Abstract
Vesicle budding in eukaryotes depends on the activity of lipid translocases (P4-ATPases) that have been implicated in generating lipid asymmetry between the two leaflets of the membrane and in inducing membrane curvature. We show that Aminophospholipid ATPase3 (ALA3), a member of the P4-ATPase subfamily in Arabidopsis thaliana, localizes to the Golgi apparatus and that mutations of ALA3 result in impaired growth of roots and shoots. The growth defect is accompanied by failure of the root cap to release border cells involved in the secretion of molecules required for efficient root interaction with the environment, and ala3 mutants are devoid of the characteristic trans-Golgi proliferation of slime vesicles containing polysaccharides and enzymes for secretion. In yeast complementation experiments, ALA3 function requires interaction with members of a novel family of plant membrane-bound proteins, ALIS1 to ALIS5 (for ALA-Interacting Subunit), and in this host ALA3 and ALIS1 show strong affinity for each other. In planta, ALIS1, like ALA3, localizes to Golgi-like structures and is expressed in root peripheral columella cells. We propose that the ALIS1 protein is a β-subunit of ALA3 and that this protein complex forms an important part of the Golgi machinery required for secretory processes during plant development.
INTRODUCTION
P-type ATPases constitute a large family of membrane-integrated pumps, which form a phosphorylated intermediate during catalysis, hence the designation P-type. P-type ATPases are divided phylogenetically into five subfamilies, among which P4-ATPases, suggested to be lipid translocases, have been identified only in eukaryotic organisms (Palmgren and Axelsen, 1998). In human, dysfunction of P4-ATPases has been associated with a number of severe diseases related to deficiencies in transmembrane translocation (flipping) of hydrophobic molecules (Paulusma and Oude Elferink, 2005). In Saccharomyces cerevisiae, the P4-ATPase subfamily contains five well-characterized members: Neo1p in the endosomal membranes, Drs2p and Dnf3p present at the trans-Golgi network, and Dnf1p and Dnf2p at the plasma membrane (PM) (Prezant et al., 1996; Chen et al., 1999; Pomorski et al., 2003). In Arabidopsis thaliana, the P4-ATPase family consists of 12 proteins, Aminophospholipid ATPase1 (ALA1) to ALA12. Only one member, ALA1, has been partially characterized to date (Gomès et al., 2000).
Transmembrane flipping of specific lipids by P4-ATPases has been proposed to be important for establishing phospholipid asymmetry between the two leaflets in the lipid bilayer. This transport activity is believed to be important for generating the initial local curvature of the membrane that precedes vesicle budding during endocytosis and exocytosis, among other processes, in eukaryotic cells (Devaux, 2000; Pomorski and Menon, 2006; Daleke, 2007). Drs2p translocates fluorescent analogs of phosphatidylserine (PS) and phosphatidylethanolamine (PE), but not of phosphatidylcholine (PC), and is needed for the formation of the PE asymmetry in post-Golgi yeast secretory vesicles (Saito et al., 2004; Alder-Baerens et al., 2006). Plant endosomal membranes also exhibit phospholipid asymmetry (Cheesbrough and Moore, 1980; Dorne et al., 1985; Tavernier and Pugin, 1995), but how this asymmetry is established is not known.
Among P4-ATPases, the best physiologically characterized are those from S. cerevisiae. Yeast Drs2p has been shown to be involved in the formation of clathrin-coated vesicles at the trans-Golgi (Chen et al., 1999; Gall et al., 2002). Yeast mutants with deletions in DRS2 produce an abnormally low number of vesicles (Gall et al., 2002) and show defects in polarized growth (Saito et al., 2004). The PM Dnf1p and Dnf2p have been suggested to play a role in endocytosis (Pomorski et al., 2003), as double mutants lacking these proteins are unable to internalize an endocytic dye at reduced temperatures. Not much evidence is available about P4-ATPases in other organisms. In Leishmania donovani parasites, a member of this family, Ld MT, has been suggested to be involved in the uptake of the drug miltefosine, an analog of PC used in the clinical treatment of leishmaniasis (Pérez-Victoria et al., 2006). In Arabidopsis, mutants having low expression levels of the putative flippase ALA1 were found to suffer from chilling sensitivity. The heterologously expressed protein was shown to be able to transport fluorescent analogs of phospholipids, but the mechanism by which this lipid translocation might relate to chilling tolerance was not investigated (Gomès et al., 2000).
Recently, members of the Cdc50p/Lem3p family in yeast were demonstrated to be involved in trafficking of P4-ATPases. These proteins are integral membrane proteins with two predicted transmembrane spans and a soluble domain, which extends into the lumen of organelles or into the extracellular space, depending on protein localization. A deletion of CDC50 results in the retention of Drs2p in the endoplasmic reticulum (ER); similarly, a deletion of DRS2 results in the retention of Cdc50p in the ER. Coexpression of both genes allows trafficking of the corresponding proteins to the Golgi membrane (Saito et al., 2004). The Cdc50p homolog Lem3p is required for the exit of Dnf1p from the ER (Saito et al., 2004), and Ld Ros3, a Lem3p homolog in L. donovani parasites, is needed for proper trafficking of the P4-ATPase Ld MT (Pérez-Victoria et al., 2006). In addition, it was recently shown that the P4-ATPase ATP8B1 from human requires a Cdc50p homolog, CDC50A, for the exit from ER and proper trafficking to the PM (Paulusma et al., 2008).
In this work, we have characterized Arabidopsis plants with mutations in a P4-ATPase, ALA3. These plants present severe defects in vesicle production at secreting peripheral columella cells of the root tip. This, together with the fact that ALA3 is located in Golgi membranes, suggests that this protein might be involved in the budding of vesicles from the trans-Golgi. We have also identified a family of Cdc50p homologs named ALIS1 to ALIS5 (for ALA-Interacting Subunit) in Arabidopsis. Interaction with members of this family was found to be a strict requirement for ALA3 function. Following expression in the heterologous host S. cerevisiae, ALIS1 and ALA3 interacted directly with each other and could be copurified in a detergent-resistant protein complex. We propose that the ALA3/ALIS1 protein complex is essential for the Golgi machinery involved in secretory processes in peripheral columella cells at the root tip.
RESULTS
ala3 Mutants Are Deficient in Root and Shoot Growth
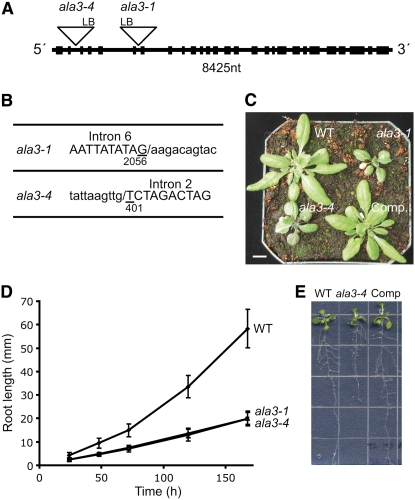
To gain more knowledge about the physiological role of the Arabidopsis P4-ATPase family, we analyzed a battery of Arabidopsis mutant lines carrying T-DNA insertions in genes coding for different P4-ATPases. In this screen, two Arabidopsis mutant lines in the ALA3 gene, SAIL_422_C12 (ala3-1) (Sessions et al., 2002) and SALK_082157 (ala3-4) (Alonso et al., 2003), were identified in the ecotype Columbia (Col-0). The structure of the ALA3 gene and the positions of the T-DNA insertions are shown in Figures 1A and 1B, respectively. Mutant plants were identified by PCR analysis (see Supplemental Figure 1 online) using the primers listed in Supplemental Table 1 online. Both wild-type and mutant plants germinated at the same time. However, compared with the wild type, ala3 plants grew much slower and presented shorter and rounder leaves (Figure 1C). Both ala3-1 and ala3-4 seedlings showed consistently short primary roots (Figures 1D and 1E). After 24 h, the mutant plants had ∼45% shorter roots than the wild type, whereas after 168 h, the difference in size was increased to 65% (Figure 1D). In addition, the vegetative and root growth phenotype could be rescued by the expression of a protein fusion of green fluorescent protein (GFP) to the N-terminal end of ALA3 (GFP:ALA3) (Figures 1C and 1E). These results indicate that ala3-1 and ala3-4 represent loss-of-function mutations that result in a general growth deficiency affecting both root and shoot tissues.
Figure 1.
ala3 Mutants Are Deficient in Root and Shoot Growth.
(A) Genomic organization of ALA3 showing the position of the T-DNA insertions in ala3-1 and ala3-4. Exons are represented by black boxes. LB, left border; nt, nucleotides.
(B) Left border sequences of the T-DNA insertion. Lowercase, T-DNA sequence; uppercase, genomic sequence.
(C) Representative example of the vegetative phenotype under short-day conditions of wild-type, ala3-1, ala3-4, and transgenic ala3-1 expressing GFP:ALA3 (Rescued) plants. Bar = 1 cm.
(D) Diagram of root growth (average ± sd) for the wild type (closed diamonds; n = 40), ala3-1 (open squares; n = 19), and ala3-4 (open triangles; n = 18).
(E) Representative examples of root growth for wild-type, ala3-4, and transgenic ala3-1 expressing GFP:ALA3 (Comp) plants. Similar results were obtained for both ala3 lines.
ALA3 Is Expressed in Key Cell Types in Shoots and Roots
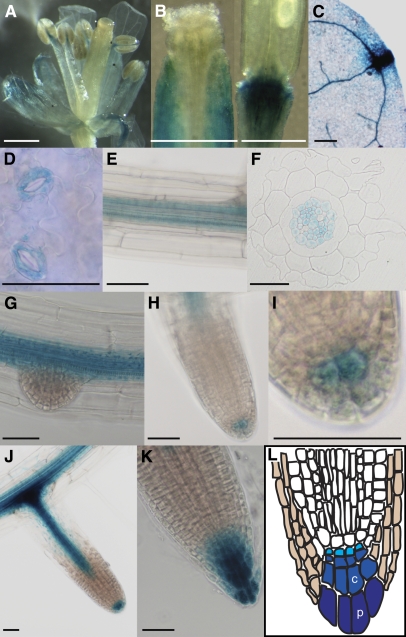
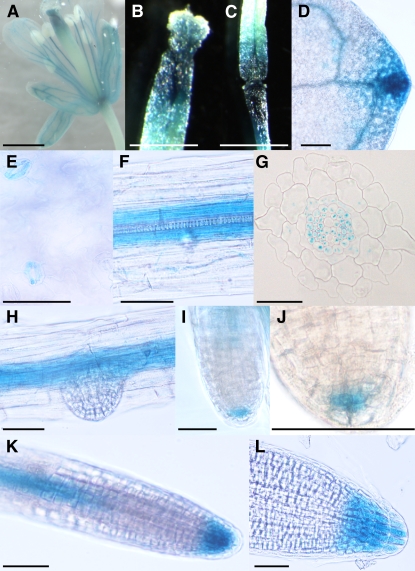
In order to learn more about the physiological role of ALA3, we investigated the tissue-specific expression of ALA3. For this purpose, a transcriptional fusion between the ALA3 promoter and the β-glucuronidase (GUS) reporter gene was generated and introduced into Arabidopsis wild-type plants. GUS expression was confirmed in sepals, petals, and the filament of the flower but not in the reproductive tissues (Figure 2A). In siliques, a strong GUS signal could be detected in the area between the seed pod and the stem, and a weak signal could be detected in the upper part of the pod but not in developing seeds (Figure 2B). Strong expression was observed in vascular shoot tissues (Figure 2C) and in stomatal guard cells (Figure 2D) of young rosette leaves. In roots, the ALA3 promoter was active in the vascular tissue in cells surrounding the xylem (Figures 2E and 2F) and in the columella root cap (Figures 2H to 2K). During the formation of lateral side roots, expression was first evident in columella root cap initials (Figures 2H and 2I) and later appeared in all cells of the columella root cap (Figures 2J and 2K).
