Abstract
To evaluate components of fruit metabolic composition, we have previously metabolically phenotyped tomato (Solanum lycopersicum) introgression lines containing segmental substitutions of wild species chromosome in the genetic background of a cultivated variety. Here, we studied the hereditability of the fruit metabolome by analyzing an additional year's harvest and evaluating the metabolite profiles of lines heterozygous for the introgression (ILHs), allowing the evaluation of putative quantitative trait locus (QTL) mode of inheritance. These studies revealed that most of the metabolic QTL (174 of 332) were dominantly inherited, with relatively high proportions of additively (61 of 332) or recessively (80 of 332) inherited QTL and a negligible number displaying the characteristics of overdominant inheritance. Comparison of the mode of inheritance of QTL revealed that several metabolite pairs displayed a similar mode of inheritance of QTL at the same chromosomal loci. Evaluation of the association between morphological and metabolic traits in the ILHs revealed that this correlation was far less prominent, due to a reduced variance in the harvest index within this population. These data are discussed in the context of genomics-assisted breeding for crop improvement, with particular focus on the exploitation of wide biodiversity.
INTRODUCTION
During the last decade, an impressive number of advances in genetics and genomics have greatly enhanced our understanding of the structural and functional aspects of plant genomes. These advances have also given us ever more powerful tools to aid in the identification of the genetic bases underlying phenotypes identified in forward genetic screens (McCallum et al., 2000; Jansen and Nap, 2001; Wesley et al., 2001; Alonso et al., 2003; Borevitz et al., 2003; Varshney et al., 2005). The adoption of quantitative trait locus (QTL) analysis of natural variation in segregating populations has become an increasingly popular approach (Frary et al., 2000; Paran and Zamir, 2003; Borevitz and Chory, 2004; Koornneef et al., 2004; Ashikari et al., 2005; Salvi and Tuberosa, 2005; Kusano et al., 2007).
Given the availability of a full genome sequence and a wide range of genetic and analytic tools, Arabidopsis thaliana is firmly established as a model system for quantitative genetics and development (Meyerowitz, 2002; Somerville and Koornneef, 2002). It has furthermore proved a very successful choice for unraveling the genetic factors underlying a range of important biological processes, including, but not limited to, seed dormancy and germination, flowering time variation, responses to light quality variation and novel atmospheres, and biotic and abiotic stress responses (for review, see Tonsor et al., 2005), as well as in the evolution of gene function (Mitchell-Olds and Schmitt, 2006). However, there are a large number of biological and societal questions that cannot be directly addressed in Arabidopsis, one of which is crop compositional quality, in particular fruit quality. Here, we examine the hereditability of metabolic traits that play an important role in tomato (Solanum lycopersicum) fruit quality by examining the levels of >70 primary metabolites in two populations of tomato resulting from an interspecific cross between S. lycopersicum and Solanum pennellii.
The improvement of crop species has been a fundamental human pursuit since cultivation began. As a result of genetic bottlenecks imposed during early domestication and modern breeding activities, cultivated varieties contain only a fraction of the variation present in the gene pool (McCouch, 2004; Doebley, 2006; Fernie et al., 2006). Because wild ancestors of most plants can still be found in their natural habitats or in germplasm centers that have been established to collect and conserve these resources (Tanksley and McCouch, 1997), the utility of these wild ancestors in future breeding strategies will be paramount. Current crop improvement strategies are focused not only on the traditional areas of yield enhancement and disease resistance but, driven by recent medical research, also on crop compositional quality for human health (Fernie et al., 2006; Harrigan et al., 2007a).
Genetic determinants of nutritional quality have long been studied. However, it is only recently that these studies have largely focused on single, or at most, a handful, of metabolites, such as carotenoid content in tomato (Liu et al., 2003a), protein content in maize (Zea mays) (Moose et al., 2004), starch content in potato (Solanum tuberosum) and rice (Oryza sativa) (Fernie and Willmitzer, 2004), and tocopherol levels in Arabidopsis (Gilliland et al., 2006). Over the last few years, however, pathway-based approaches have began to be adopted. Such studies have included detailed dissection of the pathways of glucosinolate biosynthesis (Heidel et al., 2006), seed oil synthesis (Hobbs et al., 2004), and seed-soluble oligosaccharide metabolism (Bentsink et al., 2000) in Arabidopsis as well as flavonoid biosynthesis in Populus (Morreel et al., 2006). Furthermore, within the last year, several studies have been performed at the metabolomic level in Arabidopsis, tomato, and wheat (Triticum aestivum) (Keurentjes et al., 2006; Schauer et al., 2006; Harrigan et al., 2007b; Meyer et al., 2007).
The above-mentioned studies on Arabidopsis were based on two independent recombinant inbred line populations and demonstrated wide natural variation in both primary (Meyer et al., 2007) and secondary (Keurentjes et al., 2006) metabolism. The study focused on primary metabolites interestingly revealed a metabolic signature related to biomass accumulation (Meyer et al., 2007), whereas the study focused on secondary metabolites suggested that this approach has far greater potential for dissecting the genetic control of biochemical pathways (and even the structure of the pathways themselves) than has been utilized to date (Keurentjes et al., 2006). These studies, however, being focused on Arabidopsis, made no attempt to account for the effects of environment on the metabolite content. Indeed, very few studies concerned with broad metabolite profiling of natural variance have considered this factor to date. One exception is the recent study of Harrigan et al. (2007b), who evaluated the levels of a wide range of compositional traits, including protein and oil content as well as fatty acid, amino acid, and organic acid content, in two independent maize hybrids grown at three separate locations.
As part of an ongoing project aimed at understanding the genetic basis of compositional quality in the tomato fruit, we previously demonstrated the presence of 889 QTL covering 74 metabolites in replicate harvests of interspecific (S. pennellii × S. lycopersicum) introgression lines (ILs). Subsequent studies have reported yet further QTL, both for the same metabolite (Stevens et al., 2007) and for additional metabolites (Rousseaux et al., 2005). However, it is important to note that despite finding many QTL for enhanced metabolite content in the ILs, we observed that the vast majority of cases in which metabolite content was increased were associated with a yield penalty (Schauer et al., 2006).
In this study, fruit metabolite levels were evaluated in an additional year's harvest, and the analysis was extended to lines heterozygous for the introgression of chromosomal segments from the S. pennellii genome. In doing so, it was possible to evaluate both the stability and the hereditability of the QTL that have been identified previously. Furthermore, we were able to determine their mode of inheritance, a highly important characteristic to study but one that has been overlooked in all but a handful of metabolic studies (Dhaubhadel et al., 2003; O'Reilly-Wapstra et al., 2005). We also evaluated the consequences of mode of inheritance with respect to the breeding of specific traits. Given that tomato has been demonstrated to be an ideal system in which to explore the genetic basis of heterosis (Gur and Zamir, 2004), the data set was evaluated to determine whether there was any indication of heterosis at the metabolite level. While there was little indication of heterosis at the metabolite level, these studies revealed that the strong negative association between yield and metabolite content we had characterized previously in lines homozygous for the S. pennellii introgressions (Schauer et al., 2006) was not apparent in lines heterozygous for the introgressions. The combined results are discussed within the context of the genetic regulation of primary metabolism and their impact on discussions concerning genomics-assisted crop breeding.
RESULTS
Assessment of the Hereditability of Metabolite Traits by Analysis of the Metabolite Profiles Obtained in Different Harvests of the Interspecific ILs of Tomato
We previously reported 889 single-trait QTL for metabolite accumulation following a gas chromatography–mass spectrometry (GC-MS)–based survey of a tomato IL population in which marker-defined regions of the wild species S. pennellii were replaced with homologous intervals of the cultivated variety S. lycopersicum M82 (Eshed and Zamir, 1995). This study was based on the evaluation of fruit pericarp material harvested from two independent harvests (2001 and 2003).
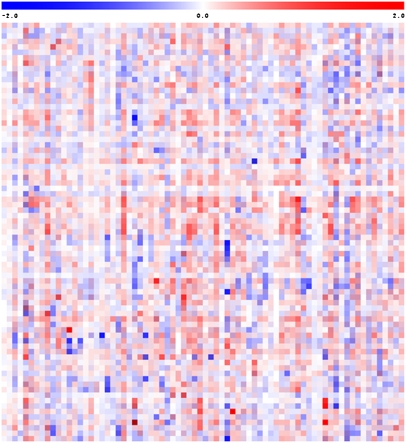
Here, we report data resulting from a third harvest (2004) (Figure 1; see Supplemental Figure 1 online for a fully annotated version). Figure 1 provides an overlay heat map in which the data from all 3 years are superimposed on one another in an additive way such that consistently large increases create a deep red color, consistently large decreases create a deep blue color, a large increase in 1 year combined with large decreases in the other 2 years create a deep bluish purple, and a large decrease in 1 year combined with large increase in the other years create a deep reddish purple. In the case of combinations of smaller changes, these provide a paler coloration or have less influence on the final coloration of the square.
Figure 1.
Overlay Heat Map of the Metabolite Profiles of Three Independent Studies of the Pericarp Metabolite Content of the ILs Compared with the Parental Control (M82).
Data come from a novel field trial performed in 2004 and are presented alongside those of 2003 and 2001 (published in Schauer et al., 2006). Large sections of the map are white or pale in color, reflecting that many of the chromosomal segment substitutions do not have an effect on the amount of every metabolite. Regions of red or blue indicate that the metabolite content is increased or decreased, respectively, after introgression of S. pennellii segments. Very dark coloring indicates that a large change in metabolite content was conserved across all three harvests, whereas purple indicates an inconsistent change in that IL relative to M82. For each harvest, GC-MS was used to quantify 74 metabolites, including amino acids, organic acids, fatty acids, sugars, sugar alcohols, and vitamins. Due to space constraints, this heat map is not annotated; however, fully annotated heat maps for the individual data sets are provided in Supplemental Figures 1 to 4 online.
