Abstract
Endosperm, a storage tissue in the angiosperm seed, provides nutrients to the embryo during seed development and/or to the developing seedling during germination. A major event in endosperm development is the transition between the syncytial phase, during which the endosperm nuclei undergo many rounds of mitosis without cytokinesis, and the cellularized phase, during which cell walls form around the endosperm nuclei. The molecular processes controlling this phase transition are not understood. In agl62 seeds, the endosperm cellularizes prematurely, indicating that AGL62 is required for suppression of cellularization during the syncytial phase. AGL62 encodes a Type I MADS domain protein that likely functions as a transcription factor. During seed development, AGL62 is expressed exclusively in the endosperm. During wild-type endosperm development, AGL62 expression is strong during the syncytial phase and then declines abruptly just before cellularization. By contrast, in mutant seeds containing defects in some FERTILIZATION-INDEPENDENT SEED (FIS) class Polycomb group genes, the endosperm fails to cellularize and AGL62 expression fails to decline. Together, these data suggest that AGL62 suppresses cellularization during the syncytial phase of endosperm development and that endosperm cellularization is triggered via direct or indirect AGL62 inactivation by the FIS polycomb complex.
INTRODUCTION
The endosperm is a fertilization product present in the seeds of angiosperms. The endosperm is an important component of the seed because it provides nutrients and other factors to the embryo during seed development and/or to the developing seedling during germination. In cereals, the endosperm comprises a large proportion of the mature seed, contains large amounts of carbohydrates and proteins, and is an important source of food, feed, and industrial raw materials (reviewed in Lopes and Larkins, 1993; Olsen, 2001; Olsen, 2004).
Several different patterns of endosperm development have been described. Nuclear is the most common pattern and is exhibited by Arabidopsis thaliana and many economically important crop plants, including maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum), soybean (Glycine max), and cotton (Gossypium hirsutum). Nuclear endosperm development consists of two main phases: an initial syncytial phase followed by a cellularized phase. During early development, the endosperm nuclei undergo multiple rounds of mitosis without cytokinesis producing a multinucleate cell, a syncytium. Each nucleus is surrounded by a sphere of cytoplasm and a radial microtubule system, comprising a nuclear-cytoplasmic domain (NCD). At the end of the syncytial phase, the endosperm consists of a peripheral layer of NCDs within a common cytoplasm that surrounds a large central vacuole. At a specific stage during seed development that varies among species, the endosperm becomes cellularized. Initially, anticlinal cell walls form between sister and nonsister nuclei, establishing a layer of alveoli along the embryo sac wall. Subsequent divisions of this cell layer occur centripetally and fill in the interior of the embryo sac cavity. The cellularized endosperm differentiates into several cell types that perform specific roles in endosperm function (reviewed in Lopes and Larkins, 1993; Olsen, 2001, 2004).
The timing of endosperm cellularization is important because it correlates with the extent of nuclear proliferation and may influence seed size, sink strength, and grain weight. In Arabidopsis and maize, in a variety of mutants (Garcia et al., 2003, 2005; Luo et al., 2005), interploidy crosses (Cooper, 1951; Scott et al., 1998; von Wangenheim and Peterson, 2004), interspecific crosses (Bushell et al., 2003), and crosses with hypomethylated strains (Adams et al., 2000; Vinkenoog et al., 2000; Xiao et al., 2006), precocious cellularization correlates with reduced nuclear proliferation and reduced seed size, and delayed cellularization correlates with increased nuclear proliferation and increased seed size. Furthermore, in maize, rice, and wheat, increased nuclear proliferation correlates with increased seed size, sink strength, and grain weight (Brocklehurst, 1977; Radley, 1978; Singh and Jenner, 1982; Reddy and Daynard, 1983; Chojecki et al., 1986; Jones et al., 1996; Liang et al., 2001; Yang et al., 2002). The molecular processes controlling the timing of endosperm cellularization are not understood.
In Arabidopsis, endosperm cellularization occurs in seeds containing transition-stage embryos (Mansfield and Briarty, 1990; Brown et al., 1999; Boisnard-Lorig et al., 2001). During the syncytial phase, Arabidopsis endosperm differentiates into three developmentally distinct regions referred to as micropylar endosperm (MCE) at the micropylar pole surrounding the embryo, chalazal endosperm (CZE) at the chalazal pole, and peripheral endosperm (PEN) between the MCE and CZE (Mansfield and Briarty, 1990; Brown et al., 1999, 2003; Boisnard-Lorig et al., 2001). Cellularization occurs first in the MCE and then in the PEN; the CZE and the PEN near the CZE do not become cellularized (Mansfield and Briarty, 1990; Brown et al., 1999; Nguyen et al., 2000; Boisnard-Lorig et al., 2001; Sorensen et al., 2002; Guitton et al., 2004). In the PEN, cellularization is initiated immediately following the eighth round of mitosis (Sorensen et al., 2002). Initial cellularization involves an unconventional cytokinetic process in which miniphragmoplasts form at the boundaries of the NCDs and transport cell plate–forming vesicles to the regions of wall formation (Otegui and Staehelin, 2000; Otegui et al., 2001).
