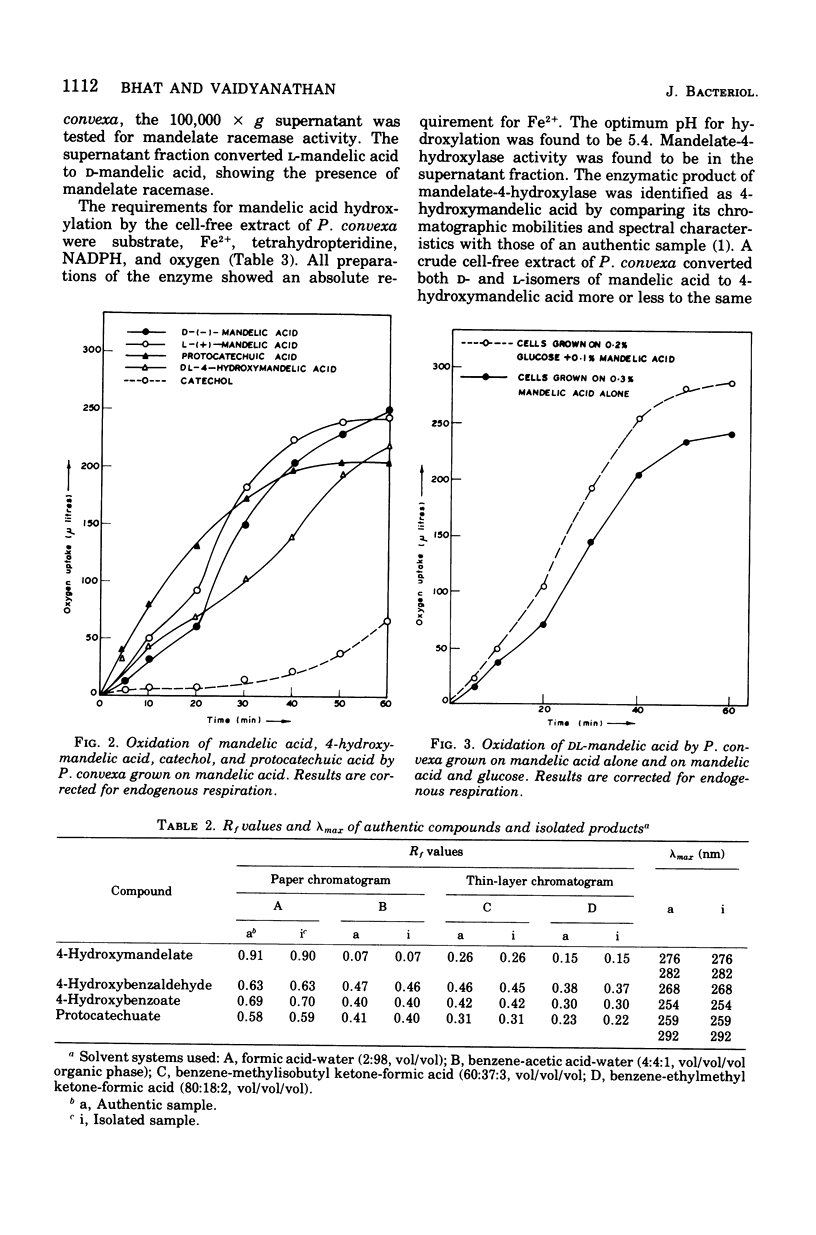
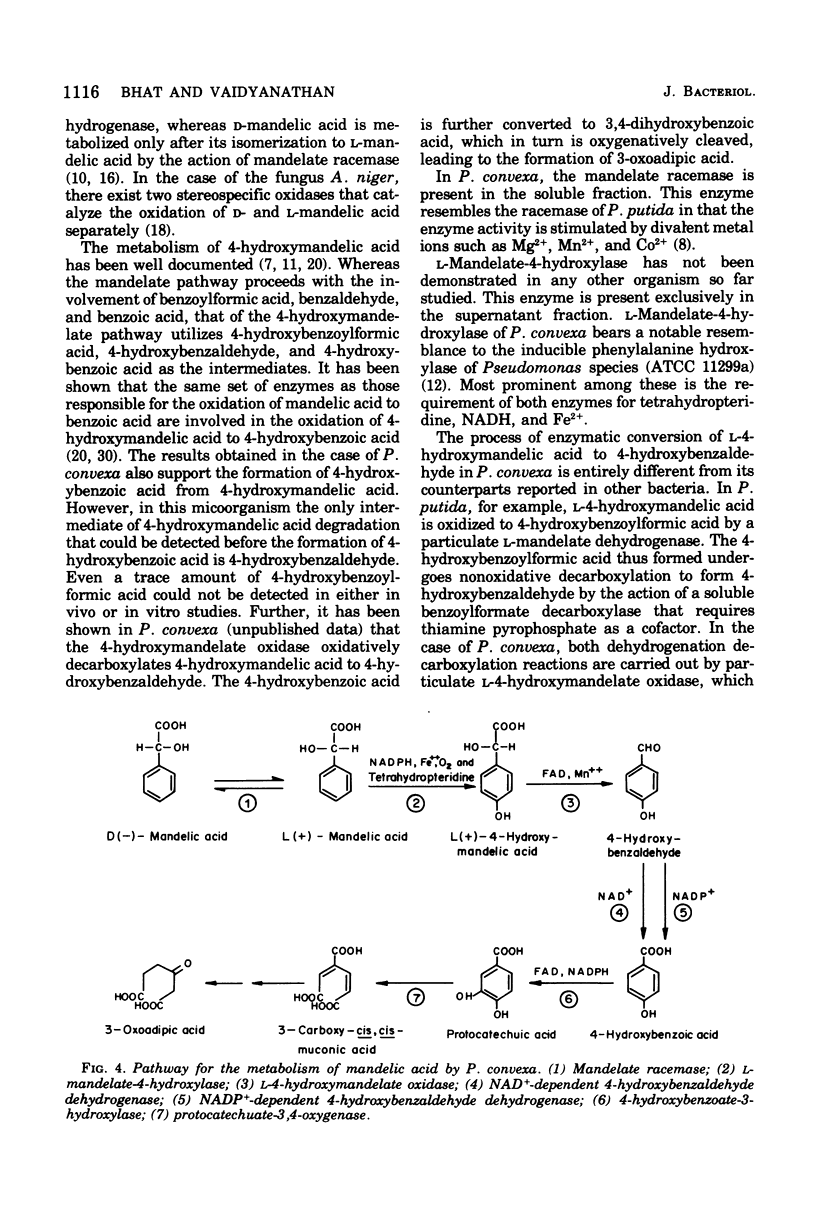
A microorganism capable of degrading DL-mandelic acid was isolated from sewage sediment of enrichment culture and was identified as Pseudomonas convexa. It was found to metabolize mandelic acid by a new pathway involving 4-hydroxymandelic acid, 4-hydroxybenzaldehyde, 4-hydroxybenzoic acid, and 3,4-dihydroxybenzoic acid as aromatic intermediates. All the enzymes of the pathway were demonstrated in cell-free extracts. L-Mandelate-4-hydroxylase, a soluble enzyme, requires tetrahydropteridine, nicotinamide adenine dinucleotide phosphate, reduced form, and Fe2+ for its activity. The next enzyme, L-4-hydroxymandelate oxidase (decarboxylating), a particulate enzyme, requires flavine adenine dinucleotide and Mn2+ for its activity. A nicotinamide adenine dinucleotide-dependent, as well as a nicotinamide adenine dinucleotide phosphate-dependent, benzaldehyde dehydrogenase has been resolved and partially purified.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRAY H. G., HUMPHRIS B. G., THORPE W. V., WHITE K., WOOD P. B. Kinetic studies of the metabolism of foreign organic compounds. III. The conjugation of phenols with glucuronic acid. Biochem J. 1952 Nov;52(3):416–419. doi: 10.1042/bj0520416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S. G., Ramanarayanan M., Vaidyanathan C. S. Mandelic acid-4-hydroxylase, a new inducible enzyme from Pseudomonas convexa. Biochem Biophys Res Commun. 1973 Jun 8;52(3):834–842. doi: 10.1016/0006-291x(73)91013-9. [DOI] [PubMed] [Google Scholar]
- CARLES J., BOURGUET M. Les acides organiques du blé. C R Seances Soc Biol Fil. 1957;151(2):383–387. [PubMed] [Google Scholar]
- Crowden R. K. Biosynthesis of the polyphenolic acid metabolites of Polyporus tumulosus Cooke. Can J Microbiol. 1967 Feb;13(2):181–197. doi: 10.1139/m67-025. [DOI] [PubMed] [Google Scholar]
- Fee J. A., Hegeman G. D., Kenyon G. L. Mandelate racemase from Pseudomonas putida. Subunit composition and absolute divalent metal ion requirement. Biochemistry. 1974 Jun 4;13(12):2528–2532. doi: 10.1021/bi00709a008. [DOI] [PubMed] [Google Scholar]
