Abstract
We develop a probabilistic method for analyzing global features of a cellular network under intrinsic statistical fluctuations, which is important when there are finite numbers of molecules. By making a self-consistent mean field approximation of splitting the variables in order to reduce the large number of degrees of freedom, which is reasonable for a not very strongly interacting network, we discovered that the underlying energy landscape of the mitogen-activated protein kinases (MAPKs) signal transduction network (with experimentally measured or inferred parameters such as chemical reaction rate coefficients in the network) is funneled toward a global minimum characterized by the nonequilibrium steady-state fixed point of the system at the end of the signal transduction process. For this system, we also show that the energy landscape is robust against intrinsic fluctuations and random perturbation to the inherent chemical reaction rates. The ratio of the slope versus the roughness of the energy landscape becomes a quantitative measure of robustness and stability of the network. Furthermore, we quantify the dissipation cost of this nonequilibrium system through entropy production, caused by the nonequilibrium flux in the system. We found that a lower dissipation cost corresponds to a more robust network. This least dissipation property might provide a design principle for robust and functional networks. Finally, we find the possibility of bistable and oscillatory-like solutions, which are important for cell fate decisions, upon perturbations. The method described here can be used in a variety of biological networks.
Keywords: funnel, stability, potential landscape, function
The ultimate goal of biology is to understand the function of specific systems. At the cell level, the function of the system is realized through the network of interactions between molecules. Unraveling the underlying mechanisms and global principles of the cellular network is the current challenge in systems biology.
There has been some experimental investigation of these networks. The experimental progress has been made through large-scale genetic mutations and screenings, gene expressions, yeast two-hybrids, pull-down, and immunoprecipitation (1–4). These experiments find that cellular networks, in general, are quite robust and perform their biological functions in the midst of environmental perturbations. Synthetic biology is aiming to follow in the footsteps of electronic design by piecing together small modular components to build a much larger network and, by doing this, study the network from a bottom-up perspective (5). This approach is just at the beginning stage and seems to have a promising future.
On the theoretical side, the topological structures of the networks have been investigated recently (6). The scale-free properties and hierarchical architectures for the networks have been shown (7–9). The highly connected nodes in the network, called hubs, might play an important role in the robustness of the network. From the engineering perspective, efforts have been made to understand the network from control perspectives with robust yet fragile natures (10). There have been a number of studies attempting to determine why networks are robust in their biological function among perturbations (11–24).
The most common way of modeling these systems is through chemical rate equations. From this, deterministic trajectories can be found, and from these trajectories inference can be drawn to the possible biological function. These trajectories can only span a small piece of the whole phase space. To determine more of the phase space and global features of this space under a set of parameters, one would need to examine all of the initial conditions, a hopeless task in most systems. To understand the network's global nature under different external conditions, one must change the inherent parameters, thus further complicating the task. Because this will generally introduce a high dimensionality into the phase space, the issues of stability, robustness, and other global properties become very hard to address and examine.
A major problem is that the typical chemical rate equations ignore inherent statistical fluctuations in the number of particles, N. This is not a problem when N ≫ 1 because fluctuations go as . There are only a finite number of molecules in the cell, typically on the order of 103 or less. The intrinsic statistical fluctuations, which are negligibly small in the bulk, become very important. Thus, it is necessary to move from a deterministic model to a statistical model. The study of the influence under external noise from the cellular environments for the MAPK system was carried out in an earlier article (19). Here, we will focus on the intrinsic noise from number fluctuation.
We can begin this statistical description by noticing that the chemical rate equations are only approximations up to the average concentration level. In the cell, there are statistical fluctuations around this concentration, coming from internal and external sources. The internal noise comes from the finite number of molecules, as we mentioned above, whereas the external noise comes from the highly dynamical and inhomogeneous environments of the interior of the cell (25–30). Both of these sources of noise are important in determining properties of the network.
Although there are huge number of states in the phase space that are hard to explore globally, the probability of each state is not uniformly distributed. It might be that most of the states only have small probabilities, but only a finite number of states have larger probabilities. The states with higher probabilities are biologically more relevant and should be the focus rather than the states with lower weights. Thus, an approach to determining the probability of each state will allow us to identify the important states for biological function and furthermore study their stability and robustness.