As can be seen in the plot, the results of the new trial were in congruence with those we reported previously. However, as would be expected, there was also considerable variance across the harvests (a point-by-point comparison of data is best performed by interrogation of the individual heat maps provided as Supplemental Figures 2 to 4 online). When the combined data set was compared, we noted, as we had done previously, a bias toward increased metabolite content in the ILs, which is best explained by the fact that the metabolite content of S. pennellii pericarp is generally greater than that of S. lycopersicum (Schauer et al., 2005b). It is difficult to display such a large data set in a truly quantitative manner. However, across the three trials, the relative difference in the content of any given metabolite ranged between a 0.02-fold and a 115-fold increase compared with the parental cultivar M82.
QTL were determined using analysis of variance (ANOVA) tests, at a significance level of 0.05, to compare statistically every IL with the common control (M82). Using this criterion, we identified only 43 single-trait QTL that were conserved across the three trials (a detailed comparison of the QTL common and unique to the various trials is provided in Supplemental Figure 5 online). This significance level was chosen to maintain consistency with our previous study; however, evaluation at other thresholds revealed the same relative drop in the number of common QTL on the addition of the third year's harvest. The QTL that are common to all three trials are also presented in Supplemental Table 1 online. They covered metabolites from all compound classes tested, and the number of metabolites per class does not appear to be enriched in any way. Analysis of the stable metabolite QTL from the perspective of their genome location, however, revealed that while they were generally well spread across the genome (with all chromosomes with the exception of 6, 10, 11, and 12 harboring QTL), there were a couple of hotspots, such as on chromosomes 4 and 7. Particularly prominent among these hotspots were the loci IL-4-4 and IL-7-2, which harbored six and five QTL, respectively, that were stable across all three harvests.
While the above data were important in confirming the validity of our previous findings, we were also keen to fully exploit the combined data acquired. For this reason, we next assessed the hereditability of the various metabolite traits by statistical analysis of the level of correlation in the combined data sets. These analyses allowed us to calculate the broad sense hereditability (H2) using an approach identical to that recently described by Semel et al. (2006). This analysis revealed that a minority of the metabolites showed strong hereditability (defined by an average H2 value of >0.4 across the field trials), with only 11 of the 75 metabolites showing this, a similar number (13) displaying low hereditability (defined by an average H2 value of <0.2 across the field trials), and the vast majority displaying intermediate hereditability (H2 values of 0.2 to 0.4) (Table 1).
Table 1.
Trait Hereditability in the Introgression Population
| Trait | 2001 H2 | 2003 H2 | 2004 H2 | Mean_H2 | 2001/2003 r | 2001/2004 r | 2003/2004 r | Mean r |
|---|---|---|---|---|---|---|---|---|
| Melezitose | 52 | 32 | 42 | 0.69 | 0.69 | |||
| Glycine | 46 | 48 | 22 | 39 | 0.60 | 0.60 | 0.51 | 0.57 |
| 5-Oxoproline | 41 | 28 | 35 | 0.56 | 0.56 | |||
| Phosphate | 33 | 35 | 29 | 32 | 0.56 | 0.67 | 0.41 | 0.55 |
| Putrescine | 31 | 41 | 21 | 31 | 0.68 | 0.42 | 0.46 | 0.52 |
| Glycerol-3-P | 41 | 49 | 21 | 37 | 0.64 | 0.44 | 0.45 | 0.51 |
| γ-Aminobutyric acid | 47 | 37 | 42 | 0.49 | 0.49 | |||
| Galacturonate | 26 | 32 | 35 | 31 | 0.51 | 0.51 | 0.45 | 0.49 |
| Citrate | 34 | 22 | 40 | 32 | 0.43 | 0.53 | 0.43 | 0.46 |
| Thr | 18 | 31 | 31 | 27 | 0.46 | 0.41 | 0.47 | 0.45 |
| S-Me-Cys | 32 | 26 | 29 | 0.42 | 0.42 | |||
| Gln | 50 | 7 | 30 | 29 | 0.51 | 0.41 | 0.32 | 0.41 |
| Ser | 45 | 40 | 45 | 43 | 0.34 | 0.55 | 0.32 | 0.40 |
| Inositol-1-P | 35 | 31 | 33 | 0.40 | 0.40 | |||
| Fructose | 44 | 40 | 48 | 44 | 0.54 | 0.47 | 0.18 | 0.40 |
| β-Ala | 42 | 41 | 20 | 34 | 0.39 | 0.44 | 0.35 | 0.39 |
| Val | 19 | 28 | 17 | 21 | 0.41 | 0.33 | 0.43 | 0.39 |
| Ile | 18 | 29 | 28 | 25 | 0.43 | 0.35 | 0.38 | 0.39 |
| Leu | 23 | 32 | 23 | 26 | 0.38 | 0.31 | 0.45 | 0.38 |
| Glucose | 55 | 43 | 40 | 46 | 0.42 | 0.44 | 0.23 | 0.36 |
| Quinate | 43 | 45 | 38 | 42 | 0.42 | 0.45 | 0.19 | 0.35 |
| Asp | 26 | 35 | 21 | 27 | 0.34 | 0.14 | 0.52 | 0.33 |
| Citramalate | 29 | 25 | 27 | 27 | 0.56 | 0.25 | 0.17 | 0.33 |
| Homoserine | 37 | 22 | 23 | 27 | 0.43 | 0.29 | 0.26 | 0.33 |
| Sucrose | 26 | 21 | 28 | 25 | 0.20 | 0.31 | 0.47 | 0.33 |
| Pro | 3 | 34 | 19 | 19 | 0.38 | 0.28 | 0.32 | 0.33 |
| Glycerate | 19 | 33 | 22 | 25 | 0.29 | 0.31 | 0.37 | 0.32 |
| Met | 22 | 34 | 22 | 26 | 0.44 | 0.30 | 0.20 | 0.31 |
| Malate | 32 | 11 | 22 | 22 | 0.11 | 0.34 | 0.45 | 0.30 |
| Dehydroascorbate | 39 | 52 | 17 | 36 | 0.27 | 0.44 | 0.16 | 0.29 |
| Glutamate | 41 | 22 | 13 | 25 | 0.38 | 0.25 | 0.23 | 0.29 |
| Rhamnose | 13 | 18 | 16 | 0.28 | 0.28 | |||
| Succinate | 36 | 36 | 21 | 31 | 0.40 | 0.14 | 0.28 | 0.27 |
| Arabinose | 33 | 26 | 30 | 0.27 | 0.27 | |||
| Cys | 26 | 18 | 43 | 29 | 0.27 | 0.22 | 0.30 | 0.26 |
| Gluconate | 58 | 29 | 18 | 35 | 0.35 | 0.11 | 0.29 | 0.25 |
| Mannitol | 46 | 12 | 29 | 0.25 | 0.25 | |||
| t4-HO-Pro | 27 | 25 | 26 | 0.25 | 0.25 | |||
| Inositol | 43 | 37 | 40 | 40 | 0.17 | 0.31 | 0.24 | 0.24 |
| Tyr | 23 | 17 | 22 | 21 | 0.22 | 0.14 | 0.36 | 0.24 |
| Ribose | 6 | 22 | 21 | 16 | 0.25 | 0.01 | 0.41 | 0.22 |
| Isocitrate | 43 | 21 | 38 | 34 | 0.29 | 0.07 | 0.30 | 0.22 |
| Phe | 21 | 22 | 25 | 23 | 0.14 | 0.32 | 0.19 | 0.22 |
| FA18:2 | 19 | 18 | 19 | 0.21 | 0.21 | |||
| 3-Phosphoglyceric acid | 61 | 54 | 28 | 48 | 0.42 | 0.03 | 0.18 | 0.21 |
| Fructose-6-P | 25 | 34 | 19 | 26 | 0.26 | 0.06 | 0.27 | 0.20 |
| FA16:0 | 65 | 31 | 48 | 0.19 | 0.19 | |||
| Ala | 20 | 30 | 19 | 23 | 0.42 | 0.06 | 0.08 | 0.19 |
| Mannose | 33 | 29 | 44 | 35 | 0.30 | 0.12 | 0.11 | 0.18 |
| Glucose-6-P | 21 | 28 | 21 | 23 | 0.21 | 0.01 | 0.27 | 0.16 |
| Uracil | 6 | 8 | 26 | 13 | 0.34 | −0.03 | 0.16 | 0.16 |
| Erythritol | 17 | 22 | 17 | 19 | 0.26 | 0.22 | −0.03 | 0.15 |
| Galactose | 25 | 25 | 34 | 28 | 0.46 | 0.03 | −0.10 | 0.13 |
| Maleate | 23 | 9 | 16 | 0.13 | 0.13 | |||
| Saccharate | 28 | 42 | 35 | 0.12 | 0.12 | |||
| Lys | 21 | 25 | 15 | 21 | 0.36 | 0.02 | −0.05 | 0.11 |
| Asn | 21 | 18 | 25 | 21 | 0.17 | 0.03 | 0.11 | 0.10 |
| Trp | 14 | 11 | 19 | 15 | 0.15 | 0.09 | 0.05 | 0.10 |
| Maltose | 17 | 20 | 16 | 18 | −0.07 | 0.19 | 0.15 | 0.09 |
| Aconitate | 6 | 19 | 12 | 0.09 | 0.09 | |||
| Arg | 33 | 32 | 36 | 34 | 0.41 | −0.05 | −0.14 | 0.07 |
| α-Tocopherol | 21 | 15 | 17 | 18 | 0.05 | 0.10 | 0.04 | 0.06 |
| 2-Oxoglutarate | 22 | 47 | 34 | 0.06 | 0.06 | |||
| Fumarate | 23 | 16 | 21 | 20 | 0.06 | −0.12 | 0.21 | 0.05 |
| FA18:0 | 59 | 33 | 46 | 0.02 | 0.02 | |||
| Sorbitol | 34 | 50 | 42 | 0.01 | 0.01 | |||
| Benzoate | 20 | 49 | 26 | 32 | 0.04 | 0.08 | −0.16 | −0.01 |
| Fucose | 6 | 22 | 28 | 19 | 0.17 | 0.10 | −0.33 | −0.02 |
| Trehalose | 60 | 40 | 11 | 37 | −0.10 | −0.02 | 0.01 | −0.04 |
| l-Ascorbate | 42 | 18 | 36 | 32 | −0.10 | 0.08 | −0.10 | −0.04 |
| Threonate | 23 | 35 | 21 | 26 | 0.18 | −0.08 | −0.26 | −0.05 |
| Maltitol | 43 | 39 | 15 | 32 | 0.02 | −0.12 | −0.16 | −0.09 |
| Shikimate | 12 | 12 | 12 | 12 | −0.10 | −0.06 | −0.13 | −0.10 |
| Isomaltose | 25 | 17 | 21 | −0.10 | −0.10 | |||
| Glycerol | 38 | 32 | 49 | 40 | −0.05 | −0.21 | −0.24 | −0.17 |
The H2 for each metabolite trait is presented for each of the independent years as well as the correlation (Pearson's test) of metabolite levels in the lines between years (r). Mean H2 shows the average hereditability, and mean r shows the average of the correlations.