- GUNSALUS C. F., STANIER R. Y., GUNSALUS I. C. The enzymatic conversion of mandelic acid to benzoic acid. III. Fractionation and properties of the soluble enzymes. J Bacteriol. 1953 Nov;66(5):548–553. doi: 10.1128/jb.66.5.548-553.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUNSALUS I. C., GUNSALUS C. F., STANIER R. Y. The enzymatic conversion of mandelic acid to benzoic acid. I. Gross fractionation of the system into soluble and particulate components. J Bacteriol. 1953 Nov;66(5):538–542. doi: 10.1128/jb.66.5.538-542.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUNTER S. E. The enzymatic oxidation of p-hydroxymandelic acid to p-hydroxybenzoic acid. J Bacteriol. 1953 Sep;66(3):341–346. doi: 10.1128/jb.66.3.341-346.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guroff G., Rhoads C. A. Phenylalanine hydroxylase from Pseudomonas species (ATCC 11299a). Purification of the enzyme and activation by various metal ions. J Biol Chem. 1967 Aug 25;242(16):3641–3645. [PubMed] [Google Scholar]
- Hegeman G. D., Rosenberg E. Y., Kenyon G. L. Mandelic acid racemase from Pseudomonas putida. Purification and properties of the enzyme. Biochemistry. 1970 Oct 13;9(21):4029–4036. doi: 10.1021/bi00823a001. [DOI] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. 3. Isolation and properties of constitutive mutants. J Bacteriol. 1966 Mar;91(3):1161–1167. doi: 10.1128/jb.91.3.1161-1167.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966 Mar;91(3):1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. II. Isolation and properties of blocked mutants. J Bacteriol. 1966 Mar;91(3):1155–1160. doi: 10.1128/jb.91.3.1155-1160.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa K., Stanier R. Y. Crystallization and properties of p-hydroxybenzoate hydroxylase from Pseudomonas putida. J Biol Chem. 1966 May 25;241(10):2453–2460. [PubMed] [Google Scholar]