Noisy conditions thus play a very important role in these chemical reaction networks and are much more realistic than the average mean concentrations. To describe the system under noisy conditions, we will define a potential energy function that is derived from the steady-state probability of the network. After this landscape is determined, the probability of each state is known and we can begin to analyze global features (19, 22, 23). This is reminiscent of equilibrium systems, where one can analyze the phase diagram and other global features. Once the network problem is put into terms of the generalized potential energy function or potential energy landscape, the issue of the global stability or robustness is much easier to address. The purpose of this article was to study the global robustness or stability against intrinsic fluctuations and random perturbation to the inherent chemical reaction rates directly from the properties of the potential energy landscape of the network. We will focus on the network landscape under the intrinsic statistical fluctuations due to the finite number of molecules in the cell.
Because the cell is an open system, the network is a far-from-equilibrium system. The dynamics is not only determined by the underlying landscape but also by nonzero flux due to the lack of detailed balance. This can lead to a more complete description with potential and the flux for the network system in analogy to the electronic circuit with voltage and current. As an open nonequilibrium system, there will be heat dissipation and entropy production in analogy from the electric circuit with voltage and current generating heat losses (23, 31, 32). This entropy production can be used to characterize the global features of the system.
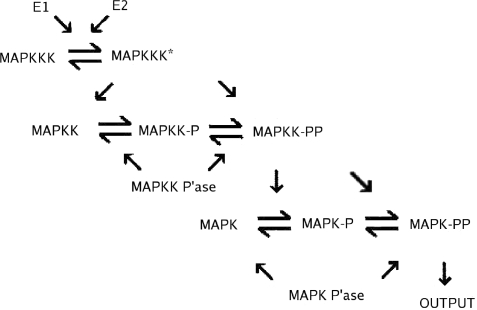
To explore the nature of the underlying potential energy landscape of the cellular networks, we will study the relatively simple yet important MAPK signal transduction network (Fig. 1). Mitogen-activated protein kinases (MAPKs) belong to a family of serine/threonine protein kinases widely conserved among eukaryotes and are involved in many cellular programs such as proliferation, differentiation, movement, and cell death. MAPK signaling cascades are organized hierarchically into the three-tiered modules. MAPKs are phosphorylated and activated by MAPK kinases (MAPKKs), which in turn are phosphorylated and activated by MAPKK kinases (MAPKKKs). The MAPKKK is in turn activated by interaction with a family of small GTPases and/or other protein kinases connecting the MAPK module to the cell-surface receptor or external stimuli (2, 3) (Fig. 1). We will examine the MAPK network by uncovering the underlying energy landscape under intrinsic statistical fluctuations through the master equations of the system.
Fig. 1.
MAPK cascade.
Results and Discussions
Once the probability distribution function, P(x, t), for the state variable x (x represents the protein concentrations in the MAPK signal transduction network case) is solved, the corresponding steady-state probability PSS(x) can be obtained by taking the long time limit. The potential energy function U(x) (14, 18–23, 33) can be related to steady-state probability:
with the partition function Z = ∫ddx exp{−U(x)}. From this, we can recognize U as a potential energy function for the network system. Once the potential energy landscape is determined, we can examine the global properties of the protein cellular networks.
The potential energy is a multidimensional function in concentration vector x space with each component of which representing the concentration of each type of proteins. For certain configurations of concentrations, the network adopts a certain potential energy (or the corresponding probability). The dimensionality of the configurational state space is huge. We are interested, first of all, in the most probable configuration that corresponds to the lowest energy state. We found that the lowest energy state or the most probable configuration is the one at the end stage (ground state) of the MAPK signal transduction, which corresponds to the fixed-point steady-state solution of the averaged chemical rate equations for the MAPK network. However, this distribution is 22-dimensional and thus very difficult to visualize or analyze directly. Because of this, we will have to consider lower dimensional projections of this free energy distribution.
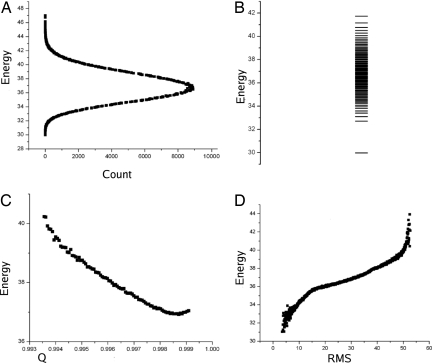
First, let us consider the 0th-dimension projection, the histogram (Fig. 2A). The minimum of this projection corresponds to the steady state of the system, which also corresponds to the traditional solution of the chemical rate equations. This correspondence comes about because all reactions only involve single reactants. Fig. 2B shows the potential energy spectrum for our system. Notice that the global minimum of the potential energy is significantly separated from the rest of the spectrum.