Figure 2.
ALA3 Is Expressed in a Variety of Cell Types in Roots and Shoots.
ProALA3:GUS studies were performed on different tissues: flower (A), silique (B), vascular tissue in young leaf (C), stomatal guard cells (D), root vascular tissue ([E] and [F]), emerging side root (G), columella root cap initials ([H] and [I]), and all cells of the columella root cap ([J] and [K]). (L) shows a schematic drawing of the root tip: tan, lateral root cap; light blue, columella initials; darker and darkest blue, columella root cap cells. c, central columella cell; p, peripheral columella cell. Bars = 0.5 mm ([A] to [C]) and 50 μm ([D] to [K]).
Border-Like Cells Remain Attached to the Root Cap in ala3 Plants
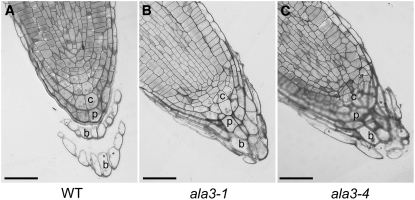
As the strongest expression for the ALA3 promoter was found to occur in the columella root cap cells, we decided to make microscopic observations of wild-type and ala3 root tips. Longitudinal semithin sections showed that the ala3 mutants were unable to release the outermost layer of cells (border-like cells) from the root tip (Figures 3B and 3C), while in wild-type plants grown under the same conditions, these cells were detached (Figure 3A).
Figure 3.
Border-Like Cells Stay Associated with the Root Cap in ala3 Mutants.
Semithin sections of 5-d-old wild-type (A), ala3-1 (B), and ala3-4 (C) roots were analyzed by light microscopy after staining with crystal violet. b, border-like cell; c, central columella cell; p, peripheral columella cell. Bar = 50 μm.
ala3 Mutants Have Defects in Vesicle Production
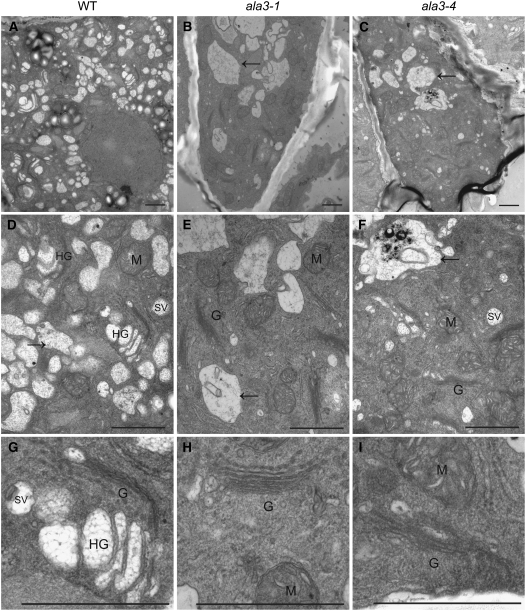
In order to further characterize the ala3 phenotype and understand the cellular processes behind the lack of release of border-like cells, the ultrastructure of different cell types at the root tip of mutant and wild-type plants was investigated using transmission electron microscopy. While peripheral columella cells (Figure 3) at the tip of wild-type roots presented a very active secretion of electron-translucent vesicles from the trans-Golgi (Figures 4A, 4D, and 4G), those of ala3 mutants showed few or none of these (Figure 4). This secretion phenotype seemed to be more severe in the ala3-1 mutant line (Figures 4B, 4E, and 4H) than in ala3-4 (Figures 4C, 4F, and 4I), in which a few vesicles resembling those of wild-type cells could still be observed. The trans-side of the Golgi apparatus in wild-type Arabidopsis is known to become hypertrophied and to accumulate slime substances in cells involved in active secretion at the root tip (Staehelin et al., 1990). Although we clearly identified quite a few of these hypertrophied vesicles at the trans-Golgi side in cells of wild-type plants under our experimental conditions (Figure 4G), none could be observed for the ala3 mutants (Figures 4H and 4I). The mutants also presented abnormally big vacuole-like structures (Figures 4B, 4C, 4E, and 4F). No differences in ultrastructure were observed in other cell types at the root cap, such as border-like cells or central columella cells (Figure 3).
Figure 4.
ala3 Mutants Show a Defect in Slime Vesicle Production and the Specialized Hypertrophied trans-Golgi Stacks in Root Tip Peripheral Columella Cells.
(A) to (C) Transmission electron microscopic overview of peripheral columella cells from the wild type, ala3-1, and ala3-4, respectively.
(D) to (F) Main cellular structures of the same cells from the wild type, ala3-1, and ala3-4, respectively.
(G) to (I) Enlarged view of Golgi stacks in peripheral columella cells of the wild type, ala3-1, and ala3-4, respectively.
Bars = 1 μm. G, Golgi; HG, hypertrophied Golgi; M, mitochondria; SV, slime vesicles. Arrows indicate vacuole-like structures.
ALA3 Localizes to the Golgi Apparatus in Planta
In order to assess the possibility that ALA3 is directly involved in secretory processes occurring in the Golgi apparatus, the intracellular localization of ALA3 in planta was investigated in plant cells expressing the GFP:ALA3 construct. The GFP:ALA3 fusion protein was active in Arabidopsis, as shown by its ability to complement the ala3-1 mutant phenotype (Figures 1C and 1E). Further support for the functionality of a GFP-tagged ALA3 was confirmed by functional complementation of a yeast P4-ATPase mutant (see below and Supplemental Figure 3 online).
Peripheral columella cells of Arabidopsis roots expressing GFP:ALA3 showed clear intracellular GFP fluorescence above background levels (Figure 5C). In contrast with a GFP-only control (Figure 5D), GFP:ALA3 fluorescence was excluded from the nucleus, was unevenly distributed in the cytoplasm, and had a punctate appearance (Figure 5C). While this pattern is consistent with a localization to intracellular membrane structures, the relatively low expression levels found in stable transgenic plants made it difficult to obtain high-resolution images of the GFP:ALA3-labeled structures.
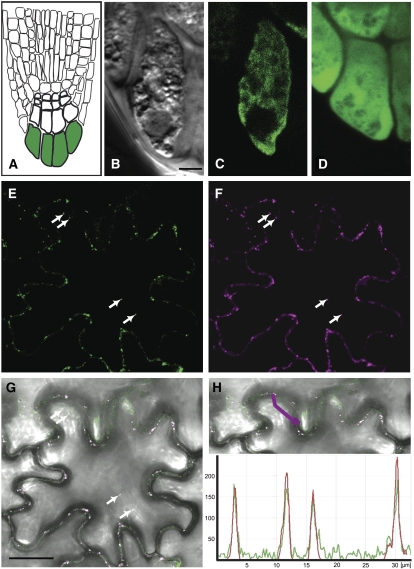
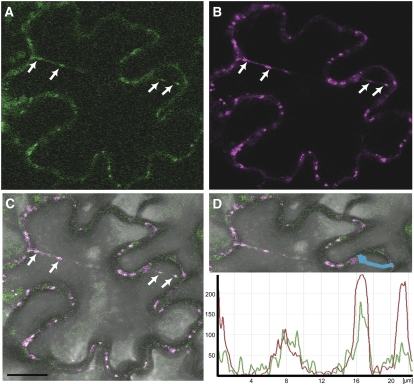
Figure 5.
ALA3 Is Localized in the Golgi Apparatus in Planta.
Fusion proteins of ALA3 and GFP were expressed, either stably in the roots of transformed Arabidopsis ala3-1 plants ([A] to [D]) or transiently in tobacco epidermal cells together with the Golgi marker ST:YFP ([E] to [H]).
(A) Diagram of the root tip. Cells in green are peripheral columella cells.
(B) Transmitted light differential interference contrast image of a peripheral columella cell from a rescued ala3-1 plant transformed with the GFP:ALA3 construct. The same cell is seen in (C).
(C) GFP signal in a peripheral columella cell from a rescued transgenic plant expressing GFP:ALA3. The signal is erratic and stronger in some places than in others, as expected for GFP expression from vesicular bodies and/or the Golgi.
(D) A control transformed ala3-1 line expressing GFP only. The GFP signal is distributed evenly throughout the cell, as expected for localization in the cytosol and nucleus.
(E) GFP fluorescence of tobacco epidermal cells coexpressing GFP:ALA3 and ST:YFP.
(F) YFP signal from the same cells (ST:YFP).
(G) Overlay on a bright-field image.
(H) Intensity plots of YFP and GFP fluorescence of the profile sketched in the merged image (magenta line). The y axis represents the intensity measured in arbitrary units.
Magenta/red, YFP; green, GFP. Arrows indicate the positions of intracellular Golgi bodies. Bars = 10 μm.
Transient expression in tobacco (Nicotiana tabacum) epidermal cells often allows for the detection of subcellular localization at a higher resolution. In single cells of tobacco leaves infiltrated with GFP:ALA3, the fusion protein was visible in small intracellular bodies (Figure 5E), as was a fusion between a rat sialyl transferase (ST) and yellow fluorescent protein (ST:YFP) (Figure 5F), known to localize to the Golgi in planta (Saint-Jore et al., 2002). Time-lapse studies demonstrated fast mobility of the intracellular bodies associated with GFP and YFP fluorescence (see Supplemental Figure 2 online), confirming that ALA3 is present in the highly mobile Golgi apparatus in plant cells. Localization of ALA3 was not influenced by the GFP fusion, as Golgi localization also was observed when GFP was fused to the C-terminal end of ALA3.
ALA3 Expressed in Yeast Fails to Complement a P4-ATPase Mutant
To understand the involvement of the Golgi-localized ALA3 in the production of secretory vesicles from this organelle, the biochemical activity of the protein had to be clarified. As direct biochemical characterization in plant membranes is troublesome due to the presence of numerous other P4-ATPase isoforms at unknown subcellular locations, we decided to express ALA3 in a heterologous system.