- Jamaluddin M., Rao P. V., Vaidyanathan C. S. Involvement of the protocatechuate pathway in the metabolism of mandelic acid by Aspergillus niger. J Bacteriol. 1970 Mar;101(3):786–793. doi: 10.1128/jb.101.3.786-793.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S. I., Fewson C. A. Enzymes of the mandelate pathway in Bacterium N.C.I.B. 8250. Biochem J. 1968 Apr;107(4):497–506. doi: 10.1042/bj1070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S. I., Fewson C. A. Metabolism of mandelate and related compounds by bacterium NCIB 8250. J Gen Microbiol. 1968 Sep;53(2):259–273. doi: 10.1099/00221287-53-2-259. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- NAIR P. M., VAIDYANATHAN C. S. A COLORIMETRIC METHOD FOR DETERMINATION OF PYROCATECHOL AND RELATED SUBSTANCES. Anal Biochem. 1964 Mar;7:315–321. [PubMed] [Google Scholar]
- Rosenberg S. L., Hegeman G. D. Genetics of the mandelate pathway in Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1270–1276. doi: 10.1128/jb.108.3.1270-1276.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg S. L. Regulation of the mandelate pathway in Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1257–1269. doi: 10.1128/jb.108.3.1257-1269.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEUBERT W. Degradation of isoprenoid compounds by micro-organisms. I. Isolation and characterization of an isoprenoid-degrading bacterium, Pseudomonas citronellolis n. sp. J Bacteriol. 1960 Mar;79:426–434. doi: 10.1128/jb.79.3.426-434.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANIER R. Y., GUNSALUS I. C., GUNSALUS C. F. The enzymatic conversion of mandelic acid to benzoic acid. II. Properties of the particulate fractions. J Bacteriol. 1953 Nov;66(5):543–547. doi: 10.1128/jb.66.5.543-547.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector T., Massey V. Interactions of substrate and non-substrate effectors with p-hydroxybenzoate hydroxylase from pseudomonas fluorescens. Biochem Biophys Res Commun. 1971 Dec 3;45(5):1219–1226. doi: 10.1016/0006-291x(71)90148-3. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Stevenson I. L., Mandelstam J. Induction and multi-sensitive end-product repression in two converging pathways degrading aromatic substances in Pseudomonas fluorescens. Biochem J. 1965 Aug;96(2):354–362. doi: 10.1042/bj0960354. [DOI] [PMC free article] [PubMed] [Google Scholar]