Endosperm cellularization is affected in a group of Arabidopsis mutants, including atfh5, knolle, hinkel, open house, runkel, pleiade, and spätzle (Sorensen et al., 2002; Ingouff et al., 2005b). Some of these genes encode proteins directly involved in the cellularization process: KNOLLE is a syntaxin that localizes to the cell plate (Lauber et al., 1997), HINKEL is a kinesin-related protein that localizes to the cell plate (Strompen et al., 2002), PLEIADE is a microtubule-associated protein that forms cross-bridges between microtubules (Muller et al., 2002), and ATFH5 is a formin-like protein that plays a role in nucleating actin and that localizes to the cell plate (Ingouff et al., 2005b). The other genes have not been identified. spätzle affects the endosperm but not the embryo, whereas the others also affect cytokinesis in the embryo, suggesting that endosperm cellularization involves some unique molecular components but that most of these components are also used during cytokinesis in other tissues.
Potential regulators of endosperm cellularization in Arabidopsis include the HAIKU (IKU) and FERTILIZATION-INDEPENDENT SEED (FIS) class genes. The IKU class genes include IKU1, IKU2, and MINI3 (Garcia et al., 2003; Luo et al., 2005). In iku1, iku2, and mini3 mutants, the endosperm cellularizes prematurely and underproliferates (Garcia et al., 2003; Luo et al., 2005). MINI3 and IKU2 encode the WRKY10 transcription factor and a Leu-rich repeat transmembrane kinase, respectively, and both genes are expressed in the syncytial endosperm (Luo et al., 2005). Although the precise roles of the IKU class genes in endosperm development remains to be determined, their mutant phenotypes suggest that they play a role in inhibiting cellularization during the syncytial phase.
The FIS class genes include FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) (Ohad et al., 1999), FIS2 (Luo et al., 1999), and MEDEA (MEA) (Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999). These genes encode polycomb group (PcG) proteins and likely function in large protein complexes, which also include MULTICOPYSUPRESSOR OF IRA1 (Kohler et al., 2003b; Guitton et al., 2004) and SWINGER (Wang et al., 2006), to repress target gene transcription (Kohler et al., 2003a). FIE, FIS2, and MEA are expressed in the syncytial endosperm (Vielle-Calzada et al., 1999; Luo et al., 2000; Yadegari et al., 2000; Wang et al., 2006). Fertilization of fis mutant female gametophytes produces seeds that abort; prior to abortion, these seeds exhibit multiple endosperm defects, including overproliferation and the absence of cellularization (Kiyosue et al., 1999; Luo et al., 2000; Vinkenoog et al., 2000; Sorensen et al., 2001; Ingouff et al., 2005a), absence of mitotic domains (Ingouff et al., 2005a), and perturbed gene expression (Sorensen et al., 2001; Kohler et al., 2003b; Ingouff et al., 2005a, 2005b). These observations suggest that the FIS genes regulate several processes during endosperm development, including cellularization.
The timing of endosperm cellularization is also affected in seeds resulting from crosses between diploid and polyploid parents. For example, fertilization of polyploid female gametophytes with diploid pollen produces seeds that cellularize precociously (Scott et al., 1998). These observations suggest that the timing of endosperm cellularization is regulated by imprinted genes. Consistent with this, reciprocal crosses between wild-type and hypomethylated strains also affect the timing of endosperm cellularization (Adams et al., 2000; Vinkenoog et al., 2000; Xiao et al., 2006). Furthermore, FIS2 and MEA are imprinted in the endosperm (Kinoshita et al., 1999; Vielle-Calzada et al., 1999; Luo et al., 2000) and are required for endosperm cellularization (discussed above).
We previously showed that the Arabidopsis AGL80 gene is required for central cell and endosperm development (Portereiko et al., 2006). AGL80 encodes a Type I MADS domain protein and is expressed in the central cell and during early endosperm development. agl80 female gametophytes have subtle defects. However, when agl80 female gametophytes are fertilized with wild-type pollen, endosperm nuclei are not present and the central cell cavity is filled with highly fluorescent material, indicating that AGL80 is required for the earliest stages of endosperm development (Portereiko et al., 2006).
MADS domain proteins typically function as homodimers and/or as heterodimers with other MADS domain proteins and may function in higher-order complexes such as tetramers of two dimers (Messenguy and Dubois, 2003; Kaufmann et al., 2005; de Folter and Angenent, 2006). For this reason, a protein–protein interactome map of the Arabidopsis MADS domain proteins was generated (de Folter et al., 2005). In this study, AGL80 was tested for interaction with all other MADS domain proteins using yeast two-hybrid assays and was found to interact with only one other protein, AGL62, which also is a Type I MADS domain protein (Parenicova et al., 2003).
The AGL80–AGL62 yeast two-hybrid interaction suggests that AGL62 may play a role in central cell and/or endosperm development. To investigate this, we examined the function of this gene during female gametophyte and seed development. We show that AGL62 is expressed during the syncytial phase of endosperm development and that agl62 endosperm undergoes precocious cellularization. We also show that AGL62 expression declines abruptly just before cellularization, that this aspect of AGL62 expression is dependent upon the activity of the FIE, FIS2, and MEA genes, and that AGL62 misexpression in fis mutant seeds is correlated with a failure to cellularize. Together, these data suggest that AGL62 is an important regulator of cellularization during endosperm development.