The ALA3 gene was cloned and expressed in a S. cerevisiae strain (Δdrs2 Δdnf1 Δdnf2) deficient in the two PM P4-ATPases, Dnf1p and Dnf2p, and in the Golgi-localized Drs2p. This provided a convenient system for studies investigating ALA3 function in a background devoid of related P4-ATPases. The ALA3 protein shows a high (61%) amino acid sequence similarity to Drs2p, which has been implicated in promoting vesicle budding in the trans-Golgi network of S. cerevisiae (Chen et al., 1999; Gall et al., 2002). To determine whether Arabidopsis ALA3 is a functional homolog of yeast Drs2p, we tested the ability of ALA3 to suppress the cold-sensitive phenotype of Δdrs2 Δdnf1 Δdnf2 yeast cells, directly related to the deletion of DRS2 (Hua et al., 2002). Unexpectedly, ALA3 did not complement the cold sensitivity of the Δdrs2 Δdnf1 Δdnf2 triple mutant (Figure 6A).
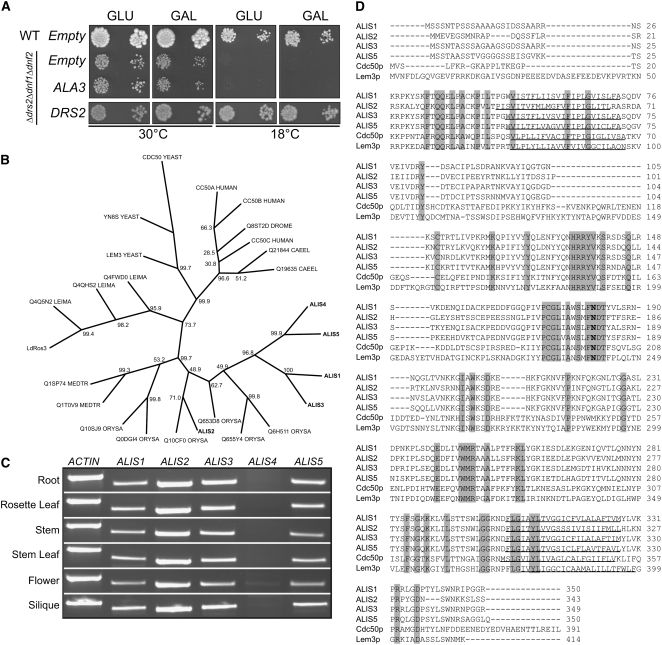
Figure 6.
Identification of a Family of Putative Subunits to ALA3.
(A) The triple yeast mutant Δdrs2 Δdnf1 Δdnf2 expressing ALA3 under the control of a galactose-inducible promoter. ALA3 cannot complement the cold-sensitive phenotype at 18°C, in contrast with plasmid-borne DRS2. Empty, vector control; GAL, galactose; GLU, glucose.
(B) Arabidopsis ALIS proteins and yeast Cdc50p belong to a family of ubiquitous eukaryotic proteins in which plant ALIS proteins evidently cluster. Unrooted phylogenetic analysis of Cdc50p homologs from Saccharomyces cerevisiae (YEAST), Homo sapiens (HUMAN), Leishmania donovani (Ld), Leishmania major (LEIMA), Drosophila melanogaster (DROME), Caenorhabditis elegans (CAEEL), Oryza sativa (ORYZA), Medicago truncatula (MEDTR), and Arabidopsis thaliana (marked in boldface). Export Protein Analysis System accession numbers are given except for those for Ld Ros3 (Q0P0L8) and Arabidopsis ALIS1 to ALIS5 (Q9LTW0, Q67YS6, Q9SLK2, Q9SA35, and Q9SAK5, respectively). Bootstrap values are expressed in percentages and placed at nodes.
(C) RT-PCR analysis of ALIS1 to ALIS5 expression in different tissues of Arabidopsis.
(D) Alignment of ALIS protein sequences derived from cloned cDNAs with those of yeast Cdc50p and Lem3p. Transmembrane domains are underlined, and a conserved predicted N-glycosylation site is marked in boldface. Conserved residues are shaded gray.
To be able to verify whether ALA3 was produced in transformed yeast cells, a hemagglutinin (HA) epitope tag was added to the N terminus of ALA3. In a total yeast membrane preparation, HA-ALA3 was readily detectable by immunoblotting (see Figure 9B), which proves that the lack of complementation is not due to a lack of expression.
Figure 9.
ALA3 in Combination with ALIS Genes Functionally Complement the Yeast drs2 Cold-Sensitive Phenotype.
The triple yeast mutant Δdrs2 Δdnf1 Δdnf2 expresses ALA3 and ALIS genes, either alone or in combination, under the control of a galactose-inducible promoter. ala3, ala3D413D; Empty, yeast cells transformed with a control plasmid; GAL, galactose; GLU, glucose.
(A) ALA3 expressed in combination with ALIS allows the growth of the cold-sensitive yeast strain at 18°C. ala3D413A in combination with ALIS1 does not complement the cold-sensitive strain at 18°C.
(B) Protein blot analysis of total yeast membranes harboring different heterologous proteins.
Identification of a Novel Family of Plant Cdc50p Homologs
If ALA3 is inactive in yeast, it could be because an unidentified factor is required for ALA3 function. Recently, Cdc50 proteins were identified in S. cerevisiae as membrane proteins essential for the proper trafficking of yeast P4-ATPases (Saito et al., 2004). A database search resulted in the identification of five Arabidopsis proteins that show high similarity to the yeast Cdc50p family (Figure 6D). The phylogenetic relationships between Cdc50p homologs in selected eukaryotes are shown in Figure 6B. Cdc50p homologs form small subfamilies depending on their organismal origin, suggesting that the isoforms diverged relatively late in the evolution of eukaryotes.
Following interaction studies (see below), the five identified Cdc50p homologs from Arabidopsis were named ALIS1 to ALIS5. Four of the corresponding cDNAs were successfully cloned: ALIS1, ALIS2, ALIS3, and ALIS5. According to the Genevestigator microarray database (https://www.genevestigator.ethz.ch), ALIS4 is expressed almost exclusively in pollen grains, but we were only able to amplify a fragment of the ALIS4 cDNA from RNA purified from flowers (Figure 6C).
The Arabidopsis ALIS proteins are 27 to 30% identical to yeast Cdc50p, and similarity ranges from 48 to 53% (Figure 6D).
ALIS1 Is Expressed at the Columella Root Cap
In order to identify a putative partner for ALA3 within the ALIS family, the tissue-specific expression of the ALIS genes was analyzed. Using RT-PCR, it could be determined that the transcripts for ALIS genes are expressed in most tissues in Arabidopsis, except ALIS4, which could only be detected in flower tissue (Figure 6C).
To assess the occurrence of ALIS in different cell types, the promoters for ALIS1 to ALIS5 were cloned, fused to GUS, and transformed into wild-type Arabidopsis plants. Analysis of GUS activity showed that one member of the family, ALIS1, presents a very similar expression pattern to ALA3 (Figure 7). The GUS signal was detected in sepals, petals, and the filament of the flower but not in the reproductive tissues (Figure 7A). Strong expression could be observed in the upper part of the seed pod and in the area between the pod and the stem (Figures 7B and 7C). In young rosette leaves, the promoter was also active in the vascular shoot tissues (Figure 7D) and in stomatal guard cells (Figure 7E). Expression in roots was detected in the vascular tissue in cells around the xylem (Figures 7F and 7G) and in the columella root cap (Figures 7I to 7L). As was the case for ALA3, expression of ALIS1 in developing lateral roots starts at the columella root cap initials (Figures 7I and 7J) and was later observed in all cells at the columella root cap (Figures 7K and 7L).
Figure 7.
ALIS1 Is Expressed in a Variety of Cell Types in Roots and Shoots.
Examination of GUS expression directed by the ALIS1 promoter was performed on different tissues: flower (A), silique ([B] and [C]), leaf vascular tissue (D), stomatal guard cells (E), root vascular tissue ([F] and [G]), emerging side root (H), columella root cap initials ([I] and [J]), and all cells of the columella root cap ([K] and [L]). Bars = 0.5 mm ([A] to [D]) and 50 μm ([E] to [L]).
ALIS1 Localizes to the Golgi Apparatus in Planta
To localize ALIS1 at the subcellular level in planta, the corresponding gene was fused to GFP and transiently expressed in tobacco leaves. Independent of the position of the fluorescent protein fusion, as for ALA3, the resulting ALIS1 fusion protein colocalized with the Golgi marker ST:YFP (Figure 8). Fluorescence of both proteins was detected in rapidly moving intracellular bodies (see Supplemental Figure 4 online), supporting the notion that ALIS1, like ALA3, is expressed in membranes of the Golgi apparatus (Saint-Jore et al., 2002). The functionality of the GFP-tagged ALIS1 was checked by functional rescue of a yeast P4-ATPase mutant (see Supplemental Figure 3 online). As the tissue- and cell-specific localization of ALA3 and ALIS1 was very similar, ALIS1 appeared as a promising candidate partner for ALA3.
Figure 8.
ALIS1 Is Localized in the Golgi Apparatus in Planta.
Fusion proteins of ALIS1 and GFP were transiently expressed in tobacco epidermal cells in concert with a Golgi marker (ST:YFP). Arrows indicate the positions of intracellular Golgi bodies.
(A) GFP fluorescence, signal of GFP:ALIS1.
(B) YFP signal (ST:YFP).
(C) Overlay on a bright-field image.
(D) Intensity plots of YFP and GFP fluorescence of the profile sketched in the merged image (blue line). The y axis represents the intensity measured in arbitrary units.
Magenta/red, YFP; green, GFP. Bar = 10 μm.
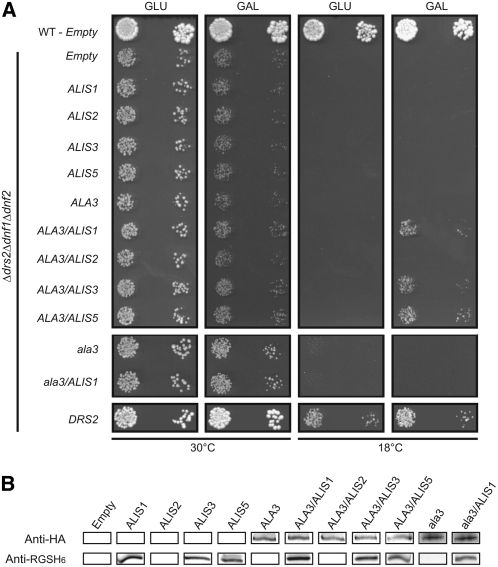
ALIS Proteins Are Essential for ALA3 Functionality
Having identified the ALIS family and confirmed that at least one of its members, ALIS1, localizes to the same tissues and subcellular compartments as ALA3, we investigated whether ALIS1 or other members of the ALIS family could be required for ALA3 function. cDNAs representing each member of the ALIS family were introduced into the Δdrs2 Δdnf1 Δdnf2 mutant strain and expressed in all possible combinations with ALA3. When expressed alone, none of the ALIS genes was capable of functionally complementing the drs2-related mutant phenotype. However, the cold-sensitive phenotype was rescued by the expression of ALA3 in combination with ALIS1, ALIS3, or ALIS5 (Figure 9A). The combination of ALA3 and ALIS2 did not lead to functional complementation (Figure 9A).