Fig. 2.
Projection of energy landscape. (A) Zero-dimensional projection: Energy histogram of network. (B) Energy spectrum of network. (C) One-dimensional projection: Energy projected across inner product Q. (D) One-dimensional projection: Energy projection across RMSD.
However, we need a quantifiable measure for this projection. To arrive at this, we will consider a dimensionless quantity, which we call the robustness ratio of the distribution, Λ = δU/ΔU, where δU = 〈U〉 − Umin, and ΔU is the half spread of the histogram. The δU is a measure of the forcing and bias toward the global minimum of the potential energy, whereas ΔU is a measure of the roughness and possibility of being locally trapped in the potential energy landscape. When Λ is significantly larger than 1, the bias toward the minimum is much stronger than the probability of local trapping; thus, the global minimum is well separated and distinct from the rest of the network potential energy spectrum. The robustness ratio for this network is 2.45. This signifies that the MAPK network is robust under intrinsic statistical fluctuations, which is not surprising because the network is required by evolutionary concerns to be robust.
Now, let us consider one-dimensional projections. The two one-dimensional coordinates we will consider are the RMS distance from global minimum, that is and a normalized inner product Q = (xmin·x)/|xmin||x|, where xmin is the minimum energy state of the system. Because this inner product is defined in the normal way for Rn, the Q value is equivalent to cos θ, where θ is the angle between these two state phase space vectors. Thus, a value of Q = 0 describes orthogonal vectors with no overlap and Q = 1 describes parallel vectors with complete overlap. From Fig. 2C, we see a downhill trend toward Q = 1, implying a tendency to align with the global minimum of the potential energy landscape. This tendency shows that there does exist a funnel in the potential energy landscape. Another coordinate, the RMS distance (RMSD), shows the overall phase space distance separation of the two states. Fig. 2D shows a similar downhill slope and overall funneled landscape toward the global minima for the RMSD projection.
It is also important to examine the parameter space. To do this, we varied the reaction rates of the system. Specifically, the reaction rates were taken from a probability distribution with a mean of the unperturbed rate, cμ, and a standard deviation given by σ = lp × cμ, where lp is the level of perturbation applied to the system. From this, we examine what happens to the system at a different location in the parameter space.
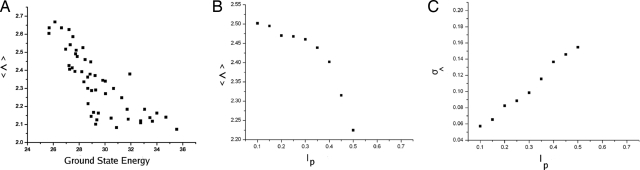
Fig. 3A shows the robustness ratio Λ of the MAPK network versus the energy of the ground state. There is a monotonic relationship between the ground state energy and the robustness ratio Λ. When Λ is larger (smaller), the landscape is more (less) robust, and the network is more (less) stable with ground state dominating (less significant). Therefore, Λ is indeed a robustness measure for the network. We observe that the system is stable under most of the perturbations through changing the rates. There are rare events that cause big changes that could significantly destabilize the system and lower both Λ and the probability of the ground state.
Fig. 3.
Landscape robustness versus perturbation levels of chemical rate constants. (A) 〈Λ〉 relationship with ground state energy. (B) 〈Λ〉 variation with change in perturbation level. (C) Standard variation versus changes in perturbation level.
Fig. 3B shows the robustness ratio Λ versus the perturbation level lp. We see again that with small variances of perturbations of the chemical reaction rates, the network is with a large Λ characterizing the funneled landscape and stability of the system. With larger variances of perturbations of chemical reaction rates, the networks typically have smaller Λ and smaller probability of ground state compared with the biological one. This means that large perturbative states are less stable than the biological one. The biological functioning network is quite different from the random ones in terms of the underlying energy landscape and stability. This shows that as the variance of the chemical rate increases, the likelihood of these rare events that cause disruption of the system becomes greater.
Fig. 3C shows the standard deviation in the robustness ratio Λ versus the perturbation level lp. We observe that the standard deviation of Λ increases with the amount of perturbation. This indicates that not only does the higher perturbation make degradation of the system more likely, but that these events become possibly more severe. This greater severity causes lower Λ to occur and thus increases the variance in the robustness ratio.