RESULTS
AGL62 Gene Structure
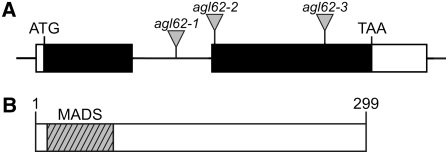
To determine the structure of the AGL62 gene, we isolated a full-length cDNA clone using RT-PCR and 5′ and 3′ rapid amplification of cDNA ends (RACE) and compared its sequence with that of the genomic sequence. Figure 1A shows that AGL62 contains two exons and one intron, a 5′ untranslated region of 35 nucleotides, and a 3′ untranslated region of 196 nucleotides.
Figure 1.
Structures of the AGL62 Gene and AGL62 Protein.
(A) AGL62 gene structure. Black boxes represent coding sequence, white boxes represent the 5′ (35 nucleotides) and 3′ (196 nucleotides) untranslated regions, and the horizontal line represents intron sequence. The insertion sites of the T-DNAs in the agl62-1, agl62-2, and agl62-3 mutants are marked by triangles. The T-DNA in agl62-1 is inserted into the intron, 474 nucleotides downstream of the start codon, and is associated with a 75-bp deletion in the intron. The T-DNA in agl62-2 is inserted in the second exon, 620 nucleotides downstream of the start codon, and is associated with a 9-bp deletion in the predicted second exon. The T-DNA in agl62-3 is inserted in the second exon, 1009 nucleotides downstream of the start codon, and is associated with a 12-bp deletion in the predicted second exon.
(B) AGL62 protein structure. AGL62 contains a MADS domain (hatched box; amino acids 6 to 66).
Figure 1B shows that AGL62 is predicted to encode a protein of 299 amino acids that contains a MADS domain toward the N terminus. ALG62 encodes a Type I MADS domain protein. As such, it lacks the intervening (I), keratin-like (K), and C-terminal (C) domains associated with MIKC-type MADS domain proteins (Parenicova et al., 2003).
AGL62 and AGL80 Interact in Yeast
In a large-scale analysis of protein–protein interaction among MADS domain proteins, AGL62 was shown to interact with AGL80 (de Folter et al., 2005). As shown in Supplemental Figure 1 online, we have confirmed this interaction using a directed yeast two-hybrid assay.
AGL62 Is Expressed during Syncytial Endosperm Development
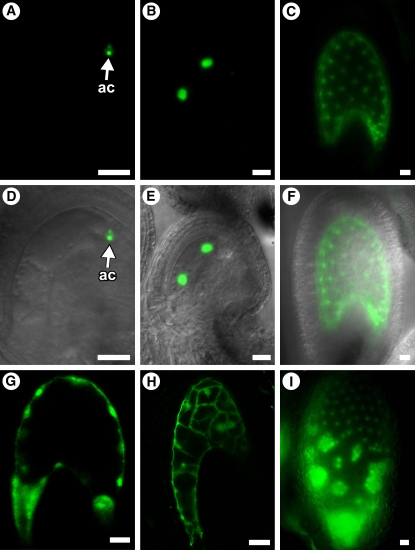
We previously showed that AGL80 is expressed in the central cell and during the 1- to 16-nucleate stages of endosperm development (Portereiko et al., 2006). To determine whether AGL62 expression overlaps with that of AGL80, we analyzed transgenic Arabidopsis plants containing a protein fusion construct, AGL62-GFP, comprising the AGL62 promoter and the entire AGL62 coding region fused with a green fluorescent protein (GFP) coding sequence. We analyzed AGL62-GFP expression at all stages of female gametophyte development. AGL62-GFP was expressed only after cellularization and exclusively in the antipodal cells (Figures 2A and 2D). These data indicate that expression of AGL62 and AGL80 does not overlap during female gametophyte development.
Figure 2.
Analysis of AGL62-GFP Expression and of agl62 Endosperm Development in GFP-Marked Endosperm.
(A) to (C) and (G) to (I) Fluorescence images.
(D) to (F) Fluorescence bright-field overlay images.
(A) and (D) Expression of AGL62-GFP in a mature female gametophyte (stage FG7). The GFP signal is associated with the antipodal cells.
(B), (C), (E), and (F) Expression of AGL62-GFP in seeds at stage II (two endosperm nuclei) ([B] and [E]) and stage VII (∼50 endosperm nuclei) ([C] and [F]) of endosperm development. Endosperm stages are described by Boisnard-Lorig et al. (2001). Expression is detected only in the endosperm nuclei.
(G) and (H) Fluorescence images of wild-type (G) and agl62-1 (H) seeds at 36 h after pollination. Fluorescence is due to endosperm expression of ProDD19:GFP. In (H), cellularization in the micropylar chamber is obscured by the embryo present in this region.
(I) Expression of AGL62-GFP in a fis2-8 seed at 6 d after pollination.
In (A) to (H), seeds are oriented with the micropylar pole to the left and the chalazal pole to the right. ac, antipodal cells. Bars = 20 μm.
Figures 2B, 2C, 2E, and 2F show that AGL62-GFP was expressed in developing seeds. Within the seed, AGL62-GFP expression was detected in the endosperm but not the embryo. During endosperm development, AGL62-GFP was expressed throughout most of the syncytial phase, from immediately following fertilization to just before cellularization (Table 1). Toward the end of the syncytial phase, AGL62-GFP expression declined and became undetectable just before cellularization (Table 1).