In order to ascertain protein expression, the N termini of the ALIS proteins were equipped with an RGSH6 tag. Yeast drs2 complementation was not affected by tagging the proteins, indicating that the addition of this small peptide did not affect protein function. Tagged ALIS1, ALIS3, and ALIS5 were readily immunodetected in yeast microsomal membranes, whereas RGSH6-ALIS2 could not be detected (Figure 9B). Lack of expressed ALIS2 protein readily explains the lack of functional complementation of drs2 by the coexpression of ALA3 and ALIS2. In addition, protein blot analyses on total yeast membranes showed that ALA3 is expressed at similar levels in both the presence and the absence of ALIS proteins.
Even though several ALIS isoforms were found to affect the function of ALA3, none was better than ALIS1. For this reason, ALIS1 is a plausible physiologically relevant partner for ALA3, and we aimed at further investigating the nature of the interaction between these two proteins.
Subcellular Localization of ALA3 in Yeast Is Independent of the Presence of ALIS1
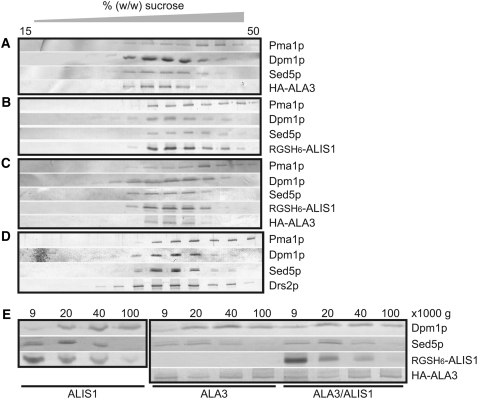
If ALA3 and ALIS1 form a functional protein complex, they would be expected to show colocalization when expressed in yeast. Total yeast microsomes harboring ALA3 and ALIS1 were subjected to sucrose density fractionation followed by protein blot analysis. Whether expressed alone or simultaneously, ALA3 and ALIS1 colocated to membranes that equilibrate at ∼28 to 38% sucrose, together with the yeast Golgi marker Sed5p (Figures 10A to 10C). Plasmid-borne Drs2p in the same yeast background equilibrated at the same density (Figure 10D).
Figure 10.
Subcellular Localization of ALA3 in Yeast Is Independent of the Presence of Colocalizing ALIS1.
Total membranes purified from yeast expressing HA-ALA3 (A), RGSH6-ALIS1 (B), HA-ALA3 and RGSH6-ALIS1 (C), and plasmid-borne DRS2 (D) were subjected to sucrose density gradient fractionation. (E) shows the differential centrifugation of yeast membranes containing RGSH6-ALIS1, HA-ALA3, or HA-ALA3 and RGSH6-ALIS1. The proteins present in each fraction were determined by protein blot analysis. Membrane marker proteins are as follows: Pma1p, PM; Dpm1p, ER; Sed5p, Golgi apparatus.
Cdc50p has been suggested to act as a chaperone required for the proper trafficking of Drs2p (Saito et al., 2004). As we could see no shift in the localization of the plant P4-ATPase in the presence of ALIS1 in our sucrose density gradients, we performed a differential centrifugation of yeast membrane homogenates harboring ALIS1 alone, ALA3 alone, or both proteins simultaneously (Figure 10E). This procedure provides a better separation of the ER and Golgi membranes. Using this strategy, we were able to localize the Sed5p cis-Golgi marker in the fractions pelleting at 9,000g and 20,000g, while the ER marker Dpm1p was enriched in the fraction pelleting at 40,000g (Figure 10E). ALA3 was present in all fractions, and the intensity of the bands in each fraction did not vary when coexpressed with ALIS1, suggesting that the localization of ALA3 in yeast membranes is independent of the presence of ALIS1.
ALA3 and ALIS1 Interact in Vivo in Yeast
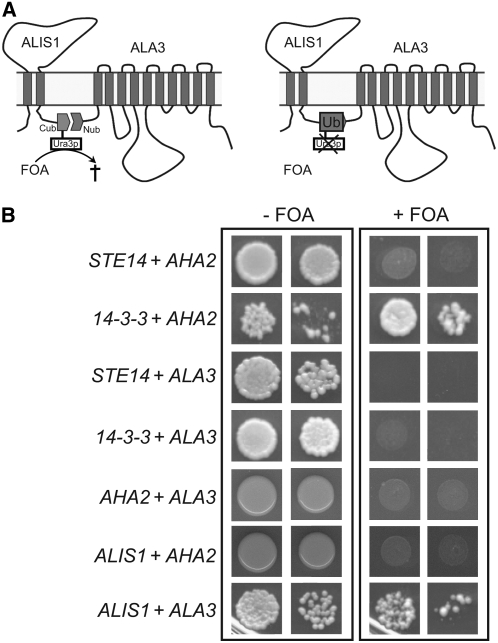
In principle, even if present in the same membrane system, ALIS1 could support ALA3 function indirectly. To ascertain a direct interaction between ALA3 and ALIS1 in vivo, we employed the yeast split-ubiquitin system (Figure 11A) (Stagljar et al., 1998), which has been used to study protein–protein interactions between membrane proteins. In this assay, interaction between two proteins carrying halves of ubiquitin leads to the formation of a complete ubiquitin, thereby causing the degradation of Ura3p. If Ura3p is present, it converts 5-fluoroorotic acid (5-FOA) to a toxic compound; if it is degraded, then no toxic compound is formed. Thus, growth of the transformed yeast on plates containing 5-FOA indicates protein–protein interaction. The clear growth obtained on selection plates for the ALA3/ALIS1 combination suggests that the two corresponding proteins are interacting in this in vivo system (Figure 11B). An interaction between ALIS1 and AHA2, an Arabidopsis P3-ATPase serving as a proton pump, was tested and found to be negative, indicating that ALIS1 does not have a general interaction with all P-type ATPases.
Figure 11.
ALA3 and ALIS1 Interact Directly in Vivo.
(A) Schematic drawing of the principle of the split-ubiquitin assay used. Interaction between two protein partners, each carrying halves of ubiquitin, leads to the formation of a full active ubiquitin, causing the degradation of the reporter protein Ura3p. If Ura3p is present, it converts 5-FOA to a toxic compound; if it is degraded, no toxic compound is formed. Thus, growth of the transformed yeast on plates containing 5-FOA indicates a positive protein–protein interaction.
(B) Cells transformed with plasmid-borne fusions of the desired proteins to either the N or C terminus of ubiquitin. As a positive control, AHA2 and its well-known interacting partner 14-3-3 (Jahn et al., 1997) were used. As a negative control, the soluble transcription factor Ste14p was tested against AHA2 and ALA3. Additionally, AHA2 was tested against ALA3 and ALIS1.
Coexpressed ALA3 and ALIS1 Form a Stable Protein Complex
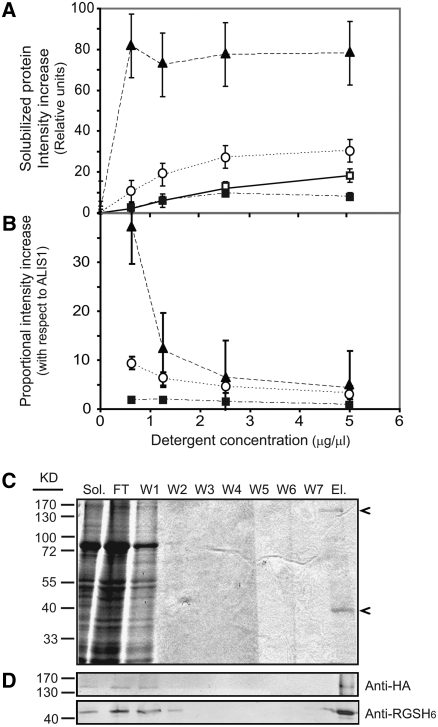
ALIS1 could exert its function by transiently interacting with ALA3, or it could establish a tight and stable interaction with its partner. In the latter case, ALIS1 should be considered a true subunit of ALA3. To test these possibilities, we first studied the solubilization patterns of ALA3 and ALIS1. Total yeast membrane protein fractions were subjected to solubilization in the presence of different concentrations of the mild detergent lysophosphatidylcholine. At low detergent concentrations, both ALA3 and ALIS1 were cosolubilized, while the PM H+-ATPase, Pma1p, and the Golgi marker, Sed5p, showed different solubilization patterns (Figures 12A and 12B). By contrast, at high detergent concentrations (>2.5 mg/mL), the proportion of ALA3 recovered with respect to the amount of ALIS1 decreased, while for the other two membrane proteins, it remained constant (Figures 12A and 12B), suggesting progressive disruption of the ALA3/ALIS1 protein complex.
Figure 12.
ALA3 and ALIS1 Cosolubilize and Can Be Copurified.
Total yeast membranes bearing HA-ALA3 and RGSH6-ALIS1 were solubilized at different concentrations of lysophosphatidylcholine, and the solubilized fractions were subjected to protein blot analysis.
(A) Intensity of immunodetected bands in the supernatant after solubilization.
(B) Intensity of each band compared with the intensity for ALIS1 at the same detergent concentration.
Closed squares, ALA3; open squares, ALIS1; closed triangles, Pma1p; open circles, Sed5p. Values shown are averages ± sd (n = 4).
(C) RGSH6-ALIS1 isolation by Ni-affinity chromatography from the detergent-solubilized fractions. Fractions collected during the affinity purification procedure were visualized by Coomassie blue staining. Bands corresponding to the molecular masses expected for ALIS1 and ALA3 are observed (arrowheads).
(D) Protein blot analysis using antibodies against the HA (ALA3) and RGSH6 (ALIS1) tags added to the proteins.
Sol., solubilized proteins; FT, flow-through; W1 to W7, washing steps; El., eluted fraction.
We further tested the ALA3/ALIS1 association by copurification of the proteins. Taking advantage of the hexahistidine group used to tag ALIS1, the protein was subjected to Ni2+-affinity chromatographic purification. Coomassie blue staining of samples collected along the purification process showed a clear band in the eluted fraction with the expected molecular mass for ALIS1 (Figure 12C). This band could be readily immunodetected with an anti-RGSH6 antibody (Figure 12D). A second protein of the expected molecular size for ALA3 was also present in the eluted fraction (Figure 12C). This could be specifically detected with an antibody against the HA tag present at the N-terminal end of ALA3 (Figure 12D). These results demonstrate that ALA3 and ALIS1 can be copurified in a tight and stable protein complex after detergent solubilization and affinity chromatography.