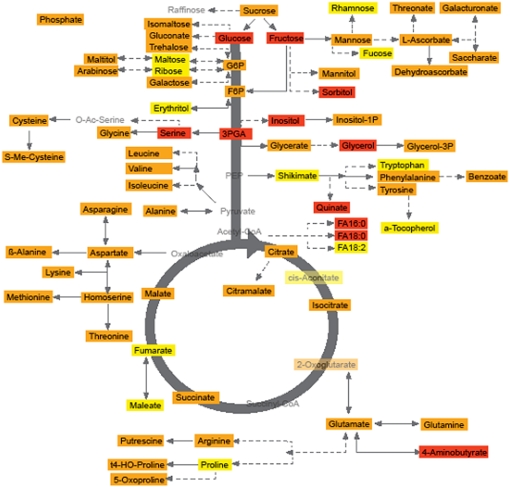
When these results are assessed from the perspective of the metabolic network (Figure 2), several trends emerge. Perhaps most prominent among these is the strong hereditability in the sugars glucose and fructose, 3-phosphoglyceric acid and its derivatives Ser, inositol, and glycerol, and the fatty acids 16:0 and 18:0 (palmitate and stearate, respectively). However, other linked metabolite pairs also display relatively high hereditability, such as the levels of the branched-chain and aspartate-derived amino acids and ascorbate-derived compounds, suggesting high robustness of the reactions catalyzed by the enzymes that interlink these metabolites. Given its importance in the human diet (Gilliland et al., 2006; Dormann, 2007), the evaluation of the hereditability of tocophenol is also of great interest. However, given that tocophenol displays low hereditability, its responsiveness to the environment suggest that it will be difficult to breed tomatoes with elevated content of this metabolite. Other metabolites with apparently low hereditability include the well-characterized stress metabolite Pro, shikimate, aconitate, fumarate, and malate and, perhaps surprisingly given the strong hereditability of palmitate and stearate, octadecadienoate.
Figure 2.
Metabolites That Display High, Moderate, and Low Hereditability as Assessed from the 3 Years of Growth Trials.
Data given in Table 1 are displayed in a pathway-based manner. Metabolites marked in red were determined to be highly hereditable, those in yellow to display low hereditability, and those in orange to be intermediate. Traits colored pale gray were not measured in this study.
In an alternative method, we evaluated the correlations of trends in metabolite levels across the population. The reason for adopting this approach is that, since H2 measures the ratio of the variation between and within the genotypes (so that high heritability means low variation within replications and high variation among genotype means), it can potentially lead to traits being artificially designated as having high heritability. In evaluating the correlation between experiments, this approach compares the averages between experiments and thus is not as likely to be biased by technical factors of data acquisition. As expected, given the previously reported technical reproducibility of the profiling method we use here (Roessner et al., 2000), analysis by this method gave highly similar results (Table 1). The correlation values for glycerol, sorbitol, and (intriguingly, in light of the above comments) palmitate and stearate were very weak, suggesting that caution should be taken with respect to their annotations as highly hereditable. By contrast, correlation analysis of the metabolites that were identified by H2 to be poorly hereditable served to confirm these diagnoses.
Analysis of Metabolite Contents in a Population Heterozygous for the S. pennellii Introgression
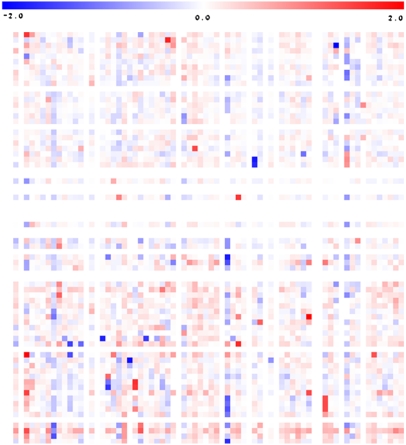
Given that the previous experiments highlighted the important genetic influence underlying the majority of the metabolite QTL, we next analyzed the metabolite content of the fruit pericarp in the standard 76 homozygous IL lines and in hybrids between ILs and M82 (ILHs; described in Semel et al., 2006) using material from the same harvest. A heat map of the metabolite profiling results of the ILHs is presented in Figure 3 (with the full data sets available in Supplemental Figures 1 to 4 online). At first glance at the entire data set, it is clear that some of the changes in metabolites are conserved in the ILs and ILHs, while others are not. Moreover, there are clear quantitative differences in those traits that are conserved. Some metabolites are present at approximately the same level in the ILH as in its parent IL, others are present at lower levels, and some are present at even higher levels.
Figure 3.
Heat Map of the Metabolite Profiles of M82 Lines Heterozygous (ILH) for Chromosomal Segmental Substitution from S. pennellii.
Results presented are pericarp metabolite content data obtained from the ILHs of the 2004 harvest. Regions of dark red or dark blue indicate that the metabolite content is increased or decreased, respectively, after introgression of S. pennellii segments. GC-MS was used to quantify 74 metabolites, including amino acids, organic acids, fatty acids, sugars, sugar alcohols, and vitamins. Due to space constraints, this heat map is not annotated; however, a fully annotated heat map including the metabolite profiles of the ILHs from the 2004 harvest is provided in Supplemental Figure 6 online.
In order to assess whether these changes are associated with a particular mode of inheritance, we subjected the combined data set to a QTL analysis in which each IL and ILH was compared with the common M82 control. If one of the lines had a significant effect (at the 1% level), it was considered as harboring a QTL. We chose a higher threshold here than in the previous analyses for two reasons. First, given that we only had data from a single harvest, it was appropriate to use a more stringent threshold, and second, for the sake of comparison with the study of Semel et al. (2006). Utilizing this approach, we resolved a total of 332 putative QTL under these conditions of statistical stringency. It is important to note that this is different from the number of QTL presented above (and in Supplemental Figure 5 online), since the current analysis is based on the 2004 data alone and combines data obtained from both the IL and ILH populations.
Assessment of the Mode of Inheritance of the Metabolic QTL
As well as allowing point-by-point analysis, the inclusion of ILHs in the analysis enables us to classify each putative wild species QTL into the following mode-of-inheritance categories: recessive, additive, dominant, or overdominant (for detailed explanation of the classification, see Semel et al. [2006]). In brief, this classification reflects a mode of inheritance in which the S. pennellii allele is compared with the M82 allele. For example, a QTL classified as dominant means that both the IL (homozygous for the S. pennellii allele) and the ILH (heterozygous) are very similar to each other and significantly different from M82. A recessive QTL is defined as one in which only the IL is significantly different from the wild type, whereas the ILH is similar to the wild type. Additivity reflects a situation in which the ILH is between its parents, which are significantly different from each other, and overdominance is inferred in situations in which the ILH is significantly higher or lower than both its parents.
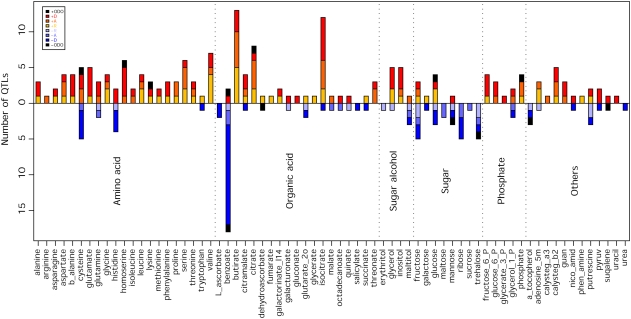
Evaluation of the results of this classification, presented in Figure 4, reveals that the vast majority of the putative wild species QTL have an increasing effect on metabolite content. However, there are a number of clear exceptions to this statement. The populations harbor slightly more decreasing than increasing QTL for His and many more decreasing than increasing QTL for benzoate, sugars, and α-tocopherol. When assessed in this way, the majority of QTL can be seen to exhibit either dominant or additive modes of inheritance. Only a minority of metabolites exhibit a considerable proportion of recessive QTL (notably, Gly, Leu, Val, γ-aminobutyric acid, glycerol, glucose, and adenosine-5-phosphate fit into this category), and overdominance is even rarer when assessed on the basis of the total number of QTL. This approach determined a total of 14 putative overdominant QTL in the following metabolites: Cys, homoserine, benzoate, citrate, dehydroascorbate, glucose, mannose, trehalose, phosphate, and squalene. It is important to note, however, that these should be interpreted with caution, since, as indicated by the application of a Bonferroni correction, the total number of putative overdominant QTL does not greatly exceed that which would be expected due to chance alone.