Table 1.
Expression of AGL62-GFP during Early Endosperm Development
| Endosperm Developmental Stagea | No. of Endosperm Nuclei | Embryo Developmental Stage | AGL62-GFP Expression |
|---|---|---|---|
| I | 1 | Zygote | ++ |
| II | 2 | Zygote | +++ |
| III | 4 | Zygote | +++ |
| IV | 8 | Elongated zygote | +++ |
| V | 14–16 | Elongated zygote, one-celled | +++ |
| VI | 26–30 | One- to two-celled | +++ |
| VII | ∼50 | One- to two-celled | +++ |
| VIII | ∼100 | Octant | + |
| IX | ∼200 | Dermatogen-globular | − |
| X | ∼300 | Heart | − |
Endosperm developmental stages defined by Boisnard-Lorig et al. (2001) and Ingouff et al. (2005a). Cellularization in the PEN is initiated during stage IX (Sorensen et al., 2002).
To determine whether the maternal and/or paternal alleles of AGL62-GFP are expressed in the endosperm, we performed reciprocal crosses between wild-type plants and plants homozygous for AGL62-GFP. Supplemental Table 1 and Supplemental Figure 2 online show that the maternal and paternal AGL62-GFP alleles were expressed equivalently, both quantitatively and temporally.
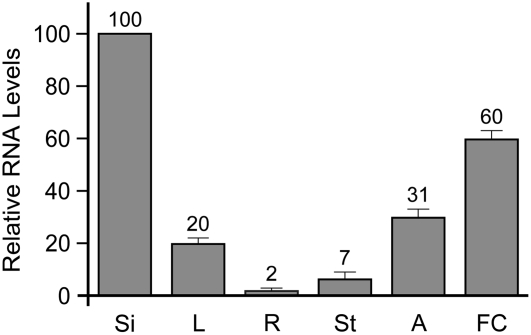
To analyze AGL62 expression elsewhere in the plant, we performed real-time RT-PCR with RNA from various organs. Consistent with expression of AGL62-GFP in the endosperm, we detected strong AGL62 expression in young siliques (Figure 3). In addition, weaker expression was detected in roots, leaves, stems, young flowers, and anthers (Figure 3). In summary, the AGL62 and AGL80 proteins interact in yeast, and the AGL62 and AGL80 genes are coexpressed during the 1- to 16-nucleate stages of endosperm development, suggesting that an AGL62-AGL80 heterodimer may function during early endosperm development.
Figure 3.
Real-Time RT-PCR Analysis of AGL62 Expression.
Real-time RT-PCR was performed with cDNAs at 1 to 3 d after pollination from siliques (Si), leaves (L), roots (R), floral stems (St), anthers (A), and flower clusters (FC). Each bar represents an average of three independent reactions, including both biological and technical replicates. In all cases, AGL62 transcript levels were normalized to ACTIN2 levels. Error bars indicate sd.
Mutations in AGL62 Affect Seed Development
To gain insight into AGL62 function, we obtained lines containing T-DNA insertions in this gene from the SALK Institute Genomic Analysis Laboratory collection (Alonso et al., 2003) (Figure 1A). We analyzed three T-DNA alleles, agl62-1 (SALK_137707), agl62-2 (SALK_022148), and agl62-3 (SALK_013792) (Figure 1A). The agl62-3 mutant exhibited semisterility and therefore is likely associated with a chromosomal rearrangement (Ray et al., 1997); this allele was not analyzed further.
To determine whether AGL62 is required for female gametophyte and/or seed development, we scored seed set in the siliques of plants heterozygous for the agl62-1 and agl62-2 mutations. With both alleles, siliques from heterozygous plants contained ∼25% defective seeds (see Supplemental Figure 3 online), suggesting that these mutations affect seed development. To test this further, we performed self-crosses with heterozygous mutant plants and scored the number of AGL62/AGL62, agl62/AGL62, and agl62/agl62 progeny. Table 2 shows that with both alleles, agl62/agl62 progeny were not observed and AGL62/AGL62 and agl62/AGL62 progeny were present in a 1:2 ratio.
Table 2.
Segregation of the agl62 Mutations
| Parental Genotypes
|
Progeny Genotypes
|
|||
|---|---|---|---|---|
| Female | Male | AGL62/AGL62 | agl62/AGL62 | agl62/agl62 |
| agl62-1/AGL62 | agl62-1/AGL62 | 32.8% (n = 84)a | 67.2% (n = 172)a | 0% (n = 0) |
| agl62-1/AGL62 | AGL62/AGL62 | 52.4% (n = 54)b | 47.6% (n = 49)b | – |
| agl62-2/AGL62 | agl62-2/AGL62 | 36.0% (n = 125)a | 64% (n = 222)a | 0% (n = 0) |
| agl62-2/AGL62 | AGL62/AGL62 | 50.3% (n = 74)b | 49.7 (n = 73)b | – |
χ2 values are not significantly different at a threshold of P = 0.01 from those expected under the hypothesis of a recessive seed-lethal phenotype (i.e., 1:2:0 segregation).
χ2 values are not significantly different at a threshold of P = 0.01 from those expected under the hypothesis of wild-type female gametophyte transmission (i.e., 1:1 segregation).