ALA3 and ALIS1 Expressed in Yeast Generate an Inward-Directed Lipid Translocase Activity
In yeast, Golgi-derived vesicles fusing with the PM have been proposed to eventually trap some of the Golgi proteins involved in vesicle production, allowing these to be transiently located to the PM. Thus, Drs2p cycles between the Golgi apparatus and the PM and is thought to contribute to the phospholipid asymmetry in the PM (Graham, 2004; Chen et al., 2006). Indeed, when PM-enriched fractions obtained from yeast cells expressing ALA3 and ALIS1 were subjected to a two-step fractionation, small amounts of ALA3 and ALIS1 could be detected in the PM (see Supplemental Figure 6 online).
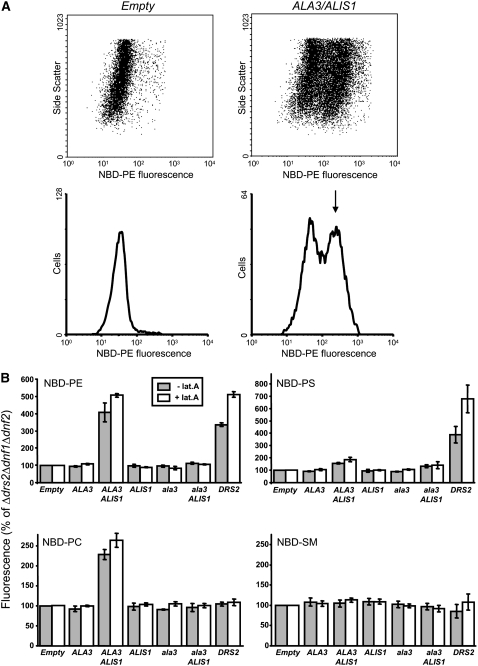
The yeast Δdrs2 Δdnf1 Δdnf2 strain used in this work provided us with an ideal tool for performing sensitive assays of phospholipid flipping, as no background from endogenous P4-ATPase activity is expected at the PM level. Thus, even trace amounts of a functional ALA3/ALIS1 protein complex eventually reaching the PM might result in detectable internalization of phospholipid analogs added to the exterior of the cells. When expressed in the yeast Δdrs2 Δdnf1 Δdnf2 background, ALA3 together with ALIS1, but neither of the proteins alone, was found to catalyze the internalization of exogenously applied [(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]-PE (NBD-PE). NBD-PS and NBD-PC were also translocated at a lower rate, whereas NBD-sphingomyelin, which is not a glycerophospholipid, was not internalized above background levels (Figure 13). As a positive control for the assay, we employed Drs2p, which was found to facilitate the internalization of the phospholipid analogs NBD-PS and NBD-PE but not NBD-PC (Figure 13B). Similar results were obtained when the experiments were performed in the presence of latrunculin A (Figure 13B), a drug that blocks endocytosis, resulting in the trapping of proteins at the yeast PM following their arrival from the Golgi (Morton et al., 2000).
Figure 13.
Coexpression of ALA3 and ALIS1 Complements the Lipid Uptake Defect of the Δdrs2 Δdnf1 Δdnf2 Yeast Mutant.
Internalization of NBD phospholipids by Δdrs2 Δdnf1 Δdnf2 mutant cells transformed with a control vector (Empty) or plasmids expressing different protein combinations. Yeast cells, preincubated with (+) or without (−) latrunculin A (lat.A), were labeled with NBD-lipid and then washed and analyzed by flow cytometry.
(A) Coexpression of ALA3 and ALIS1 resulted in a population of cells with increased NBD-lipid uptake (arrow). Representative histograms of NBD-PE–labeled cells are shown.
(B) Accumulation of NBD lipids is shown as a percentage of fluorescence intensity relative to control Δdrs2 Δdnf1 Δdnf2 mutant cells (Empty). One hundred percent corresponds to 37 ± 8 and 35 ± 5 arbitrary units (NBD-PE), 84 ± 20 and 72 ± 23 arbitrary units (NBD-PS), 29 ± 1 and 26 ± 1 arbitrary units (NBD-PC), and 83 ± 37 and 52 ± 23 arbitrary units (NBD-SM) in the absence and presence of latrunculin A, respectively. Results for NBD-PS are averages ± se from at least three independent experiments; all other data represent averages ± range of two independent experiments. ala3, ala3D413A.
To ascertain that lipid internalization was due to an active ALA3/ALIS1 protein complex, we generated a mutated version of ALA3 in which Asp-413 in the core sequence DKTGT was replaced with Ala. This residue is conserved in all P-type pumps and is phosphorylated and dephosphorylated during the catalytic cycle (Møller et al., 1996). The ala3D413A mutant in combination with ALIS1 failed to generate any activity that promoted the internalization of phospholipid analogs (Figure 13B) and further failed to complement the cold-sensitive phenotype of the yeast triple mutant (Figure 9A). This demonstrates that in order for phospholipid translocation to take place, catalytically active ALA3 is required.
DISCUSSION
ALA3 Plays an Important Role in Root Development
A marked phenotype of ala3 mutant lines is a deficiency in the growth of roots, which corresponds well with the prominent expression of ALA3 in key cell types of this organ. On the basis of functional complementation experiments, as well as lipid translocation studies in yeast and localization studies in planta, we have provided evidence that ALA3 is a plant equivalent of Drs2p, a yeast P4-ATPase, suggested to be involved in the production of vesicles at the trans-Golgi, a key feature of the secretory pathway (Chen et al., 1999; Gall et al., 2002; Graham, 2004). That ALA3 is localized in the Golgi apparatus of Arabidopsis is, in addition, supported by a recent large-scale proteomic study (Dunkley et al., 2006). We conclude that the deficiency in the growth of the ala3 mutants most likely is due to a defect in secretion at the peripheral columella cells of the root tip.
To understand the role of ALA3 in root development, it is important to review the role of individual types of root cap cells for plant growth. Every cell at the very tip of the root goes through different developmental stages during root growth, turning from a meristematic cell into a central columella cell, then into a peripheral columella cell, and finally into a border cell. Peripheral columella cells are specialized in secreting diverse substances to modulate the environment around the growing root tip and protect plant health (Brighman et al., 1995). These cells are released from the root cap, usually as independent units, but in Arabidopsis they can remain attached to each other, forming an organized layer (Vicré et al., 2005), and thus are called border-like cells. In ala3 plants, the border-like cells are not released from the root tip. Such an event might directly inhibit normal root development.
While central columella cells are involved in gravity perception using starch-filled amyloplasts (Kiss et al., 1989), peripheral columella cells are specialized in slime secretion (Sack and Kiss, 1989). The Golgi apparatus in Arabidopsis central columella cells is a polarized organelle usually formed by approximately seven membranous stacks: two cis, two medial, and two or three trans stacks. During the transition from central columella cell to peripheral columella cell, the Golgi apparatus undergoes extensive changes. The trans-Golgi acquires extra stacks and becomes hypertrophied as it accumulates polysaccharides (Staehelin et al., 1990). Slime vesicles released from the trans-Golgi then carry the cargo across the cell and fuse with the PM for the formation and organization of the cell wall (Bolwell, 1988; Moore et al., 1991; Zhang and Staehelin, 1992; Høj and Fincher, 1995; Ito and Nishitani, 1999; Cosgrove, 2000). Dependent upon the mode of preparation, these vesicles are electron-translucent (our preparation) or electron-dense (after high-pressure freezing and freeze substitution) (Staehelin et al., 1990; Vicré et al., 2005). Previous cytochemical studies have shown that these vesicles contain polysaccharides (Rougier, 1971).
In mutant ala3 plants, hypertrophied trans-Golgi structures in the columella peripheral cells are missing and there is virtually no secretory activity of slime vesicles. This suggests that polysaccharides and enzymes, which are important for the correct formation and subsequent degradation of the cell wall, might not reach the PM as they fail to leave the trans-Golgi. This could readily explain the inability of ala3 root tips to release border-like cells, which would remain attached to the root tip as long as the cell wall that connects them is not degraded.
Molecular Mechanism of the ALA3/ALIS1 Protein Complex
When coexpressed in yeast with ALIS1, ALA3 was found to be capable of contributing to transmembrane flipping of a fluorescent PE analog. Lipid translocation between the two leaflets of a biological membrane increases membrane curvature and has been suggested previously to be one of the starting steps in vesicle formation (Pomorski and Menon, 2006).
ALA3-dependent translocation of lipids in the Golgi membrane could be needed to generate extra curvature for the formation of the hypertrophied trans-Golgi structures and eventually result in the formation of vesicles. This would explain why the absence of the flippase in ala3 plants results in a lack of hypertrophied structures and defective secretory activity.
How lipid translocation relates to vesicle production is not clear in any system. At least two possibilities can be considered. First, flippase activity might lead to accumulation in the cytosolic leaflet of the membrane of specific lipids important for the recruitment of GTPases and coat proteins that are directly responsible for vesicle formation (Jürgens, 2004). Second, flippase action might be directly responsible for this process by moving lipid mass from one leaflet to the other, in this way creating a tension driving vesicle formation. Subsequent attachment of GTPases and coat proteins, in this scenario, might serve a role in stabilizing vesicles as they bud out.
ALIS Proteins as β-Subunits of Plant P4-ATPases
The fact that ALA3 and at least ALIS1 interact directly in vivo, forming a stable and strong association, which is a prerequisite for the formation of a functional unit, provides compelling evidence for classifying ALIS proteins as true β-subunits of ALA3. To our knowledge, this is the first example of a β-subunit for a plant P-type pump. On the basis of colocalization and immunoprecipitation experiments, it has been shown that members of the Cdc50p/Lem3p subfamily may act as subunits of the P4-ATPases in the yeast S. cerevisiae (Saito et al., 2004), in the human parasite L. donovani (Pérez-Victoria et al., 2006), and in humans (Paulusma et al., 2008), suggesting that this might be a common feature of this type of pump in all eukaryotic organisms. In contrast with ALA3, Arabidopsis ALA1 does not seem to require an additional protein for activity when expressed in yeast (Gomès et al., 2000). Whether the activity measured for ALA1 was the result of an effective interaction of this protein with endogenous yeast Cdc50/Lem3 proteins or a consequence of specific biochemical features remains to be elucidated. In this context, it is intriguing that there are 12 P4-ATPases in Arabidopsis but only 5 ALIS proteins. A similar situation is found in humans, in which 14 P4-ATPases are present in contrast with only 3 Cdc50p homologs. Coming challenges will be to identify which ALA protein interacts with which ALIS protein and to determine whether some P4-ATPases are able to act alone.