Figure 4.
Distribution of the QTL Mode of Inheritance for Metabolite Accumulation.
Each vertical bar represents the number of QTL for a specific trait, colored according to mode-of-inheritance categories: A, additive; D, dominant; ODO, overdominant; R, recessive. The bars above the 0 line represent the number of increasing QTL, whereas the negative bars represent the number of decreasing QTL relative to M82.
When the distribution of the mode of inheritance is compared across the different compound classes, some clear differences can be observed. As mentioned above, sugars and organic acids displayed more negative behavior with respect to their parental IL than metabolites of the other compound classes. χ2 tests also revealed significant differences across compound types in the level of both positive and negative dominant modes of inheritance (Table 2). Increasing dominant QTL were a prominent mode in amino acids, sugar alcohols, and phosphorylated intermediates, being less prominent in organic acids and miscellaneous compounds and a minority mode of inheritance in sugars. However, the situation was mirrored for negative dominance, which was considerable in sugars, organic acids, and miscellaneous compounds but minor in amino acids, sugar alcohols, and phosphates.
Table 2.
Qualitative Distribution of Mode of Inheritance Showing the Numbers of QTL That Were Classified in Each Category for Each Chemical Compound Class
| Mode | Amino Acids (22 Traits) | Organic Acids (22 Traits) | Sugars (12 Traits) | Sugar Alcohols (5 Traits) | Phosphates (5 Traits) | Others (12 Traits) | P (χ2) |
|---|---|---|---|---|---|---|---|
| +Overdominant | 3 (3) | 2 (2) | 1 (3) | 0 (0) | 1 (6) | 0 (0) | 0.71 |
| −Overdominant | 0 (0) | 3 (3) | 2 (5) | 0 (0) | 0 (0) | 2 (5) | 0.19 |
| +Dominant | 50 (48) | 29 (28) | 4 (10) | 14 (44) | 9 (50) | 11 (30) | 0.0003 |
| −Dominant | 9 (9) | 25 (25) | 13 (33) | 1 (3) | 1 (6) | 8 (22) | 0.0003 |
| +Additive | 17 (16) | 17 (17) | 1 (3) | 2 (6) | 4 (22) | 5 (14) | 0.15 |
| −Additive | 2 (2) | 4 (4) | 7 (18) | 1 (3) | 1 (6) | 0 (0) | 0.0013 |
| +Recessive | 21 (20) | 18 (18) | 5 (13) | 9 (28) | 2 (11) | 7 (19) | 0.59 |
| −Recessive | 2 (2) | 4 (4) | 6 (15) | 5 (16) | 0 (0) | 4 (11) | 0.0046 |
| Total | 104 (100) | 102 (100) | 39 (100) | 32 (100) | 18 (100) | 37 (100) |
The numbers in parentheses represent the percentage of this category among all QTL in that group. The signs that precede the mode of inheritance indicate whether it is an increasing (+) or decreasing (−) QTL relative to M82. A statistical comparison between the different metabolite groups was conducted in each mode of inheritance using a χ2 test (with 1 df) by classifying the QTL into those that belong to this mode of inheritance and those that do not belong for each group.
Together, these data suggest that the proportion of metabolic traits that were dominant was not greatly influenced by the compound class of the metabolite trait. While both positive additive and positive recessive inheritance were independent of compound class, the negative modes of both types of inheritance displayed clear compound class–dependent differences. In the case of the negative additives, this large variance was due to the high proportion of sugars displaying this mode of inheritance as well as the low proportion of sugar alcohols displaying additive behavior. By contrast, the sugar phosphates displayed a higher proportion of (putative) negative recessive mode-of-inheritance QTL than any other compound class, with the exception of the sugars, while phosphorylated intermediates displayed very little recessive behavior.
Detailed Evaluation of the Mode of Inheritance of the Metabolic QTL
In order to assess whether the distribution of the mode of inheritance was influenced by the quantitative influence of a loci in the determination of a given trait, we next performed the same evaluation but only took into consideration the more major QTL. However, the resultant mode-of-inheritance distribution (which is presented as Supplemental Table 2 online) was generally the same as that for the full list of putative QTL, indicating that this pattern was generally independent of the magnitude of contribution toward a given trait. However, when the QTL that were reported above as stable across all 3 years of field trials were evaluated (see Supplemental Data Set 1 online), the mode of distribution was somewhat different, since, although the majority (∼57%) of the QTL for which we assigned modes of inheritance were classified as dominant, a far greater percentage displayed a recessive mode of inheritance (∼29%).
As a second approach, we compared the IL and ILH metabolite content by evaluating the correlations between the values of a given metabolite in the ILs versus their respective ILH progeny (see Supplemental Data Set 1 online). This revealed that for 50 of 78 traits (64%), this correlation is significant. Interestingly, the list of metabolites that were not greatly influenced by the zygosity of the introgression was overrepresented by phosphorylated intermediates and organic acids, while those that were influenced appeared to be overrepresented by sugars and sugar alcohols.
Given the apparent influence of compound class on the mode of inheritance, we next evaluated whether the putative mode-of-inheritance QTL of the various metabolites were colocalized to those of other metabolites that were chemically similar. We hoped that this would provide information on the genetics of the enzymes catalyzing the reactions that link the metabolic nodes of the network. We carried this out by examining the locations of all 332 QTL (see Supplemental Data Set 2 online). Several interesting observations resulted from this analysis, with 11 of the 74 ILs harboring at least one metabolite pair that display the same mode-of-inheritance QTL. Among the 13 metabolite pairs, all of the major inheritance modes were represented with pairs alternatively exhibiting dominant, additive, and recessive inheritance. Although four of these were glucose–fructose pairs (IL-2-2, IL-2-6-5, IL-9-3-1, and IL-10-1-1), each pair displayed a different inheritance type; in addition, there was a glucose 6-phosphate–fructose 6-phosphate pair (IL-1-2), a Gly–Ser pair (IL-1-2), a Gln–Glu pair (IL-1-4-18), a Leu–Val pair (IL-3-2), two Ile–Leu pairs (IL-8-3 and IL-12-3), an Asn–β-Ala pair (IL-11-9-1), a succinate–fumarate pair (IL12-3), and a homoserine–Lys pair (IL-12-4).
Unfortunately, analysis of the map positions of the genes of primary metabolism that have been reported for tomato (Causse et al., 2004) and the highly syntenic potato (Chen et. al., 2001) did not reveal the presence of candidate genes at these positions. However, given the paucity of information on metabolism-associated genes (with the exception of QTL for sugar metabolism), we cannot conclude much from this comparison. In the case of the sugars, genes for their import into the fruit and their subsequent metabolism have been mapped, and while one hexokinase gene is associated with one of the putative QTL we mentioned above (IL-2-2), the other putative QTL do not colocalize with any of the expected candidates. However, it remains to be seen where the genes that encode the biosynthetic (and degradative) machinery of other pathways of primary metabolism are localized before the generality of this finding can be assessed.
Metabolite–Morphology Associations in the ILs and ILHs
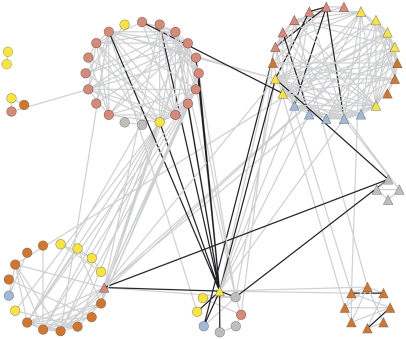
In our previous study (Schauer et al., 2006), we observed that many of the metabolite traits that we identified were associated with morphological traits, and the harvest index (HI) was identified as a major pleiotropic hub. The nature of the ILs makes such multitrait analysis possible and thus allows the identification and potential dissection of functional relationships between traits. In our previous study, we were able to demonstrate a highly robust relationship between HI and metabolite composition. In order to evaluate whether this relationship also holds true in the ILHs, we performed correlation analysis between the metabolite traits reported here and the morphological traits of the ILHs that were reported previously for plant material of the same harvest. In order to obtain the most reliable comparison, we also recalculated values for the ILs from the 2004 harvest (Figures 5 and 6).
Figure 5.
Cartographic Representation of the Combined Metabolic and Morphological Networks of the ILs.
Each morphological trait (node) is represented by a triangle and color as follows: red, fruit-associated; orange, flower-associated; yellow, yield-associated; gray, seed morphology; light blue, seed yield. Each metabolic trait (node) is represented by a circle and color as follows: red, amino acids; orange, sugars and sugar alcohols; yellow, organic acids; gray, phosphorylated intermediates; light blue, miscellaneous metabolites. Interactions are indicated with lines: gray represents positive correlations and black represents negative correlations. A fully annotated version of this figure is available as Supplemental Figure 8 online.
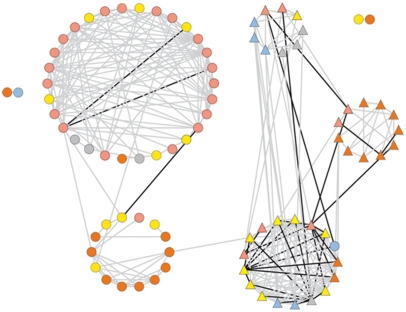
Figure 6.