To determine whether a female gametophyte defect contributes to this phenotype, we crossed heterozygous mutant plants as female parents with wild-type males and scored the number of AGL62/AGL62 and agl62/AGL62 progeny. Table 2 shows that with both alleles, AGL62/AGL62 and agl62/AGL62 progeny were present in a 1:1 ratio, indicating that the female gametophyte is not affected. Consistent with this, the siliques resulting from these crosses exhibited full seed set (see Supplemental Figure 3 online). Together, these data indicate that the agl62-1 and agl62-2 mutations confer recessive seed-lethal phenotypes.
Molecular Complementation of the agl62-1 Allele
To confirm that the seed-lethal defect is attributable to defects in AGL62, we introduced AGL62-GFP into the agl62-1 mutant. We crossed plants homozygous for AGL62-GFP with agl62-1/AGL62 mutants. In the F1 generation, we identified plants hemizygous for AGL62-GFP and heterozygous for agl62-1 and allowed these plants to self-pollinate. In the F2 generation, we identified plants heterozygous for agl62-1 and homozygous for AGL62-GFP; these plants had full seed set, indicating that disruption of AGL62 is responsible for the seed-lethal defect in agl62-1 mutants and that the AGL62-GFP fusion protein is functional.
Mutations in AGL62 Affect Early Endosperm Development
To characterize the seed-lethal phenotype of agl62 mutants, we analyzed the phenotype of agl62 seeds using several approaches. We first characterized the timing of seed abortion. To do so, we used light microscopy to observe the siliques of self-pollinated heterozygous plants at 1 to 10 d after pollination. Supplemental Figure 4 online shows that with both alleles, defective seeds were observed as early as 2 d after pollination. Supplemental Figure 4 online also shows that defective seeds were collapsed, suggesting that endosperm development was affected (Neuffer and Sheridan, 1980).
To determine the basis of the seed lethality, we self-pollinated agl62-1/AGL62 and agl62-2/AGL62 flowers, waited 0 to 48 h, and analyzed developing seeds using confocal laser scanning microscopy (CLSM). At the terminal developmental stage of female gametophyte development (stage FG7), all female gametophytes within heterozygous pistils were wild type. These data suggest that the agl62 mutations do not affect female gametophyte development, which is consistent with the genetic data presented above (Table 2; see Supplemental Figure 3 online). By contrast, both alleles exhibited defects in seed development. agl62-1 and agl62-2 seeds had similar phenotypes. Here, we present a detailed description of the agl62-1 allele.
Using CLSM, defective seeds were not apparent at time points earlier than 24 h after pollination. At 24 h after pollination, agl62-1 seeds had subtle defects. As with the wild type (Figure 4A), the agl62-1 seeds had embryos at the zygote stage and four to eight endosperm nuclei that were positioned along the embryo sac wall (Figures 4B and 5). However, in contrast with the wild type, the agl62-1 NCDs were elongated in an axis perpendicular to the embryo sac wall (Figure 4B); this morphology resembles that of wild-type NCDs just before cellularization (Brown et al., 1999), suggesting that agl62-1 seeds were initiating cellularization at this time point.
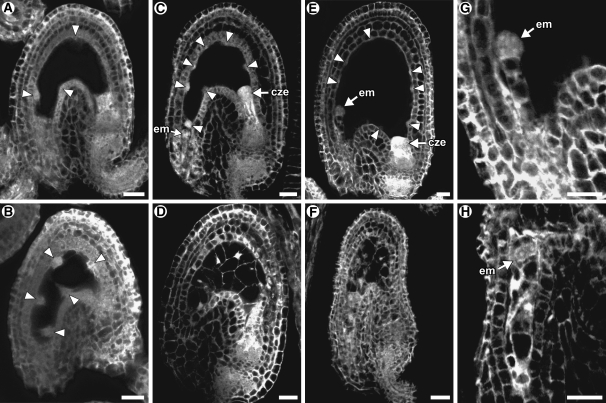
Figure 4.
Microscopy Analysis of Wild-Type and agl62-1 Seeds.
All panels are CLSM images of unstained tissue. The confocal microscope detects autofluorescence. In these images, cytoplasm is gray, vacuoles are black, and nucleoli are white. Arrowheads point to syncytial nuclei. In all panels, the seeds are oriented with the micropylar pole to the left and the chalazal pole to the right. cze, chalazal endosperm; em, embryo. Bars = 20 μm.
(A) Wild-type seed at 24 h after pollination. At this time point, the endosperm is uncellularized and contains four to eight nuclei, and the embryo is at the zygote stage.
(B) agl62-1 seed at 24 h after pollination. At this time point, the endosperm is uncellularized and typically contains four to eight nuclei, and the embryo is at the zygote stage.
(C) Wild-type seed at 36 h after pollination. At this time point, the endosperm is uncellularized and typically contains 16 to 30 nuclei, and the embryo typically is at the two-celled proembryo stage.
(D) agl62-1 seed at 36 h after pollination. At this time point, the endosperm is cellularized and typically contains 14 nuclei, and the embryo is at the one- or two-celled proembryo stage.
(E) Wild-type seed at 48 h after pollination. At this time point, the endosperm is uncellularized and contains >50 nuclei, and the embryo typically is at the quadrant stage.