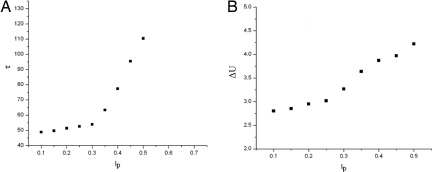
We can also examine a measure of the characteristic kinetic time τ it takes to reach steady state. We will do this by considering the same initial conditions while perturbing the rates at different variance levels of the system. From Fig. 4A, the average τ does increase with an increase in the perturbation level. This is also matched by an increase in the energy spread of the system, 〈ΔU〉 (Fig. 4B). This increase in 〈ΔU〉 indicates that there is more local trapping in the system, which indicates a longer time being “stuck” in the local roughness of the system and thus the increase in τ.
Fig. 4.
Characteristic time and landscape roughness versus perturbation level of chemical rate constant. (A) Variation of characteristic time to reach steady state with perturbation level. (B) ΔU versus perturbation level. This indicates an increase in local trapping of the system.
The MAPK system is open and reaches a nonequilibrium state. The steady-state solution of the system is not an equilibrium state. This come about because the flux (Fijsteady-state = −TijPisteady-state + TjiPjsteady-state), where Tij is the transition probability from state i to state j, of the system at steady state is not zero. In an equilibrium state, the flux would reach zero. Therefore, to describe a nonequilibrium network system, we need the steady-state probability or the underlying landscape as well as the flux. Because of this flux, the nonequilibrium steady state will dissipate energy, resulting in heat loss. This dissipation will be equal to the entropy production rate (23, 31, 32) in steady state. The entropy production rate for the whole network (Ṡ) includes the contribution from the system (Ṡsys) (equal to zero in steady state) and the dissipation from environments (Ṡdis):
 |
Because this entropy production is a feature of the global properties of the network, we can use it to analyze global features of the network, such as stability and robustness.
In Fig. 5A, we plotted the entropy production (per unit time) or the dissipation cost of the network in steady state, Ṡ, versus Λ for certain variance of the chemical reaction rates. We can see the entropy production rate decreases as Λ increases. Thus, we can suggest that the most robust systems produce the least amount of entropy. The fact that robustness is linked with the entropy production rate may reflect the fact that fewer fluctuations and perturbations in rates lead to a more robust and stable network—and more energy saving—and therefore fewer costs in the mean time. This might provide us with a design principle of optimizing the connections of the network with minimum dissipation cost for the network.
Fig. 5.
Normalized entropy production versus robustness ratio (A) or perturbation level (B). Entropy production was normalized so that the unperturbed system has an entropy production value of 1.
In Fig. 5B, we can see that the entropy production rate increases as the variances of the inherent chemical reaction rates increase. This implies that the lesser the variation in the underlying chemical reactions rates, the more robust the network and the less entropy production or heat loss for the network. This can be very important for the network design. This implies that nature might evolve such that the network is robust against internal (intrinsic) and environmental perturbations, and performs specific biological functions with minimum dissipation cost. In our study, this is also the equivalent of optimizing the robustness or stability of the network.
For most of the perturbations, we observed the strong funnel unchanging and thus a monostable solution. However, under some of the perturbations there did arise bistable or oscillatory solutions. For the bistable solutions, there was generally a strong minima and a weak minima, with the weak minima being at a higher energy and the weak minima occupying a much smaller region in phase space. (See Fig. 6 for a two-dimensional projection of a bistable state.) We speculate that this bistability is formed by the different rates being such values as to form a feed-forward-like connection between the middle of the cascade and the end. Under the set of perturbations that we ran, we found that monostability occurred 84% of the time, whereas bistability and oscillatory-like solutions occurred 12% and 4% of the time. Most of the perturbations that cause the change to the global features are related to sections of the network that connect different subsections of the network. Notice also that that these important nodes, which connect different subsections, are also the hubs of the network. We must be careful in calling some of these solutions oscillatory; because we are working in a high-dimensional space, it is possible that these oscillatory-looking solutions are actually strange attractors. This is why we classify the solutions as oscillatory-like. Thus, under most perturbations, the system is in a strong robust format. The possible bistability is important toward making cell fate decisions (34). This bistability is especially important because it occurs without imposing an artificial feedback loop on the system.
Fig. 6.
Two-dimensional projection of the bistable state. This two-dimensional projection is onto the plane formed by the two minima. The axes are normalized so that each minima corresponds to a distance of 1 from the origin.