Cartographic Representation of the Combined Metabolic and Morphological Networks of the ILHs.
Each morphological trait (node) is represented by a triangle and color as follows: red, fruit-associated; orange, flower-associated; yellow, yield-associated; gray, seed morphology; light blue, seed yield. Each metabolic trait (node) is represented by a circle and color as follows: red, amino acids; orange, sugars and sugar alcohols; yellow, organic acids; gray, phosphorylated intermediates; light blue, miscellaneous metabolites. Interactions are indicated with lines: gray represents positive correlations and black represents negative correlations. A fully annotated version of this figure is available as Supplemental Figure 9 online.
The network analysis of the 2004 IL data yielded a remarkably similar cartography to that which we documented previously for the 2001 and 2003 data (Figure 5) (Schauer et al., 2006), suggesting that it is highly stable in this network across harvests. That for the ILHs, however, was markedly different from both of these (Figure 6). However, the most valid comparison is with the IL network of 2004, since the comparison of this network with that resulting from the ILH lines is highly controlled, given that the lines were grown, harvested, and evaluated following the same randomization procedure. This comparison reveals that, in the case of the ILHs, there was a clear reduction in the number of correlations between metabolic and morphological traits. There were only 5 strongly correlating trait pairs between the different phenotyping studies, as opposed to 30 strongly correlating trait pairs in the ILs. When stringent correlations of P < 0.001 were applied, the number of correlations using permissive conditions were 15 and 93 for the ILH and IL network, respectively.
While surprising, the evaluation of the HI distribution in the populations revealed that the ILs displayed a significantly broader variation in this trait than the ILHs (see Supplemental Figure 7 online), suggesting that problems of sterility in the ILs may be the primary course of this effect. Close analyses of these figures and the underlying data (presented in Supplemental Data Set 1 online) revealed that, in addition to the changed pattern of metabolite–morphological correlations in the ILHs, there are additionally a large number of the metabolite–metabolite correlations that are specific either to the IL or the ILH population. This underscores the complexity of the hereditability of the metabolite traits that we determined.
DISCUSSION
In recent years, there has been much renewed interest in the possibility of breeding not only higher yielding but also better quality crops. One potential approach to this end is the combined use of metabolite profiling and introgression breeding. In our previous work (Schauer et al., 2006), we demonstrated that, when used in tandem, these approaches were able to rapidly identify a large number of metabolite accumulation QTL. While there has been much interest in influencing fruit size and shape as well as improving the organoleptic properties of tomato (Frary et al., 2003; van der Knaap et al., 2004; Chaib et al., 2006), nutritional quality has largely been overlooked in tomato breeding programs. However, the compositional quality of crops is receiving increasing interest, particularly given the results of recent studies highlighting the nutritional importance of lycopene, flavonoids, and chlorogenic acid in the human diet (Davuluri et al., 2005; Dixon, 2005; Niggeweg et al., 2006; Rein et al., 2006). Such improvements are particularly important in tomato, since, in this species (with the exception of lycopene-derived volatiles), the flavor components associated with nutrition have been depleted through breeding (Goff and Klee, 2006; Morris and Sands, 2006). One such approach to identify genetic material suitable for reintroducing these traits is the introgression approach, whereby wild allelic variance is introduced back into cultivated species by marker-assisted selection of single chromosome segment substitutions (Zamir, 2001; Giovannoni, 2006).
In our previous study, we evaluated the metabolite profiles of two independent harvests. Here, we report the results of an additional year's profiling that allows us greater confidence in the statistical analysis of hereditability and thus indirectly the influence of environment on the metabolic variance recorded. While the crop was grown on the same experimental farm in three different years, environmental influences clearly still exist. Perhaps unsurprisingly, the majority of metabolites appear to be influenced by a mixture of genetic and environmental factors. More surprising was the relatively low number of metabolites deemed to be environmentally determined, with Pro and shikimate being the only metabolites classified as exhibiting low hereditability that have been consistently documented in the literature as changing under conditions of stress (Hare and Cress, 1997; Yoshiba et al., 1997; Bartels and Sunkar, 2005; Bohnert et al., 2006).
Moreover, our results are somewhat contrary to those recently reported by Harrigan et al. (2007a), who analyzed three different wheat inbred lines grown at three different sites and found very high environmental differences in sugars, organic acids, and amino acids between the sites. However, a direct comparison between that study and our own is not entirely appropriate, since the studies are of very different structure, with our study evaluating >70 genotypes across multiple harvests from the same field as opposed to the more limited genetic variance but greater environmental variance of the wheat study. The increased variability seen by Harrigan et al. (2007a) suggests a strong environmental component due to soil composition for the metabolites analyzed, which may have been minimized in using the same location for each of the tomato trials. By contrast, the use of broader genetic variance in our study would likely have maximized the genetic differences.
Given that so few studies to date have used such broad genetic variance as that offered by the ILs, it is currently difficult to place these findings in a broader context. Regardless, it is clear, from both the conservation of QTL between the harvests (Figure 1) and the evaluation of hereditability effects themselves (Table 1, Figure 2), that the content of a considerable proportion of the metabolites tested is relatively consistent within the genotype across several harvests. This suggests that the proposed use of this approach for improving nutritional quality is valid, since among the metabolites that display strong hereditability are hexose sugars and unsaturated fatty acids, while among those displaying reasonable hereditability are the vitamin ascorbate and the essential amino acids Met, Thr, Val, and Ile. It should be noted that the contents of several other metabolites are more responsive to environmental factors (Figure 2). A notable feature of the 43 QTL that were conserved across all three harvests was that the majority of them were represented by a relatively small number of loci. Evaluation of the yield-associated traits affected in the same lines revealed that the two loci exhibiting the greatest metabolic changes also exhibited large morphological changes, reinforcing the conclusions we made in our previous study concerning the association between yield-associated and chemical composition traits.
It will be highly interesting to determine whether the multiharvest trends observed here also hold true in studies profiling the levels of other metabolites, since several recently established methods are being applied to phenotype broad genetic variance in the Solanaceae (Tikunov et al., 2005; Fraser et al., 2007). However, it is worth pointing out that the levels of volatile compounds, although varying across different harvests, were relatively constant with respect to the levels found in the cultivated control (Tieman et al., 2006). A similar pattern has also been reported for carotenoid levels (Liu et al., 2003b). Moreover, results from a subset of those metabolites analyzed here are broadly consistent with previous measurements made on the ILs (Causse et al., 2002; Rousseaux et al., 2005), allowing us high confidence in our data set. While mapping genes in tomato is still an arduous procedure, it will likely soon be greatly facilitated by the release of data from the nascent international genome sequencing project (Mueller et al., 2005).
While the above studies allow us confidence in our ability to determine the genomic regions underlying the altered chemical composition in the tomato ILs, in order to assess the practical application of these findings we thought it imperative to better understand the mechanisms of their inheritance and their interaction with yield-associated traits. Evaluation of some of the interesting findings of our previous study (Schauer et al., 2006) revealed similarities and differences between the ILs and ILHs. For example, IL4-4 has been demonstrated to be characterized by an upregulation of the pathway linking citrate to γ-aminobutyric acid and Pro (Schauer and Fernie, 2006; Schauer et al., 2006). Given that these metabolites are not highly correlated across the population and that the results of another study revealed upregulation of this pathway at the level of transcription (Baxter et al., 2005), this result suggests that this upregulation is likely mediated by an alteration in the function of a regulatory gene. Interestingly, results of this study suggest that the pathway that is upregulated also includes the polyamines spermidine and putrescine and, more importantly, that the upregulation is conserved in the heterozygous line, albeit to a much lesser extent. The well-characterized Brix9-2-5 locus (Fridman et al., 2002, 2004) harbored the sugar QTL in both heterozygous and homozygous states. However, the minor QTL for malate in IL7-5 was not reproduced in this study. Unfortunately, we were also not able to assess the effect of the chromosomal segment of IL6-3, which harbors self-pruning SP, in the heterozygous form, since this did not bear fruits suitable for analysis in the 2004 harvest. As discussed previously, the S. pennellii ILs represent an excellent resource for the analysis of heterosis, since they are a nearly isogenic population that allows the partitioning of QTL into defined genomic regions, eliminating a major part of the genome-wide epistasis (Semel et al., 2006). As an extension of a previous phenomic study, we identified 332 putative QTL for metabolite accumulation (using a stringent statistical cutoff of P < 0.01). In the previous study, Semel et al. (2006) were able to demonstrate that reproductive and nonreproductive traits were generally characterized by different patterns of inheritance (Semel et al., 2006; Lippman et al., 2007). Perhaps unsurprisingly, the distribution of the putative metabolite QTL we report here is more similar to that of the nonreproductive morphological traits reported previously in that very few overdominant QTL were reported; however, for all other modes of inheritance there was a strong bias toward increasing metabolite content, particularly in the case of the dominant QTL (Figure 4).
Previous QTL mapping in the ILs has suggested that alleles that came from the S. pennellii parent tend to affect the trait in the direction relative to the S. pennellii value. For example, most of the QTL for the trait fruit weight are decreasing in the ILs, since the fruit weight of S. pennelli is very small compared with that of the other parent, M82 (Semel et al., 2006). By contrast, the majority of the QTL for the trait Brix are increasing, since S. pennelli has much higher Brix than M82 (Fridman et al., 2000, 2004). It is likely that this principle is behind a number of the changes noted in the metabolite data; see, for example, the increased contents of the fatty acids stearate, palmitate, and octadecadienoate. However, this is not universally the case, since a large number of amino acids are increased in the ILs despite being predominantly lower in the wild species (Schauer et al., 2005b). The exact reasons for the transgressive behavior in amino acid content cannot be explained in this study. That said, it is an interesting observation that is worthy of further experimentation.