Only a few other P-type pumps require additional subunits, namely P1A-ATPases, a small family of bacterial K+ pumps (Altendorf et al., 1998), and the family of P2D-ATPases, which comprise the animal Na+/K+- and H+/K+-ATPases. Na+/K+- and H+/K+-ATPases have a catalytic α-subunit with P-type ATPase signature sequences and a heavily glycosylated β-subunit (Geering, 2001). The β-subunit has a short cytoplasmic N terminus and a large C-terminal extracellular domain connected by a single transmembrane span, which has direct contact to two transmembrane segments of the α-subunit (Morth et al., 2007).
ALIS proteins structurally resemble the β-subunit of P2D-ATPases in a number of ways. First, they are predicted to have a large ectodomain comprising ∼75% of the total polypeptide. However, in contrast with P2D-ATPase β-subunits, ALIS proteins appear to have two, instead of one, transmembrane spans. Second, ALIS proteins are predicted to have a well-conserved N-glycosylation site in the ectodomain. This Asn is conserved between Cdc50p homologs from both yeast (Figure 6D) and humans (Katoh and Katoh, 2004). Even though it remains to be shown whether ALIS proteins are glycosylated in planta, this seems plausible since at least Lem3p is glycosylated in vivo (Kato et al., 2002).
Molecular Function of the β-Subunit
The β-subunit of Na+/K+- and H+/K+-ATPases serves as a chaperone for the newly synthesized α polypeptide, being essential for correct folding and proper membrane insertion, and plays an important role in trafficking pump heterodimers to the PM (Geering, 2001). Similarly, genetic evidence has been provided that an association of the P4-ATPase with Cdc50p/Lem3 proteins is a prerequisite for proper trafficking to their membrane destination (Saito et al., 2004; Pérez-Victoria et al., 2006). In this work, we were unable to observe any mobility shift for ALA3 due to the presence of the subunit in fractionated yeast microsomal membranes using two independent methods. This was also the case in planta, in which both proteins were detected in the Golgi apparatus of transiently expressing tobacco epidermal leaves. However, we cannot rule out the possibility that ALIS1 is required for the trafficking of ALA3 in Arabidopsis, as both yeast and tobacco possess Cdc50p homologs that might interact with ALA3 and facilitate its exit from the ER.
Previous reports on yeast and parasites and, more recently, on humans were unable to clarify the role of the β-subunit in the catalytic activity of the complex formed with the P4-ATPase, as the P4-ATPase was retained in the ER when expressed alone and never reached the PM, where activity was measured (Saito et al., 2004; Pérez-Victoria et al., 2006; Paulusma et al., 2008). In our case, although ALA3 is localized to the same membranes when expressed alone or together with ALIS1 in yeast, no lipid transport activity could be detected in the absence of the subunit. This points to a requirement of the β-subunit to form a functional complex responsible for lipid transport activity.
As seems to be the case for the β-subunit of P2D-ATPases (Geering, 2001), the different ALIS proteins could play a role in modulating the transport properties of the final protein complex. In contrast with P4-ATPases, all other P-type pumps transport small cations that are occluded in the interior of the transmembrane domain during transport. As such a transport pathway is difficult to envisage for a large phospholipid, flipping might occur in the interface between the catalytic subunit of P4-ATPases and a supporting β-subunit. A dumbbell-shaped density, which could correspond to a phospholipid head group, is observed between the transmembrane spans of the α- and β-subunits in the crystal structure of the sodium-potassium pump (Morth et al., 2007). This supports the notion that a phospholipid can intercalate at the interface between membrane-embedded subunits of a P-type pump. Alternatively, P4-ATPases might not translocate lipids directly but rather influence such a transport system with flippase activity indirectly (e.g., by providing it with an essential metal cofactor). Future biochemical and structural analyses of the purified ALA3/ALIS1 protein complex are likely to resolve the function of P4-ATPases and the specific role of ALIS proteins in the translocation mechanism. The fact that several ALIS isoforms support the functionality of ALA3 provides us with a perfect future opportunity to study the substrate specificity and biochemical properties of each independent protein complex.
Conclusion
In summary, we have shown the requirement of a plant P-type ATPase for a β-subunit and shown the involvement of the protein complex in vesicle production in actively secreting plant cells. This knowledge will facilitate future characterization of P4-ATPases, which, despite being the largest subfamily of P-type ATPases in eukaryotes, remains the least characterized group of primary pumps in all systems.
METHODS
Database Search
Identification of homologs to Cdc50p, protein similarity, and identity studies were performed at the National Center for Biotechnology Information. For protein sequences, see Supplemental Sequences 1 online.
Phylogenetic Analysis
Multiple alignments were done in ClustalW (Thompson et al., 1994) (see Supplemental Alignment 1 and Supplemental Data Set 1 online). Gaps were removed (see Supplemental Alignment 2 online) before the phylogenetic analysis was completed using the Phylip 3.5c package with the Fitch-Margoliash method (Felsenstein, 1989) via a Web-based server (http://bioweb.pasteur.fr/). One thousand trees were used to generate the bootstrap values. For N-glycosylation site prediction, the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) was used. For transmembrane helices prediction, we used the TMHMM server version 2.0 (Sonnhammer et al., 1998).
RT-PCR
Total RNA was isolated from wild-type Arabidopsis thaliana ecotype Col-0 using the RNeasy plant mini kit (Qiagen). Reverse transcription was performed with Moloney murine leukemia virus reverse transcriptase (New England Biolabs), and the cDNA preparation was used as template in a standard PCR. ACTIN3 was used as a control; see Supplemental Table 2 online for primers.
Cloning of ALA3 and ALIS Genes
For the primers used, see Supplemental Table 3 online. Plasmids are listed in Supplemental Table 4 online. A PCR strategy using Phusion High-Fidelity DNA Polymerase (Finnzymes) was used for initial cloning. ALIS1, ALIS2, ALIS3, and ALIS5 were amplified from an Arabidopsis total cDNA preparation. BamHI and SacI artificial restriction sites were introduced at the 5′ and 3′ ends of the genes and used for cloning into pRS423-GAL1-10 (Burgers, 1999). A full-length version of ALA3 (At1g59820) was PCR-amplified from clone RAFL07-17-H11 (Seki et al., 1998, 2002), from RIKEN, and cloned into pENTR/D-TOPO (Invitrogen) using the pENTR/D-TOPO cloning kit. By overlapping PCR with this plasmid as template, an ala3 mutant version containing a D413A point mutation was generated. A KpnI site was introduced in order to identify ala3D413A mutants. This PCR product was cloned into pENTR/D-TOPO. DRS2 and DNF1 were also cloned in this plasmid after PCR amplification from pRS313-DRS2 (Chen et al., 1999) and wild-type Saccharomyces cerevisiae DNA, respectively. Transfer of the genes to expression vectors was performed using Gateway technology (Invitrogen). The yeast expression vector pRS423-GAL1-10 was converted to this system by cloning Gateway Reading Frame Cassette A into the unique HincII site. To generate fusions of the different proteins of interest to GFP, the corresponding genes were transferred to plasmid pMDC43 or pMDC84 (Curtis and Grossniklaus, 2003). For tissue-specific expression studies, 1.5 kb of the 5′ untranslated region for ALA3 and ALIS1 were PCR-amplified, cloned between restriction sites NcoI and BamHI (ALA3) or KpnI (ALIS1) in plasmid pENTR11 (Invitrogen), and subsequently transferred to the plant binary plasmid pMDC162 (Curtis and Grossniklaus, 2003) as before. The clones used to make stable transgenic Arabidopsis lines were constructed using a PCR-based cloning strategy. ALA3 cDNA was PCR-amplified using polymerase Ex-Taq (Takara) and the pSPORT cDNA library (Invitrogen) as template. The artificial restriction sites AscI and NotI were added to the 5′ and 3′ ends, respectively, to allow subcloning into a derivative of the pGreenII plant vector (Hellens et al., 2000), giving rise to a 5′ end GFP-tagged version of ALA3 under the control of the cauliflower mosaic virus 35s promoter. All obtained clones were fully sequenced.
Yeast Strains and Media
Functional complementation assays were performed employing S. cerevisiae mutant strain ZHY709 (MATα his3 leu2 ura3 met15 dnf1Δ dnf2Δ drs2∷LEU2; Hua et al., 2002), with strain BY4741 (MATα his3 leu2 ura3 met15; EUROSCARF) as the wild type. Cells were grown in standard rich medium with glucose or galactose or in selective synthetic medium SD or SG (Rose and Broach, 1990) at 30°C. To solid media was added 2% agar (Villalba et al., 1992).
Yeast Transformation and Growth
Yeast cells were transformed by the lithium acetate method (Gietz and Woods, 2002). Transformants were incubated in liquid SG medium for 4 h and then diluted with water to OD600 = 0.1, 0.01, and 0.001. Drops of 5 μL were spotted onto plates and incubated at 18°C for 6 to 8 d or at 30°C for 2 to 3 d. All experiments were repeated independently at least three times.
Preparation of Yeast Membrane Fractions
Cellular membranes for protein expression analysis were prepared as described previously (Villalba et al., 1992). Sucrose gradient centrifugation was performed essentially as described by Serrano and Villalba (1995), but cells were resuspended into a final volume of 2 mL of buffer per gram fresh weight and pepstatin A (1 μg/mL) was used instead of chymostatin. Microsomal fractions (1 mg of total protein) were loaded onto continuous 18 to 53% (w/w) sucrose gradients. Sucrose was quantified in each fraction using a PAL1 refractometer (Atago). Differential centrifugation of yeast homogenates was performed as described by Vehring et al. (2007) using 1 μg/mL pepstatin A and 0.1 mM phenylmethylsulfonyl fluoride (PMSF) as protease inhibitors and with the addition of 1 mM DTT. For the generation of spheroplasts, cells were incubated with 0.3 mg/mL Lyticase (L4025; Sigma-Aldrich) at 30°C for 2 h. Sucrose density fractionation of PM-enriched fractions was performed on 43% and 53% (w/w) sucrose step gradients.
Protein Immunodetection
Protein samples were quantified by the method of Bradford using γ-globulin as a standard. For protein blot analysis of total membrane fractions, 50 μg of total protein was precipitated with trichloroacetic acid (TCA) and loaded onto SDS-PAGE gels (Villalba et al., 1992). For sucrose gradients, 200 μL of each fraction was TCA-precipitated and resuspended in 40 μL of 1× loading buffer. Ten microliters of this solution was subjected to protein blotting (Villalba et al., 1992). Immunodetection of HA-ALA3 and RGSH6-ALIS proteins was performed using the rabbit anti-HA polyclonal antibody SG77 (Zymed Laboratories) and RGS-His antibody, BSA-free (Qiagen), respectively. Drs2p was immunodetected using a rabbit polyclonal antibody (Chen et al., 1999), Dpm1p with anti-dolichol phosphate mannose synthase antibody (Molecular Probes), Sed5p with affinity-purified anti-Sed5p (Sapperstein et al., 1996), and Pma1p with a polyclonal antibody raised against its C terminus (Monk et al., 1991). Bands were visualized with the 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT) color development substrate (Promega).