Conclusions
We used the experimentally inferred rate parameters to prove that the network is funneled in configurational space of protein concentrations toward the ground nonequilibrium steady-state fixed point under the intrinsic statistical fluctuations. As already mentioned, the parameter space of the cellular network is huge, in fact exponential (MN, where M is the number of specific type of proteins and N is the number of different protein types). It is impossible to search all of the parameter space to explore the global stability or robustness with dynamical equations. Our study provides a self-consistent mean field method to reduce the dimensionality to polynomial (M × N) so that large networks can be studied.
We show that natural evolution might only select certain parameter space with the funneled underlying energy landscape. The other part of the parameter space that generates the rough potential landscape cannot guarantee the global robustness and therefore is not able to appropriately perform the specific biological function required for efficient transformation of the information signals. They are more likely to phase out from evolution. The funneled landscape, therefore, may be a realization of the Darwinian principle of natural selection at the cellular network level for efficient transformation of the information (signal transduction). As we see, the funneled landscape provides an optimal criterion to select the suitable parameter subspace of cellular networks, guarantee the robustness, cost the least dissipations, and perform specific biological functions. This will lead to an algorithm for the network connections that is potentially useful for network design.
Under perturbations, the system can be made into a bistable system. It is also important to note that this is done without directly introducing a feed-back or feed-forward interaction into the network. This bistability will be important in the determination of the cell fate decisions important for this network.
It is worth pointing out that the approach described here is general and can be applied to many cellular networks such as signaling transduction networks (2), metabolic networks (35), cell cycle networks (11), and gene regulatory networks (12, 15, 21, 22).
Methods
Most theoretical models of chemical networks involve the use of chemical rate equations. As mentioned before, there are problems with this approach.
We will first start with a description of the MAPK cascade (see Fig. 1). In the first step, the activation of MKKK is catalyzed by an enzyme E1. In the second step, the reverse is catalyzed by an enzyme E2. The third and fourth steps represent the two-collision, nonprocessive mechanism for the double phosphorylation of MKK; these steps are catalyzed by MKKK-P, whereas their reverses are catalyzed by a phosphatase KKPase. Likewise, the seventh and eighth steps represent the double phosphorylation of MAPK catalyzed by the active MKK-PP, and the reverse steps are catalyzed by a phosphatase KPase.
Based on the Michaelis–Menten enzyme kinetic equation, one can derive a set of differential equations that describe the average rate process for concentrations in the cascade (2, 3). The associated chemical reaction rate coefficients are measured or inferred from the experiments in Xenopus oocyte (2, 3).
The probabilistic evolution of this network due to the intrinsic fluctuations from a finite number of molecules follows the master equation in high dimensional protein concentration space. Thus, the basic equations of model become dP(xi)/dt = Σμaμ(xi − Δxi,μ)P(xi − Δxi,μ) − aμ(xi)P(xi), where aμ is the reaction rate for reaction μ, and Δxi,μ is the change of xi during the reaction. There are some basic assumptions with this model, primarily that the system is made of Markov processes.
This is still very difficult if there are many different types of particles, since the number of equations of this form will grow as n1 × n2 × n3 × … We will be making a self-consistent mean field approximation (12, 21, 22), that is P(xi, xj, …) = ΠiPi(xi). This reduces the number of equations to n1 + n2 + …
Because the individual probability distributions are now independent, we can describe each Pi(xi) by a series of moments: 〈xi〉, 〈xi2〉, …, 〈xin〉, … Thus, we can convert from a series of differential equations describing the probability to a series of moment differential equations. For example, we can calculate the first moment to infer the approximate Poisson distribution, we can calculate the second moment to infer the approximate Gaussian distribution for individual probability P(Xi). The moment equations are listed in supporting information (SI) Appendix. We notice that these moment equations are themselves closed (they do not depend on higher order moments) for MAPK, so they can be solved exactly. We did so numerically up to second order moment to infer the underlying distribution. We only present the results in the main text.
Supplementary Material
Acknowledgments.
This work was partially supported by a National Science Foundation Career Award, the American Chemical Society Petroleum Research Fund, and the National Science Foundation of China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708708105/DC1.