(F) agl62-1 seed at 48 h after pollination. At this time point, the endosperm is cellularized and typically contains 14 nuclei, and the seed is partially collapsed.
(G) Wild-type embryo at 48 h after pollination. The embryo is at the quadrant stage.
(H) agl62-1 embryo at 48 h after pollination. The embryo is at the two-celled proembryo stage.
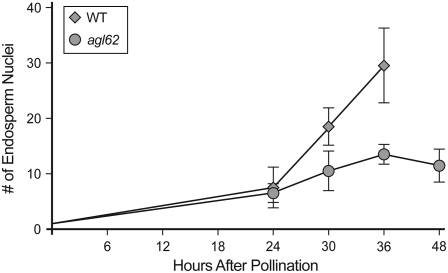
Figure 5.
Number of Endosperm Nuclei in Wild-Type and agl62-1 Seeds.
Each point represents the average of nine seeds. Wild-type seeds at 48 h after pollination were not scored but typically contain >60 endosperm nuclei. Error bars indicate sd.
At 36 and 48 h after pollination, defective seeds were readily apparent. In wild-type seeds, the endosperm was uncellularized at both time points and typically consisted of 30 endosperm nuclei at 36 h after pollination (Figures 4C and 5) and >50 endosperm nuclei at 48 h after pollination (Figure 4E). In agl62-1 seeds at both time points, the endosperm was cellularized (Figures 4D and 4F) and the number of nuclei was reduced (Figure 5). In wild-type seeds, endosperm cellularization did not occur until 5 to 6 d after pollination. Endosperm cellularization in agl62-1 seeds does not appear to be similar to wild-type endosperm with regard to spatial and temporal events: cellularization of agl62-1 occurs rapidly and encompasses the entire embryo sac chamber (Figures 4D and 4F) in contrast with that of the wild type, which occurs progressively and initially in a single peripheral cell layer (Olsen, 2004).
Embryo development was also abnormal in agl62-1 seeds. The wild-type seeds typically had quadrant-stage embryos at 48 h after pollination (Figure 4G). By contrast, agl62-1 embryos typically were at the one- or two-celled proembryo stage at 48 h after pollination (Figures 4H). At 36 h after pollination, embryo development was similar in wild-type and agl62-1 seeds.
To further characterize the endosperm defect in agl62-1 seeds, we used fluorescence microscopy to analyze development of GFP-marked endosperm. The ProDD19:GFP construct drives GFP expression in the central cell and endosperm (Figure 2G) in the wild type (Steffen et al., 2007). Figure 2H shows that this construct was also expressed in agl62-1 endosperm, suggesting that the agl62 defect is not due to misspecification.
We self-pollinated plants homozygous for ProDD19:GFP and heterozygous for agl62-1, waited 24 to 48 h, and observed developing seeds using fluorescence microscopy. Using this approach, defective seeds could not be detected at 24 h after pollination. At 36 and 48 h after pollination, defective seeds resembled those discussed above: the endosperm was cellularized and the number of nuclei was reduced (Figures 2G and 2H). Together, these data indicate that AGL62 is required for suppression of cellularization and promotion of nuclear proliferation during early endosperm development.
AGL62 Is Temporally Misexpressed in fis Seeds
In crosses of fis/FIS females with wild-type males, 50% of the progeny seeds are genotype fis/FIS, and these develop abnormally due to paternal imprinting and/or maternal effects. fis/FIS seeds exhibit several endosperm defects, including the absence of cellularization and overproliferation (Kiyosue et al., 1999; Luo et al., 2000; Vinkenoog et al., 2000; Sorensen et al., 2001; Ingouff et al., 2005a). This phenotype could result from AGL62 misexpression in fis mutant endosperm. To test this, we analyzed expression of AGL62-GFP in fie, fis2, and mea seeds.
We crossed plants homozygous for AGL62-GFP as males with fie-1/FIE (Ohad et al., 1999), fis2-8/FIS2 (Wang et al., 2006), and mea-3/MEA (Kiyosue et al., 1999) females and scored the number of seeds expressing AGL62-GFP at 3 to 7 d after pollination. In parallel, we analyzed control seeds resulting from crosses of AGL62-GFP males with wild-type females. The results of this analysis are shown in Figure 6.
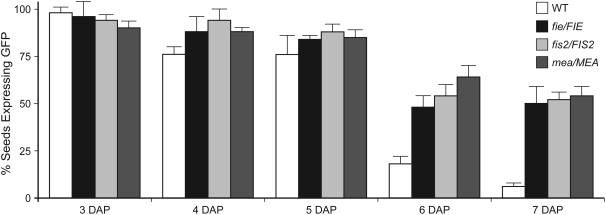
Figure 6.
Expression of AGL62-GFP Expression in fis Seeds.
Percentage of seeds expressing AGL62-GFP in siliques resulting from crosses of homozygous AGL62-GFP males with the wild type (white bars), fie-1/FIE (black bars), fis2-8/FIS2 (light-gray bars), and mea-3/MEA (dark-gray bars) females. Error bars indicate sd.
In wild-type plants, AGL62-GFP was expressed in most seeds at 3 to 5 d after pollination and in few seeds by 7 d after pollination (Figure 6). Those seeds expressing AGL62-GFP at 7 d after pollination were at relatively young developmental stages (typically, seeds containing endosperm at stage VIII) and had weak GFP signals that were equivalent to those reported in Table 1.