The findings of our study provide no support for the proposed biochemical mechanisms of hybrid vigor (Milborrow, 1998). However, they are in keeping with a recent phenomic study of the exact same lines revealing that reproductive traits, but not nonreproductive morphological traits, displayed a high number of increasing overdominant QTL (Semel et al., 2006). The authors of that study suggest that the alliance of these overdominant QTL with higher reproductive fitness was selected for in evolution and was domesticated by humans to improve crop yields. Our analysis of metabolite QTL suggested that they share a similar mode of inheritance to the other nonreproductive traits and, as such, is consistent with the previous hypothesis. While a somewhat negative finding, the essential lack of heterosis in metabolite content in the ILs remains a valuable one, since it adds to the debate regarding whether there is indeed a metabolic basis underlying heterosis (Causse et al., 1994; Titok et al., 2005; Dong et al., 2006).
A second interesting observation that arose in detailed evaluations of the mode of inheritance was that a reasonable number of pairs of metabolites that are metabolically proximal to one another displayed similar modes of inheritance. This finding suggests that the putative QTL responsible are likely to affect the efficiency of the enzyme-catalyzed reactions that link the metabolites in question. While comparison with the map positions of those few metabolically associated genes that have been mapped to date (Chen et al., 2001; Causse et al., 2004) revealed that the glucose–fructose pairs are unlikely to be linked to alterations in structural genes for sugar metabolism, the paucity of mapping data yet available renders the analysis of the other pairs and even a definitive analysis of the glucose–fructose pairs impossible at present. While this comparison suggests that the inheritance of the glucose–fructose pairs is unlikely to be due to the IL harboring a different allele of a structural gene of sugar metabolism, it does not preclude the possibility that the IL harbors allelic variance in a regulatory gene affecting the underlying metabolic pathways of sugar metabolism.
Indeed, for none of the examples presented above can we yet state a mechanism. The patterns observed could be due to variation in the levels or kinetic characteristics of either enzymes linking the metabolites in question or key enzymes operating upstream or downstream of these metabolites. Furthermore, we currently cannot exclude the possibility that the effects are due to genetic variance in regulatory genes. This reasoning suggests that employing a combination of enzyme profiling as applied for the characterization of natural variance in Arabidopsis (Cross et al., 2006; Sulpice et al., 2007) and broad transcript profiling on the IL population could prove an effective way of elucidating the mechanisms that underlie them. In addition to merely increasing our understanding of the stability and inheritability of QTL for tomato fruit metabolite composition, the findings of this study are highly informative for future breeding strategies.
Intriguingly, the evaluation of the heterozygous ILHs revealed that, in the majority of cases, traits of primary metabolism are either dominantly or additively inherited. This finding is of great importance with respect to attempts to breed crops with improved chemical composition, since many attempts to metabolically engineer plants to produce elevated levels of a given metabolite were able to meet this goal, but only at the cost of compromising yield (Fernie and Willmitzer, 2004; Van Camp, 2005). Evaluation of the trait network of the heterozygous ILHs revealed that morphological parameters were less tightly associated to metabolic traits than they were in the homozygous IL population and, furthermore, that while a minority of metabolites were clearly morphogically associated in both populations, the majority appeared to be linked only in the homozygous ILs. While further harvests of the ILHs need to be analyzed similarly in comparison with the IL population in order to confirm the putative QTL reported here, the data in this study allow several important conclusions to be made. Importantly, evaluation of the distribution of various traits suggested that while the distribution of the metabolite traits was broadly similar between the homozygous IL and heterozygous ILH populations, the distribution in HI was significantly broader in the homozygous IL population.
The most significant finding reported here is that it is possible to uncouple enhanced metabolite content from penalties with respect to plant performance and fertility. The suggested redevelopment of hybrid genetic material based on natural biodiversity could prove an important milestone in the use of genomics-driven breeding approaches. Introgression breeding can be used for the metabolic engineering of crop species in a similar manner to its successful application in breeding for disease resistance and herbicide and salinity tolerance (Zamir, 2001; McCouch, 2004). Our study suggests that the underlying reason for the uncoupling of the metabolic and morphological traits may well be the fact that the ILHs harbor far fewer fertility problems and a far narrower range of fruit sizes than the ILs. Interestingly, close analyses revealed that the most stably inherited QTL displayed a more similar distribution of mode-of-inheritance type to that observed previously for the morphological traits (Semel et al., 2006) than to that obtained for the entire list of putative QTL.
Our study also confirmed that even traits (in this instance, metabolites) exhibiting low hereditability could be valuable targets for breeding, since it demonstrated several instances in which specific lines can display consistent trait effects relative to the control. It thus highlights the importance of both global and genotype-by-genotype analyses in studies using genome-scale populations such as that described here. Considered alongside the current generation (and morphological phenotyping) of a far greater number of sublines, large-scale tomato transcript profiling experiments, and the ongoing tomato genome sequencing project, the findings of this study offer great promise for the utilization of natural biodiversity in future crop improvement strategies.
METHODS
Growth Conditions
The metabolite data set presented is based on field-grown tomato (Solanum lycopersicum) ILs (and their respective heterozygous counterparts) (Semel et al., 2006) from 2004. The field trials were conducted at the Western Galilee Experimental Station in Akko, Israel. Plants were grown in a completely randomized design with one plant per square meter. Seedlings were grown in greenhouses for 35 to 40 d and then transferred to the field. Twelve seedlings of each homozygous IL and heterozygous ILH (IL × M82) were transplanted as well as 70 seedlings of M82. Eight ILs were not included in the analysis because of poor germination (ILH2-4, IL3-1, ILH3-4, ILH6-2, ILH6-2-2, ILH6-4, ILH7-2, and ILH9-3-2). Fruit was harvested when 80 to 100% of the tomatoes were red (Eshed and Zamir, 1995). The field was irrigated with 320 m3 of water per 1000 m2 of field area throughout the season. Morphological and reproductive traits have been described previously for this harvest (Semel et al., 2006). Data obtained were compared with those published previously for metabolite content and morphological parameters from material harvested in Akko in the 2001 and 2003 seasons (Schauer et al., 2006).
Metabolite Profiling
Relative metabolite contents were determined using an established GC-MS protocol allowing the quantification, in methanol extracts, of sugars, sugar alcohols, organic and amino acids, and the vitamins tocopherol and ascorbate essentially as described previously (Roessner et al., 2001; Lisec et al., 2006). The only modifications to these procedures were that they were optimized for tomato fruit as described by Roessner-Tunali et al. (2003) and that we utilized mass-spectral libraries (Schauer et al., 2005a; Kopka et al., 2005) for peak identification. Use of these libraries allowed us to detect a slightly larger number of metabolites than in our previous study (Schauer et al., 2006).
Statistics
Statistical analyses were performed using the JMP IN 5.1 software package (SAS Institute), R statistical software (www.r-project.org), or Microsoft Excel 7.0. Using these programs, we performed ANOVA tests as well as χ2 tests and determined Pearson's correlation coefficients. The distribution of metabolic changes was calculated using a Gaussian distribution with a ratio of 0.95.
IL Mapping
To map the metabolites, a two-way ANOVA was used to partition metabolic variation into genotype, environment, and genotype × environmental interaction effects, regarding the possibility of all three effects as random for the purpose of this analysis. The ANOVA method has been commonly applied to transcriptional analysis and shows excellent robustness (Schauer et al., 2006). Effects were assigned only when data were available for a parameter in all 3 years with at least four replicates of each determination within a year. As an additional criterion, we estimated the P value both for the combined effect and for the independent effects exerted by environment and genotype independently.
H2 (σ2G/σ2G+E) was calculated for each trait for which the genotype was identified as a factor with a random effect, and the genetic variation was calculated as the percentage of the total variation (genetic × environmental). For the QTL mapping, each IL or ILH was compared (by t test) with M82 as well as with each other. If either of them was significantly different (at the 1% level) from the reference genotype M82, the introgression was considered as harboring a QTL. Correlation analysis was additionally performed across the entire population by means of the Pearson correlation coefficient in order to determine possible technical artifacts.
Heat Maps
Heat maps were calculated using the heat map module of the statistical software environment R, version 1.9. False-color imaging was performed on the log10-transformed metabolite data. We scaled data internally on a column basis to have a mean of 0 and a sd of 1. Metabolite data were taken only in instances in which metabolite content was determined in all six replicates of an IL (in any given harvest).
Mode-of-Inheritance Classification
The phenotypic effect of a QTL was considered to be the effect of the significant line (IL or ILH) and was presented as a percentage of M82. Positive values are used for increasing QTL in which the introgression was higher than in M82, and negative values are used for decreasing QTL. If both the IL and the ILH had significant effects in the same direction, the higher value was considered the QTL phenotypic effect. If both the IL and ILH were significant but in opposite directions relative to M82, the introgression was considered as harboring two QTL: one increasing and the other decreasing.
The mode of inheritance of a QTL was determined according to a decision tree (Semel et al., 2006). In cases in which the IL was significantly different from M82 and the ILH phenotype was between the IL and M82, there were three possibilities: (1) if the ILH was significantly different from the IL but not from M82, it was considered recessive; (2) if the ILH differed from both parents or did not differ from either of them, it was considered additive; and (3) if the ILH differed from M82 but not from the IL, the QTL was assigned as dominant. The last possibility is that the ILH was significantly higher or lower than both its parents, in which case it was considered overdominant.