References
- 1.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1673. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 2.Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA. 1996;93:10078–10082. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kholodenko BN. Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur J Biochem. 2000;267:1583–1593. doi: 10.1046/j.1432-1327.2000.01197.x. [DOI] [PubMed] [Google Scholar]
- 4.Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–933. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 5.Basu S, Gerchman Y, Collins C, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 6.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 7.Barabasi AL, Bonabeau E. Scale-free networks. Sci Am. 2003;288(5):60–69. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- 8.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 9.Milo R, et al. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 10.Morohashi M, et al. Robustness as a measure of plausibility in models of biochemical networks. J Theor Biol. 2002;216:19–30. doi: 10.1006/jtbi.2002.2537. [DOI] [PubMed] [Google Scholar]
- 11.Tyson JJ, Chen K, Novak B. Integrative analysis of cell cycle control in budding yeast. Nat Rev Mol Cell Biol. 2001;2:908–916. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasai M, Wolynes PG. Stochastic gene expression as a many body problem. Proc Natl Acad Sci USA. 2003;100:2374–2379. doi: 10.1073/pnas.2627987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, Long T, Lu Y, Ouyang Q, Tang C. The yeast cell cycle network is robustly designed. Proc Natl Acad Sci USA. 2004;101:4781–4786. doi: 10.1073/pnas.0305937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ao P. Potential in stochastic differential equations: Novel construction. J Phys A Math Gen. 2004;37:L25–L30. [Google Scholar]
- 15.Zhu XM, Lan L, Hood L, Ao P. Calculating biological behaviors of epigenetic states in the phage lambda life cycle. Funct Integr Genomics. 2004;4:188–195. doi: 10.1007/s10142-003-0095-5. [DOI] [PubMed] [Google Scholar]
- 16.Qian H, Bear DA. Thermodynamics of stoichiometric biochemical networks far from equilibrium. Biophys Chem. 2005;114:213–220. doi: 10.1016/j.bpc.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Qian H, Reluga TC. Nonequilibrium thermodynamics and nonlinear kinetics in a cellular signaling switch. Phys Rev Lett. 2005;94 doi: 10.1103/PhysRevLett.94.028101. 028101. [DOI] [PubMed] [Google Scholar]
- 18.Hornos JEM, et al. Self-regulating gene: An exact solution. Phys Rev E. 2005;72 doi: 10.1103/PhysRevE.72.051907. 051907. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Huang B, Xia XF, Sun ZR. Funneled landscape leads to robustness of cellular network: MAP kinase signal transduction. Biophys J Lett. 2006;91:L54–L56. doi: 10.1529/biophysj.106.086777. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Wang J, Huang B, Xia XF, Sun ZR. Funneled landscape leads to robustness of cell networks: Yeast cell cycle. PLoS Comput Biol. 2006;2:1385–1394. doi: 10.1371/journal.pcbi.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K, Lepzelter D, Wang J. Single molecule dynamics and statistical fluctuations of gene regulatory networks: A repressilator. J Chem Phys. 2007;126 doi: 10.1063/1.2424933. 034702. [DOI] [PubMed] [Google Scholar]
- 22.Kim K, Wang J. Potential landscape and robustness of a gene regulatory network: Toggle switch. PLoS Comput Biol. 2007;3:565–577. doi: 10.1371/journal.pcbi.0030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han B, Wang J. Quantifying robustness and dissipation cost of yeast cell cycle network: The funneled energy landscape perspectives. Biophys J. 2007;92:3755–3763. doi: 10.1529/biophysj.106.094821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lepzelter D, Kim K, Wang J. Dynamics and intrinsic statistical fluctuations of a gene switch. J Phys Chem B. 2007;111:10239. doi: 10.1021/jp071735u. [DOI] [PubMed] [Google Scholar]
- 25.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 27.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilar JMG, Guet CC, Leibler S. Modeling network dynamics: The lac operon, a case study. J Cell Biol. 2003;161:471–476. doi: 10.1083/jcb.200301125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 31.Schnakenberg J. Network theory of microscopic and macroscopic behavior of master equation systems. Rev Mod Phys. 1976;48:571–585. [Google Scholar]
- 32.Qian H. Mesoscopic nonequilibrium thermodynamics of single macromolecules and dynamic entropy-energy compensation. Phys Rev E. 2001;65 doi: 10.1103/PhysRevE.65.016102. 016102. [DOI] [PubMed] [Google Scholar]
- 33.Van Kampen NG. Stochastic Processes in Physics and Chemistry. Amsterdam: Elsevier Science; 1992. [Google Scholar]
- 34.Xiong W, Ferrell J. A positive-feedback-based bistable “memory module” that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 35.Torres NV. Modeling approach to control of carbohydrate metabolism during citric acid accumulation by Aspergillus niger: I. Model definition and stability of the steady state. Biotech Bioeng. 1994;44:104–110. doi: 10.1002/bit.260440115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.