In the fie-1/FIE, fis2-8/FIS2, and mea-3/MEA crosses, expression of AGL62-GFP was similar to that in the wild type at 3 to 5 d after pollination: most seeds expressed AGL62-GFP and did so at equivalent quantitative levels, indicating no effect of the fie, fis2, or mea mutations at these time points. However, in contrast with the wild type, ∼50% of the seeds continued to express AGL62-GFP at ≥6 d after pollination (Figure 6). Those seeds expressing AGL62-GFP at 6 to 7 d after pollination had strong GFP signals (Figure 2I), which is in contrast with the weak GFP signals associated with the few seeds expressing AGL62-GFP in the control crosses. Furthermore, those seeds expressing AGL62-GFP at 6 to 7 d after pollination exhibited multiple endosperm defects associated with fis mutant seeds, including the absence of cellularization, overproliferation, and the presence of large nuclear clusters (nodules) near the chalazal pole (Figure 2I), suggesting that they were derived from fertilization of fie, fis2, and mea female gametophytes. These data suggest that the FIS PcG complex is required for suppression of AGL62 expression at the end of the syncytial phase of endosperm development.
DISCUSSION
AGL62 Encodes a Type I MADS Domain Protein
AGL62 is a member of the MADS box gene family. Proteins in this family contain a structurally conserved MADS domain that functions in DNA binding (Hayes et al., 1988; Treisman, 1992; Tilly et al., 1998), and many MADS domain proteins have been shown to function as transcriptional regulators (Irish, 2003; Robles and Pelaz, 2005). Thus, it is likely that AGL62 also functions as a transcriptional regulator.
MADS box genes are divided into two groups referred to as Type I and Type II. The Arabidopsis genome contains ∼61 Type I genes and ∼46 Type II genes. AGL62 falls within the Type I group (Parenicova et al., 2003). The Type II genes have been investigated extensively and functions have been determined for >20 Type II genes (Kofuji et al., 2003; Martinez-Castilla and Alvarez-Buylla, 2003; Parenicova et al., 2003; Verelst et al., 2007). By contrast, functional information is available for only three other Type I genes. AGL37/PHE1 (Kohler et al., 2003a) and AGL80 (Portereiko et al., 2006) play a role in central cell and/or endosperm development. Loss-of-function information is not available for AGL28; however, overexpression of this gene causes precocious flowering (Yoo et al., 2006).
AGL62 Promotes Nuclear Proliferation and Suppresses Cellularization during Syncytial Endosperm Development
agl62 seeds have three defects: precocious endosperm cellularization (Figures 4D and 4F), reduced number of endosperm nuclei (Figure 5), and abnormal embryo development (Figure 4H). AGL62 expression is not detected in the developing embryo (Figures 2B, 2C, 2E, and 2F), suggesting that the embryo development defect results from the endosperm development defects. In principle, reduced nuclear proliferation could result from early cellularization or vice versa. However, multiple rounds of cell division occur after initial cellularization during endosperm development in the wild type (Mansfield and Briarty, 1990; Brown et al., 1999), which indicates that cellularization per se does not preclude mitosis, and mutants defective in nuclear proliferation do not necessarily cellularize prematurely (Menand et al., 2002). Thus, most likely, AGL62 is required independently for both nuclear proliferation and suppression of cellularization.
Several other mutants that undergo premature endosperm cellularization have been identified, including iku1, iku2, and mini3 (Garcia et al., 2003; Luo et al., 2005). These three mutants cellularize ∼24 h prematurely, at 72 h after pollination and produce viable seed. Thus, the cellularization defects in these three mutants is much less severe than that of agl62, which initiates endosperm cellularization at ∼24 h after pollination (Figure 4B) and does not produce viable seeds (Figure 4F; see Supplemental Figures 3 and 4 online). Genetic analysis suggests that the IKU1, IKU2, and MINI3 genes function in a single pathway (Luo et al., 2005). The genetic relationship between AGL62 and this pathway remains to be determined.
The timing of endosperm cellularization is also known to be affected by the balance between the maternal (m) and the paternal (p) genomes, which is generally 2m:1p and in seeds resulting from reciprocal crosses between hypomethylated and wild-type parents. In Arabidopsis, increased maternal dosage (4m:1p or 6m:1p) or fertilization of wild-type embryo sacs with hypomethylated pollen causes precocious cellularization and increased paternal dosage (2m:2p or 2m:3p) or fertilization of hypomethylated embryo sacs with wild-type pollen has the opposite effect (Scott et al., 1998; Adams et al., 2000; Xiao et al., 2006). These observations suggest that imprinted genes play a role in controlling the timing of cellularization during endosperm development (Scott et al., 1998; Adams et al., 2000). AGL62 itself does not appear to be imprinted based on the segregation patterns of the agl62 mutations (Table 2) and on expression of the maternally and paternally derived alleles of AGL62-GFP (see Supplemental Table 1 and Supplemental Figure 2 online). This suggests that AGL62 may be regulated directly or indirectly by imprinted genes, of which genes in the FIS PcG complex are the most obvious candidates.