Network Analysis
Network analysis was performed exactly as described by Schauer et al. (2006). In brief, correlation between all trait (metabolite plus morphological) pairs was tested using IL or ILH mean values (n = 76 lines). Because the 83 traits yield 3403 pairs, we choose strict significance levels (0.0001). The pairs that resulted were considered as a network in which a vertex corresponds to a trait and a link between two vertices corresponds to significant correlations between these two traits. This network was then subjected to the same cartography algorithm used in our previous study (Schauer et al., 2006). Essentially, this algorithm divides the network into modules or groups of vertices that are more connected between themselves than to nodes from other modules, yielding a cartographic representation of a complex network. In implementing this algorithm, negative correlations were considered to be equal to positive correlations.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Annotated Overlay Heat Map of the Metabolite Profiles of the ILs in Comparison with That of the Parental Control (M82) from the Individual Data Sets of 2001, 2003, and 2004.
Supplemental Figure 2. Annotated Heat Map of the Metabolite Profiles of the ILs in Comparison with That of the Parental Control (M82) from the Individual Data Set of 2001.
Supplemental Figure 3. Annotated Heat Map of the Metabolite Profiles of the ILs in Comparison with That of the Parental Control (M82) from the Individual Data Set of 2003.
Supplemental Figure 4. Annotated Heat Map of the Metabolite Profiles of the ILs in Comparison with That of the Parental Control (M82) from the Individual Data Set of 2004.
Supplemental Figure 5. Venn Diagram Illustrating the Number of Shared QTL in the 2001, 2003, and 2004 Trials.
Supplemental Figure 6. Annotated Heat Map of the Metabolite Profiles of S. lycopersicum Lines Homozygous (IL) or Heterozygous (ILH) for Chromosomal Segmental Substitution from S. pennellii.
Supplemental Figure 7. HI Comparison between Homozygous and Heterozygous ILs.
Supplemental Figure 8. Cartographic Representation of the Combined Metabolic and Morphological Networks of the ILs.
Supplemental Figure 9. Cartographic Representation of the Combined Metabolic and Morphological Networks of the ILHs.
Supplemental Table 1. QTL Conserved across the Field Trials from All 3 Years.
Supplemental Table 2. Qualitative Distribution of Mode of Inheritance Showing the Numbers of Major QTL That Were Classified in Each Category for Each Chemical Compound Class.
Supplemental Data Set 1. Correlation Analyses of IL and ILH Data Sets.
Supplemental Data Set 2. QTL Detected at a Stringency of 1%, Sorted by IL.
Supplementary Material
Acknowledgments
This research was supported in part by a grant from the German–Israeli Cooperation Project and in part by the European Union SOL Integrated Project FOOD-CT-2006-016214.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alisdair R. Fernie (fernie@mpimp-golm.mpg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., Stepanova, A.N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F.M., and Ecker, J.R. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari, M., Sakakibara, H., Lin, S.Y., Yamamoto, T., Takashi, T., Nishimura, A., Angeles, E.R., Qian, Q., Kitano, H., and Matsuoka, M. (2005). Cytokinin oxidase regulates rice grain production. Science 309 741–745. [DOI] [PubMed] [Google Scholar]
- Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. Review. Crit. Rev. Plant Sci. 24 23–58. [Google Scholar]
- Baxter, C.J., Sabar, M., Quick, W.P., and Sweetlove, L.J. (2005). Comparison of changes in fruit gene expression in tomato introgression lines provides evidence of genome-wide transcriptional changes and reveals links to mapped QTLs and described traits. J. Exp. Bot. 56 1591–1604. [DOI] [PubMed] [Google Scholar]
- Bentsink, L., Alonso-Blanco, C., Vreugdenhil, D., Tesnier, K., Groot, S.P., and Koornneef, M. (2000). Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol. 124 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert, H.J., Gong, Q., Li, P., and Ma, S. (2006). Unraveling abiotic stress tolerance mechanisms—Getting genomics going. Curr. Opin. Plant Biol. 9 180–188. [DOI] [PubMed] [Google Scholar]
- Borevitz, J.O., and Chory, J. (2004). Genomics tools for QTL analysis and gene discovery. Curr. Opin. Plant Biol. 7 132–136. [DOI] [PubMed] [Google Scholar]
- Borevitz, J.O., Liang, D., Plouffe, D., Chang, H.S., Zhu, T., Weigel, D., Berry, C.C., Winzeler, E., and Chory, J. (2003). Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causse, M., Duffe, P., Gomez, M.C., Buret, M., Damidaux, R., Zamir, D., Gur, A., Chevalier, C., Lemaire-Chamley, M., and Rothan, C. (2004). A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. J. Exp. Bot. 55 1671–1685. [DOI] [PubMed] [Google Scholar]
- Causse, M., Saliba-Colombani, V., Lecomte, L., Duffe, P., Rousselle, P., and Buret, M. (2002). QTL analysis of fruit quality in fresh market tomato: A few chromosome regions control the variation of sensory and instrumental traits. J. Exp. Bot. 53 2089–2098. [DOI] [PubMed] [Google Scholar]
- Causse, M.A., et al. (1994). Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138 1251–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaib, J., Lecomte, L., Buret, M., and Causse, M. (2006). Stability over genetic backgrounds, generations and years of quantitative trait locus (QTLs) for organoleptic quality in tomato. Theor. Appl. Genet. 112 934–944. [DOI] [PubMed] [Google Scholar]
- Chen, X., Salamini, F., and Gebhardt, C. (2001). A potato molecular-function map for carbohydrate metabolism and transport. Theor. Appl. Genet. 102 284–295. [Google Scholar]
- Cross, J.M., von Korff, M., Altmann, T., Bartzetko, L., Sulpice, R., Gibon, Y., Palacios, N., and Stitt, M. (2006). Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 142 1574–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri, G.R., et al. (2005). Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 23 890–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaubhadel, S., McGarvey, B.D., Williams, R., and Gijzen, M. (2003). Isoflavonoid biosynthesis and accumulation in developing soybean seeds. Plant Mol. Biol. 53 733–743. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A. (2005). Engineering of plant natural product pathways. Curr. Opin. Plant Biol. 8 329–336. [DOI] [PubMed] [Google Scholar]
- Doebley, J. (2006). Plant science. Unfallen grains: How ancient farmers turned weeds into crops. Science 312 1318–1319. [DOI] [PubMed] [Google Scholar]
- Dong, J., Wu, F.B., Jin, Z.Q., and Huang, Y.Q. (2006). Heterosis for yield and some physiological traits in hybrid cotton Cikangzal. Euphytica 151 71–77. [Google Scholar]
- Dormann, P. (2007). Functional diversity of tocochromanols in plants. Planta 225 269–276. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., and Zamir, D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie, A.R., Tadmor, Y., and Zamir, D. (2006). Natural genetic variation for improving crop quality. Curr. Opin. Plant Biol. 9 196–202. [DOI] [PubMed] [Google Scholar]
- Fernie, A.R., and Willmitzer, L. (2004). Carbohydrate metabolism. In The Handbook of Plant Biotechnology, P. Christou and H.K. Klee, eds (Chichester, UK: Wiley), pp. 1230–1262.