Split-Ubiquitin in Vivo Protein Interaction
The split-ubiquitin system by Stagljar and coworkers (1998) was used. Gateway Reading Frame Cassette A was introduced into the prey pRS314-CUP-Nub plasmid (Wittke et al., 1999) at the unique SmaI site. ALA3 and AHA2 were transferred to this plasmid. The ALIS1 gene and the gene encoding the 14-3-3 isoform φ were cloned into the Ste14-Cub-rUra bait plasmid (Wittke et al., 1999) using the unique EcoRI and SalI sites (see Supplemental Table 3 online). Yeast strain JD47-13C (MATα his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52; Dohmen et al., 1995) transformed with the constructions was grown on selection medium containing 1 mg/mL 5-FOA (Sigma-Aldrich).
Solubilization of Membrane Protein Fractions and Data Analysis
Total yeast membranes were diluted to 5 mg protein/mL in 20% glycerol, 50 mM Tris-HCl, pH 9.0, 2 mM DTT, 0.2 mM PMSF, 2 μg/mL pepstatin A, and l-α-lysophosphatidylcholine from egg yolk (Sigma-Aldrich) at the desired concentration. Equal volumes of protein and detergent samples (in a total of 200 μL) were mixed gently and incubated at room temperature for 10 min and centrifuged in a Beckman Airfuge centrifuge (130,000g, 10 min). Supernatants were TCA-precipitated and subjected to SDS-PAGE and immunoblotting. Bands were visualized with the color development substrate (Promega), and the total intensity of immunodetected bands was measured and normalized to the background using Imagequant software version 5.0. The results are averages of four independent solubilization experiments.
Ni2+ Affinity Chromatography
The procedure for solubilization was as described above except that it was performed at 4°C in a total volume of 50 mL and the samples were centrifuged at 40,000 rpm (Beckman 50.2Ti rotor) for 30 min followed by the addition of KCl (10 mM). The affinity matrix (500 μL of Ni-nitrilotriacetic acid agarose; Qiagen) was washed twice with wash buffer (20% glycerol, 50 mM Tris-HCl, pH 9.0, 1 mM DTT, 0.1 mM PMSF, 1 μg/mL pepstatin A, and 0.005% lysophosphatidylcholine) supplemented with 10 mM KCl and subsequently mixed with the solubilized sample. After gentle shaking at 4°C for 18 h, the matrix was washed four times with wash buffer with 10 mM KCl, once with wash buffer with 10 mM KCl and 2 mM imidazole, once with wash buffer with 2 mM imidazole, and once with wash buffer without additives. For elution, the matrix was incubated twice for 5 min with 0.5 M imidazole in wash buffer. Eluates were pooled and TCA-precipitated along with 1-mL samples from the binding and washing steps.
NBD-Lipid Uptake and Flow Cytometry
Palmitoyl-(NBD-hexanoyl)-phosphatidylserine (NBD-PS), myristoyl-(NBD-hexanoyl)-phosphatidylethanolamine (NBD-PE), myristoyl-(NBD-hexanoyl)-phosphatidylcholine (NBD-PC), and 6-NBD-hexanoyl-sphingosine-1-phosphocholine (NBD-sphingomyelin; NBD-SM) were from Avanti Polar Lipids. All NBD-lipid stocks (10 mM) were prepared in DMSO. Uptake experiments were performed essentially as described previously (Pomorski et al., 2003). Flow cytometry of NBD-labeled cells was performed on a Becton Dickinson FACS equipped with an argon laser using Cell Quest software. One microliter of 1 mg/mL propidium iodide in water was added to 107 cells in 1 mL of PBS just before FACS analysis. Twenty thousand cells were analyzed without gating during the acquisition. A histogram of the red fluorescence (propidium iodide) was used to set the gate that excluded dead cells from the analysis. After scatter gating to exclude cells of abnormal morphology typically observed for the deletion strain (Δdrs2 Δdnf1 Δdnf2), the green fluorescence (NBD) of living cells was plotted on a histogram and the mean fluorescence intensity was calculated (Figure 13A).
Plant Transformation
Agrobaterium tumefaciens strain GV3101 containing the helper plasmid pSoup was used to make the stable transgenic lines of the ala3-1 mutant. GFP:ALA3 (ps1019) was introduced into Agrobacterium by electroporation, and transformants were selected on 2× YT plates (1.6% tryptone, 1% yeast extract, 0.5% NaCl, and 1.5% agar) supplemented with 50 μg/mL kanamycin, 15 μg/mL tetracycline, and 50 μg/mL gentamycin. For introduction of all other constructs into wild-type Arabidopsis or tobacco (Nicotiana tabacum) leaves, Agrobacterium strain C58C1 (Koncz and Schell, 1986) was transformed by electroporation and transformants were selected on YEP plates (1% yeast extract, 2% peptone, and 1.5% agar) containing 25 μg/mL gentamycin and 50 μg/mL kanamycin. Transient expression in tobacco epidermal cells was performed as described by Sparkes et al. (2006). For stable transformation of wild-type and ala3 mutant plants, plants (ecotype Col-0 background) were transformed by floral dipping (Clough and Bent, 1998). Seeds were selected on 0.5× MS plates (Murashige and Skoog basal medium [M5519 or M0654 and M0529; Sigma], 0.5 g/L MES, pH 5.7, and 1% agar). Depending on the selection marker, the medium was supplemented with 25 μg/mL hygromycin B and 200 μg/mL carbenicillin or 25 μg/mL hygromycin B. See Supplemental Table 1 online for generated plant lines.
Plant Growth and Complementation by ALA3
Plants were grown for 10 d on 0.5× MS medium plates containing 0.5% sucrose under 24-h light conditions and transplanted to soil [Sunshine Mix 2 Basic/LB2 soil (SunGro Horticulture), fertilized with 1/10 Hoagland's solution No. 2 + 5 μM Fe(EDDHA) (Becker Underwood)] in a growth chamber under short-day conditions (8 h of light/16 h of dark, 19°C, 70% humidity, ∼645 fc light intensity). The root growth assay was done using segregating seeds from heterozygous ala3-1 and ala3-4. Seeds were stratified at 4°C for 96 h before germination at room temperature under 24 h of light. Four days after germination, plates were rotated 90° to set time point 0. Root length was measured after 24, 48, 72, 120, and 168 h. Plants were then genotyped and root growth data were matched to the genotype. For the ala3-rescuing analysis, T2 plants segregating from a hygromycin-resistant T1 parent were used.
GUS Assay and Image Processing
Three independent plants for each GUS-expressing line were grown for 25 d under short-day conditions (8 h of light) on 0.5× MS plates containing hygromycin B before transplanting to soil. Root and shoot samples were GUS-stained as described by Haritatos et al. (2000). After 3 weeks under short-day conditions, transplanted plants were transferred to 16 h of light. After an additional 2 weeks, flowers and siliques were GUS-stained. Microscopes and cameras were as follows: Nikon Eclipse 80i light microscope; Nikon Digital Sight DS L1 image-capture system; Leica MZFLIII fluorescence stereomicroscope; Leica DC camera; and Leica Image Manager 500.
Fixation, Embedding, and Staining of Arabidopsis Roots
GUS-stained roots were fixed in 3% glutaraldehyde, dehydrated in ethanol, and embedded in LR White resin as described by Haritatos et al. (2000). Subsequently, 2 μm cross sections of the roots were prepared. Plants for electron microscopic studies were grown for 5 d under 24-h light conditions on 0.5× MS plates. Roots for transmission electron microscopy were prepared essentially as described previously (Schulz et al., 1998), except that chemical fixation was performed in 100 mM cacodylate buffer without vacuum. Semithin sections (1 μm) were prepared and stained with 0.03% crystal violet for light microscopy. Ultrathin sections (∼50 nm) were used for transmission electron microscopy.
Confocal Microscopy
A Leica TCS SP2/MP confocal laser-scanning microscope with a 63×/1.2 numerical aperture water-immersion objective was used. GFP was excited at 488 nm, and emissions were recovered in the interval 495 to 512 nm. YFP was excited at 514 nm, and fluorescent emissions were measured at 525 to 540 nm. Sequential scanning between lines was used to follow both fluorescent proteins at once. No fluorescence bleed-through was detected under our experimental conditions (see Supplemental Figure 5 online). To detect GFP expression in stable transgenic lines of Arabidopsis, an Olympus 1x81 microscope with a 60× oil-immersion lens was used. GFP was again excited at 488 nm, but emission was recovered in the 500- to 535-nm range.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: ALA3 (At1g59820), ALIS1 (At3g12740), ALIS2 (At5g46150), ALIS3 (At1g54320), ALIS4 (At1g16360), ALIS5 (At1g79450), and ACTIN3 (At2g37620).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PCR Analysis of ala3 Mutant Lines.
Supplemental Figure 2. Movement of Intracellular Bodies Containing ALA3.
Supplemental Figure 3. Functional Complementation Test for GFP-Tagged ALA3 and ALIS1.
Supplemental Figure 4. Movement of Intracellular Bodies Containing ALIS1.
Supplemental Figure 5. Bleed-Through Analysis of GFP and YFP Fluorescence under Standard Experimental Conditions.
Supplemental Figure 6. Localization of ALA3 and ALIS1 to the PM in Yeast.
Supplemental Table 1. Primers Used for ala3 Identification and Seed Stock Information.
Supplemental Table 2. Primers Used for RT-PCR.
Supplemental Table 3. Primers Used for Cloning.
Supplemental Table 4. Plasmids Constructed in This Work.
Supplemental Data Set 1. Alignment of ALIS Proteins and Yeast Cdc50p and Lem3p.
Supplemental Alignment 1. Full-Length ClustalW Multiple Alignment.
Supplemental Alignment 2. ClustalW Multiple Alignment without Gaps.
Supplementary Material
Acknowledgments
We thank Peter Burgers, Todd R. Graham, Hans Dieter Schmitt, Federico Valverde, and Chris Hawes for generously providing plasmids, antibodies, markers, and yeast strains. Hanka Paulitschke's contribution to the lipid transport experiments is greatly appreciated. This work was supported by the European Union Framework V Program (the FLIPPASES Project), by the Danish National Research Foundation (the PUMPKIN Center), and by Deutsche Forschungsgemeinschaft Grant Po748-4 (to T.P.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michael Gjedde Palmgren (palmgren@life.ku.dk).
Online version contains Web-only data.
References
- Alder-Baerens, N., Lisman, Q., Luong, L., Pomorski, T., and Holthuis, J.C.M. (2006). Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol. Biol. Cell 17 1632–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Altendorf, K., Gassel, M., Puppe, W., Mollenkamp, T., Zeeck, A., Boddien, C., Fendler, K., Bamberg, E., and Drose, S. (1998). Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol. Scand. Suppl. 643 137–146. [PubMed] [Google Scholar]
- Bolwell, G.P. (1988). Synthesis of cell wall components: Aspects of control. Phytochemistry 27 1235–1253. [Google Scholar]
- Brighman, L.A., Woo, H.H., and Hawes, M.C. (1995). Root border cells as tools in plant cell studies. Methods Cell Biol. 49 377–387. [DOI] [PubMed] [Google Scholar]
- Burgers, P.M. (1999). Overexpression of multisubunit replication factors in yeast. Methods 18 349–355. [DOI] [PubMed] [Google Scholar]
- Cheesbrough, T.M., and Moore, T.S. (1980). Transverse distribution of phospholipids in organelle membranes from Ricinus communis L. var. Hale endosperm. Plant Physiol. 65 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y., Ingram, M.F., Rosal, P.H., and Graham, T.R. (1999). Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Wang, J., Muthusamy, B.-P., Liu, K., Zare, S., Andersen, R.J., and Graham, T.R. (2006). Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 7 1503–1517. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2000). Loosening of plant cell walls by expansins. Nature 407 321–326. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke, D.L. (2007). Phospholipid flippases. J. Biol. Chem. 282 821–825. [DOI] [PubMed] [Google Scholar]
- Devaux, P.F. (2000). Is lipid translocation involved during endo- and exocytosis? Biochimie 82 497–509. [DOI] [PubMed] [Google Scholar]
- Dohmen, R.J., Stappen, R., McGrath, J.P., Forrova, H., Kolarov, J., Goffeau, A., and Varshavsky, A. (1995). An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 270 18099–18109. [DOI] [PubMed] [Google Scholar]
- Dorne, A.-J., Joyard, J., Block, M.A., and Douce, R. (1985). Localization of phosphatidylcholine in outer envelope membrane of spinach chloroplast. J. Cell Biol. 100 1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley, T.P.J., et al. (2006). Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 103 6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5 164–166. [Google Scholar]
- Gall, W.E., Geething, N.C., Hua, Z., Ingram, M.F., Liu, K., Chen, S.I., and Graham, T.R. (2002). Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr. Biol. 12 1623–1627. [DOI] [PubMed] [Google Scholar]
- Geering, K. (2001). The functional role of β subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 33 425–438. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350 87–96. [DOI] [PubMed] [Google Scholar]
- Gomès, E., Jakobsen, M.K., Axelsen, K.B., Geisler, M., and Palmgren, M.G. (2000). Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell 12 2441–2454. [PMC free article] [PubMed] [Google Scholar]
- Graham, T.R. (2004). Flippases and vesicle-mediated protein transport. Trends Cell Biol. 14 670–677. [DOI] [PubMed] [Google Scholar]
- Haritatos, E., Ayre, B.G., and Turgeon, R. (2000). Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol. 123 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Høj, P.B., and Fincher, G.B. (1995). Molecular evolution of plant beta-glucan endohydrolases. Plant J. 7 367–369. [DOI] [PubMed] [Google Scholar]
- Hua, Z., Fatheddin, P., and Graham, T.R. (2002). An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell 13 3162–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., and Nishitani, K. (1999). Visualization of EXGT-mediated molecular grafting activity by means of a fluorescent-labeled xyloglucan oligomer. Plant Cell Physiol. 40 1172–1176. [Google Scholar]
- Jahn, T., Fuglsang, A.T., Olsson, A., Brüntrup, I.M., Collinge, D.B., Volkmann, D., Sommarin, M., Palmgren, M.G., and Larsson, C. (1997). The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens, G. (2004). Membrane trafficking in plants. Annu. Rev. Cell Dev. Biol. 20 481–504. [DOI] [PubMed] [Google Scholar]
- Kato, U., Emoto, K., Fredriksson, C., Nakamura, H., Ohta, A., Kobayashi, T., Murakami-Murofushi, K., Kobayashi, T., and Umeda, M. (2002). A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J. Biol. Chem. 277 37855–37862. [DOI] [PubMed] [Google Scholar]
- Katoh, Y., and Katoh, M. (2004). Identification and characterization of CDC50A, CDC50B and CDC50C genes in silico. Oncol. Rep. 12 939–943. [PubMed] [Google Scholar]
- Kiss, J.Z., Hertel, R., and Sack, F.D. (1989). Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta 177 198–206. [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Møller, J.V., Juul, B., and le Maire, M. (1996). Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta 1286 1–51. [DOI] [PubMed] [Google Scholar]
- Monk, B.C., Montesinos, C., Ferguson, C., Leonard, K., and Serrano, R. (1991). Immunological approaches to the transmembrane topology and conformational changes of the carboxyl-terminal regulatory domain of yeast plasma membrane H+-ATPase. J. Biol. Chem. 266 18097–18103. [PubMed] [Google Scholar]
- Moore, P.J., Swords, K.M.M., Lynch, M.A., and Staehelin, L.A. (1991). Spatial organization of the assembly pathways of glycoproteins and complex polysaccharides in the Golgi apparatus of plants. J. Cell Biol. 122 589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morth, J.P., Pedersen, B.P., Toustrup-Jensen, M.S., Sørensen, T.L.M., Petersen, J., Andersen, J.P., Vilsen, B., and Nissen, P. (2007). Crystal structure of the sodium-potassium pump. Nature 450 1043–1049. [DOI] [PubMed] [Google Scholar]
- Morton, W.M., Ayscough, K.R., and McLaughlin, P.J. (2000). Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat. Cell Biol. 2 376–378. [DOI] [PubMed] [Google Scholar]
- Palmgren, M.G., and Axelsen, K.B. (1998). Evolution of P-type ATPases. Biochim. Biophys. Acta 1365 37–45. [DOI] [PubMed] [Google Scholar]
- Paulusma, C.C., Folmer, D.E., Ho-Mok, K.S., de Waart, D.R., Hilarius, P.M., Verhoeven, A.J., and Oude Elferink, R.P. (2008). ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane flippase activity. Hepatology 47 268–278. [DOI] [PubMed] [Google Scholar]
- Paulusma, C.C., and Oude Elferink, R.P. (2005). The type 4 subfamily of P-type ATPases, putative aminophospholipid translocases with a role in human disease. Biochim. Biophys. Acta 1741 11–24. [DOI] [PubMed] [Google Scholar]
- Pérez-Victoria, F.J., Sánchez-Cañete, M.P., Castanys, S., and Gamarro, F. (2006). Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J. Biol. Chem. 281 23766–23775. [DOI] [PubMed] [Google Scholar]
- Pomorski, T., Lombardi, R., Riezman, H., Devaux, P.F., van Meer, G., and Holthuis, J.C.M. (2003). Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell 14 1240–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomorski, T., and Menon, A.K. (2006). Lipid flippases and their biological functions. Cell. Mol. Life Sci. 63 2908–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezant, T.R., Chaltraw, W.E., Jr., and Fischel-Ghodsian, N. (1996). Identification of an overexpressed yeast gene which prevents aminoglycoside toxicity. Microbiology 142 3407–3414. [DOI] [PubMed] [Google Scholar]
- Rose, A.B., and Broach, J.R. (1990). Propagation and expression of cloned genes in yeast: 2-microns circle-based vectors. Methods Enzymol. 185 234–279. [DOI] [PubMed] [Google Scholar]
- Rougier, M. (1971). Etude cytochemique de la secretion des polysaccarides vegetaux a l'aide d'un material de choix: Les cellules de la coiffe de Zea mays. J. Microsc. (Paris) 10 67–82. [Google Scholar]
- Sack, F.D., and Kiss, J.Z. (1989). Root cap structure in wild-type and in a starchless mutant of Arabidopsis. Am. J. Bot. 76 454–464. [PubMed] [Google Scholar]
- Saint-Jore, C.M., Evins, J., Batoko, H., Brandizzi, F., Moore, I., and Hawes, C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29 661–678. [DOI] [PubMed] [Google Scholar]
- Saito, K., Fujimura-Kamada, K., Furuta, N., Kato, U., Umeda, M., and Tanaka, K. (2004). Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 15 3418–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein, S.K., Lupashin, V.V., Schmitt, H.D., and Waters, M.G. (1996). Assembly of the ER to Golgi SNARE complex requires Uso1p. J. Cell Biol. 132 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, A., Kühn, C., Reismeier, J.W., and Frommer, W.B. (1998). Ultrastructural effects in potato leaves due to antisense inhibition of the sucrose transporter indicate an apoplastic mode of phloem loading. Planta 206 533–543. [Google Scholar]
- Seki, M., Carninci, P., Nishiyama, Y., Hayashizaki, Y., and Shinozaki, K. (1998). High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15 707–720. [DOI] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296 141–145. [DOI] [PubMed] [Google Scholar]
- Serrano, R., and Villalba, J.M. (1995). Expression and localization of plant membrane proteins in Saccharomyces. Methods Cell Biol. 50 481–496. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., von Heijne, G., and Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6 175–182. [PubMed] [Google Scholar]
- Sparkes, I.A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protocols 1 2019–2025. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., Giddings, T.H., Jr., Kiss, J.Z., and Sack, F.D. (1990). Macromolecular differentiation of Golgi stacks in root tips of Arabidopsis and Nicotiana seedlings as visualized in high pressure frozen and freeze-substituted samples. Protoplasma 157 75–91. [DOI] [PubMed] [Google Scholar]
- Stagljar, I., Korostensky, C., Johnsson, N., and te Heesen, S. (1998). A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 95 5187–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernier, E., and Pugin, A. (1995). Transbilayer distribution of phosphatidylcholine and phosphatidylethanolamine in the vacuolar membrane of Acer pseudoplatanus cells. Biochimie 77 174–181. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehring, S., Pakkiri, L., Schröer, A., Alder-Baerens, N., Herrmann, A., Menon, A.K., and Pomorski, T. (2007). Flip-flop of fluorescently labeled phospholipids in proteoliposomes reconstituted with Saccharomyces cerevisiae microsomal proteins. Eukaryot. Cell 6 1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicré, M., Santaella, C., Blanchet, S., Gateau, A., and Driouich, A. (2005). Root border-like cells of Arabidopsis. Microscopical characterization and role in the interaction with rhizobacteria. Plant Physiol. 138 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba, J.M., Palmgren, M.G., Berberián, G.E., Ferguson, C., and Serrano, R. (1992). Functional expression of plant plasma membrane H+-ATPase in yeast endoplasmic reticulum. J. Biol. Chem. 267 12341–12349. [PubMed] [Google Scholar]
- Wittke, S., Lewke, N., Müller, S., and Johnsson, N. (1999). Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell 10 2519–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G.F., and Staehelin, L.A. (1992). Functional compartmentation of the Golgi apparatus in plant cells: Immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol. 99 1070–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.