- Frary, A., Doganlar, S., Frampton, A., Fulton, T., Uhlig, J., Yates, H., and Tanksley, S. (2003). Fine mapping of quantitative trait loci for improved fruit characteristics from Lycopersicon chmielewskii chromosome 1. Genome 46 235–243. [DOI] [PubMed] [Google Scholar]
- Frary, A., Nesbitt, T.C., Grandillo, S., Knaap, E., Cong, B., Liu, J., Meller, J., Elber, R., Alpert, K.B., and Tanksley, S.D. (2000). fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289 85–88. [DOI] [PubMed] [Google Scholar]
- Fraser, P.D., Enfissi, E.M., Goodfellow, M., Eguchi, T., and Bramley, P.M. (2007). Metabolite profiling of plant carotenoids using the matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Plant J. 49 552–564. [DOI] [PubMed] [Google Scholar]
- Fridman, E., Carrari, F., Liu, Y.S., Fernie, A.R., and Zamir, D. (2004). Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305 1786–1789. [DOI] [PubMed] [Google Scholar]
- Fridman, E., Liu, Y.S., Carmel-Goren, L., Gur, A., Shoresh, M., Pleban, T., Eshed, Y., and Zamir, D. (2002). Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol. Genet. Genomics 266 821–826. [DOI] [PubMed] [Google Scholar]
- Fridman, E., Pleban, T., and Zamir, D. (2000). A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. USA 97 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland, L.U., Magallanes-Lundback, M., Hemming, C., Supplee, A., Koornneef, M., Bentsink, L., and Dellapenna, D. (2006). Genetic basis for natural variation in seed vitamin E levels in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 103 18834–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni, J.J. (2006). Breeding new life into plant metabolism. Nat. Biotechnol. 24 418–419. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., and Klee, H.J. (2006). Plant volatile compounds: Sensory cues for health and nutritional value? Science 311 815–819. [DOI] [PubMed] [Google Scholar]
- Gur, A., and Zamir, D. (2004). Unused natural variation can lift yield barriers in plant breeding. PLoS Biol. 2 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare, P.D., and Cress, W.A. (1997). Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 21 79–102. [Google Scholar]
- Harrigan, G.G., et al. (2007. a). Impact of genetics and environment on nutritional and metabolite components of maize grain. J. Agric. Food Chem. 55 6177–6185. [DOI] [PubMed] [Google Scholar]
- Harrigan, G.G., et al. (2007. b). Metabolite analyses of grain from maize hybrids grown in the United States under drought and watered conditions during the 2002 field season. J. Agric. Food Chem. 55 6169–6176. [DOI] [PubMed] [Google Scholar]
- Heidel, A.J., Clauss, M.J., Kroymann, J., Savolainen, O., and Mitchell-Olds, T. (2006). Natural variation in MAM within and between populations of Arabidopsis lyrata determines glucosinolate phenotype. Genetics 173 1629–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs, D.H., Flintham, J.E., and Hills, M.J. (2004). Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiol. 136 3341–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R.C., and Nap, J.P. (2001). Genetical genomics: The added value from segregation. Trends Genet. 17 388–391. [DOI] [PubMed] [Google Scholar]
- Keurentjes, J.J., Fu, J., de Vos, C.H., Lommen, A., Hall, R.D., Bino, R.J., van der Plas, L.H., Jansen, R.C., Vreugdenhil, D., and Koornneef, M. (2006). The genetics of plant metabolism. Nat. Genet. 38 842–849. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., and Vreugdenhil, D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55 141–172. [DOI] [PubMed] [Google Scholar]
- Kopka, J., et al. (2005). GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 21 1635–1638. [DOI] [PubMed] [Google Scholar]
- Kusano, M., Fukushima, A., Kobayashi, M., Hayashi, N., Jonsson, P., Moritz, T., Ebana, K., and Saito, K. (2007). Application of a metabolomic method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenotyping of natural variants in rice. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 855 71–79. [DOI] [PubMed] [Google Scholar]
- Lippman, Z.B., Semel, Y., and Zamir, D. (2007). An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr. Opin. Genet. Dev. 17 545–552. [DOI] [PubMed] [Google Scholar]
- Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., and Fernie, A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protocols 1 387–396. [DOI] [PubMed] [Google Scholar]
- Liu, J., Cong, B., and Tanksley, S.D. (2003. a). Generation and analysis of an artificial gene dosage series in tomato to study the mechanisms by which the cloned quantitative trait locus fw2.2 controls fruit size. Plant Physiol. 132 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.S., Gur, A., Ronen, G., Causse, M., Damidaux, R., Buret, M., Hirschberg, J., and Zamir, D. (2003. b). There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol. J. 1 195–207. [DOI] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A., and Henikoff, S. (2000). Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 123 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch, S. (2004). Diversifying selection in plant breeding. PLoS Biol. 2 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, R.C., Steinfath, M., Lisec, J., Becher, M., Witucka-Wall, H., Torjek, O., Fiehn, O., Eckardt, A., Willmitzer, L., Selbig, J., and Altmann, T. (2007). The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz, E.M. (2002). Plants compared to animals: The broadest comparative study of development. Science 295 1482–1485. [DOI] [PubMed] [Google Scholar]
- Milborrow, B.V. (1998). A biochemical mechanism for hybrid vigour. J. Exp. Bot. 49 1063–1071. [Google Scholar]
- Mitchell-Olds, T., and Schmitt, J. (2006). Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441 947–952. [DOI] [PubMed] [Google Scholar]
- Moose, S.P., Dudley, J.W., and Rocheford, T.R. (2004). Maize selection passes the century mark: A unique resource for 21st century genomics. Trends Plant Sci. 9 358–364. [DOI] [PubMed] [Google Scholar]
- Morreel, K., Goeminne, G., Storme, V., Sterck, L., Ralph, J., Coppieters, W., Breyne, P., Steenackers, M., Georges, M., Messens, E., and Boerjan, W. (2006). Genetical metabolomics of flavonoid biosynthesis in Populus: A case study. Plant J. 47 224–237. [DOI] [PubMed] [Google Scholar]
- Morris, C.E., and Sands, D.C. (2006). The breeder's dilemma—Yield or nutrition? Nat. Biotechnol. 24 1078–1080. [DOI] [PubMed] [Google Scholar]
- Mueller, L.A., et al. (2005). The SOL genomics network. A comparative resource for Solanaceae biology and beyond. Plant Physiol. 138 1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggeweg, R., Kocher, T., Gentzel, M., Buscaino, A., Taipale, M., Akhtar, A., and Wilm, M. (2006). A general precursor ion-like scanning mode on quadrupole-TOF instruments compatible with chromatographic separation. Proteomics 6 41–53. [DOI] [PubMed] [Google Scholar]
- O'Reilly-Wapstra, J.M., Potts, B.M., McArthur, C., Davies, N.W., and Tilyard, P. (2005). Inheritance of resistance to mammalian herbivores and of plant defensive chemistry in an Eucalyptus species. J. Chem. Ecol. 31 357–375. [DOI] [PubMed] [Google Scholar]
- Paran, I., and Zamir, D. (2003). Quantitative traits in plants: Beyond the QTL. Trends Genet. 19 303–306. [DOI] [PubMed] [Google Scholar]
- Rein, D., Schijlen, E., Kooistra, T., Herbers, K., Verschuren, L., Hall, R., Sonnewald, U., Bovy, A., and Kleemann, R. (2006). Transgenic flavonoid tomato intake reduces C-reactive protein in human C-reactive protein transgenic mice more than wild-type tomato. J. Nutr. 136 2331–2337. [DOI] [PubMed] [Google Scholar]
- Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Linke, T., Willmitzer, L., and Fernie, A. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner, U., Wagner, C., Kopka, J., Trethewey, R.N., and Willmitzer, L. (2000). Simultaneous analysis of metabolites in potato tuber by gas chromatography mass spectrometry. Plant J. 23 131–142. [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali, U., Hegemann, B., Lytovchenko, A., Carrari, F., Bruedigam, C., Granot, D., and Fernie, A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133 84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux, M.C., Jones, C.M., Adams, D., Chetelat, R., Bennett, A., and Powell, A. (2005). QTL analysis of fruit antioxidants in tomato using Lycopersicon pennellii introgression lines. Theor. Appl. Genet. 111 1396–1408. [DOI] [PubMed] [Google Scholar]
- Salvi, S., and Tuberosa, R. (2005). To clone or not to clone plant QTLs: Present and future challenges. Trends Plant Sci. 10 297–304. [DOI] [PubMed] [Google Scholar]
- Schauer, N., and Fernie, A.R. (2006). Plant metabolomics: Towards biological function and mechanism. Trends Plant Sci. 11 508–516. [DOI] [PubMed] [Google Scholar]
- Schauer, N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24 447–454. [DOI] [PubMed] [Google Scholar]
- Schauer, N., Steinhauser, D., Strelkov, S., Schomburg, D., Allison, G., Moritz, T., Lundgren, K., Roessner-Tunali, U., Forbes, M.G., Willmitzer, L., Fernie, A.R., and Kopka, J. (2005. a). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579 1332–1337. [DOI] [PubMed] [Google Scholar]
- Schauer, N., Zamir, D., and Fernie, A.R. (2005. b). Metabolic profiling of leaves and fruit of wild species tomato: A survey of the Solanum lycopersicum complex. J. Exp. Bot. 56 297–307. [DOI] [PubMed] [Google Scholar]
- Semel, Y., Nissenbaum, J., Menda, N., Zinder, M., Krieger, U., Issman, N., Pleban, T., Lippman, Z., Gur, A., and Zamir, D. (2006). Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. USA 103 12981–12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C., and Koornneef, M. (2002). A fortunate choice: The history of Arabidopsis as a model plant. Nat. Rev. Genet. 3 883–889. [DOI] [PubMed] [Google Scholar]
- Stevens, R., Buret, M., Duffe, P., Garchery, C., Baldet, P., Rothan, C., and Causse, M. (2007). Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiol. 143 1943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice, R., Tschoep, H., Von Korff, M., Bussis, D., Usadel, B., Hohne, M., Witucka-Wall, H., Altmann, T., Stitt, M., and Gibon, Y. (2007). Description and applications of a rapid and sensitive non-radioactive microplate-based assay for maximum and initial activity of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Cell Environ. 30 1163–1175. [DOI] [PubMed] [Google Scholar]
- Tanksley, S.D., and McCouch, S.R. (1997). Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277 1063–1066. [DOI] [PubMed] [Google Scholar]
- Tieman, D.M., Zeigler, M., Schmelz, E.A., Taylor, M.G., Bliss, P., Kirst, M., and Klee, H.J. (2006). Identification of loci affecting flavour volatile emissions in tomato fruits. J. Exp. Bot. 57 887–896. [DOI] [PubMed] [Google Scholar]
- Tikunov, Y., Lommen, A., de Vos, C.H.R., Verhoeven, H.A., Bino, R.J., Hall, R.D., and Bovy, A.G. (2005). A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 139 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titok, V.V., Yurenkova, S.I., Titok, M.V., and Khotyljova, L.V. (2005). Characterisation of energy metabolism during ontogenesis of fibre flax in heterosis. Russ. J. Genet. 41 539–544. [Google Scholar]
- Tonsor, S.J., Alonso-Blanco, C., and Koornneef, M. (2005). Gene function beyond the single trait: Natural variation, gene effects, and evolutionary ecology in Arabidopsis thaliana. Review. Plant Cell Environ. 28 2–20. [Google Scholar]
- Van Camp, W. (2005). Yield enhancement genes: Seeds for growth. Curr. Opin. Biotechnol. 16 147–153. [DOI] [PubMed] [Google Scholar]
- van der Knaap, E., Sanyal, A., Jackson, S.A., and Tanksley, S.D. (2004). High-resolution fine mapping and fluorescence in situ hybridization analysis of sun, a locus controlling tomato fruit shape, reveals a region of the tomato genome prone to DNA rearrangements. Genetics 168 2127–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, R.K., Graner, A., and Sorrells, M.E. (2005). Genomics-assisted breeding for crop improvement. Trends Plant Sci. 10 621–630. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Yoshiba, Y., Kiyosue, T., Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1997). Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 38 1095–1102. [DOI] [PubMed] [Google Scholar]
- Zamir, D. (2001). Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2 983–989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.