AGL62 Interacts with AGL80 during Early Endosperm Development
AGL62 (Figures 2B and 2E, Table 1) and AGL80 (Portereiko et al., 2006) are coexpressed during the 1- to 16-nucleate stages of endosperm development, and the encoded proteins interact in yeast (see Supplemental Figure 1 online) (de Folter et al., 2005). These data suggest that AGL62 may function as a heterodimer with AGL80 during the earliest stages of endosperm development. However, genetic analysis has not revealed a role of AGL80 in controlling endosperm cellularization and/or proliferation due to early defects in central cell development (Portereiko et al., 2006).
The absence of AGL80 expression after the 16-nucleate stage (Portereiko et al., 2006) suggests that AGL62 interacts with itself or other MADS domain proteins during the remainder of the syncytial phase. Consistent with this, AGL62 also interacts with seven other MADS domain proteins: AGL36, AGL37/PHE1, AGL38/PHE2, AGL86, AGL90, AGL92, and AGL97 (de Folter et al., 2005). Furthermore, all of the genes encoding the AGL62-interacting proteins are expressed during seed development (Parenicova et al., 2003; de Folter et al., 2004). Experiments are in progress to determine which of these colocalize with AGL62 during syncytial endosperm development.
FIE, FIS2, and MEA Are Required for Suppression of AGL62 Expression
In wild-type seeds, AGL62 expression is high throughout the syncytial phase and then declines abruptly just before cellularization (Table 1). In fis mutant seeds, AGL62 expression does not decline and continues until the seeds collapse (Figure 6). These results suggest very strongly that the FIS PcG complex mediates repression of AGL62 expression at the end of the syncytial phase. In fie, fis2, and mea seeds, endosperm fails to cellularize and overproliferates (Kiyosue et al., 1999; Luo et al., 2000; Vinkenoog et al., 2000; Sorensen et al., 2001; Ingouff et al., 2005a). Given that AGL62 suppresses cellularization and promotes proliferation, it is likely that temporal extension of AGL62 expression contributes to the phenotype of fis mutant seeds.
The FIS PcG complex also regulates the expression of many other genes during endosperm development, including AGL37/PHE1 (Kohler et al., 2003a, 2005; Makarevich et al., 2006), ATFH5 (Sorensen et al., 2001; Ingouff et al., 2005b), and several genes marked in enhancer-trap lines (Ingouff et al., 2005a). As with AGL62, AGL37/PHE1 and several enhancer-trap markers (KS22, N9319, and M11) are transiently expressed during the syncytial phase of endosperm development, and expression is temporally extended in fis mutant seeds (Kohler et al., 2003a; Ingouff et al., 2005a). These observations suggest that the phenotype of fis mutant seeds results from misexpression of many genes in addition to AGL62. Consistent with this, reduction of AGL37/PHE1 expression in mea seeds reduces seed abortion (Kohler et al., 2003a). Collectively, these observations suggest that a major function of the FIS PcG complex during endosperm development is to regulate the timing of endosperm cellularization and that it does so by repressing (directly or indirectly) the expression of a battery of genes at the end of the syncytial phase.
Currently, the pathway by which the FIS PcG complex suppresses AGL62 expression is unknown. This regulation could be direct, as with PcG regulation of PHE1 (Kohler et al., 2003a), or indirect. Also, it currently is unclear how AGL62 suppression is achieved specifically at the end of the syncytial phase. All of the FIS genes are expressed at earlier time points of syncytial endosperm development (Vielle-Calzada et al., 1999; Luo et al., 2000; Yadegari et al., 2000; Wang et al., 2006), and yet AGL62 expression is not suppressed until stage VII. These observations suggest that the FIS PcG complex incorporates additional components in its regulation of AGL62.
Model for AGL62 Function during Endosperm Development
The data presented above suggest that AGL62 functions as a transcription factor within the gene regulatory network that controls the timing of cellularization during endosperm development. The AGL62 expression pattern (Figures 2B, 2C, 2E, and 2F, Table 1) and mutant phenotype (Figures 4B, 4D, and 4F) suggest that AGL62 functions during the syncytial phase to suppress the expression of genes required for endosperm cellularization. Furthermore, the AGL62 expression pattern in fis mutant seeds (Figure 6) suggests that endosperm cellularization is triggered by suppression of AGL62 at the end of the syncytial phase and that this suppression is mediated by the FIS PcG complex.
Identification and analysis of genes downstream of AGL62 should identify genes required for endosperm cellularization and/or proliferation, and identification of factors necessary for AGL62 expression in the endosperm should provide insight into the gene regulatory circuitry specifying the syncytial developmental pattern. Such studies should ultimately lead to an understanding of the gene regulatory network controlling the timing of cellularization during endosperm development.
METHODS
Plant Material and Plasmids
T-DNA insertion mutants agl62-1 (SALK_137707), agl62-2 (SALK_022148), and agl62-3 (SALK_013792) were obtained from the Salk Institute Genomic Analysis Laboratory collection (Alonso et al., 2003). Ramin Yadegari (University of Arizona) provided the fie-1, fis2-8, and mea-3 mutants and the pBI-GFP(S65T) plasmid.
Plant Growth Conditions
Seeds were sterilized in chlorine gas and germinated on plates containing 0.5× Murashige and Skoog salts (Sigma-Aldrich; M-9274), 0.05% MES, 0.5% sucrose, and 0.8% Phytagar (Life Technologies). Ten-day-old seedlings were transferred to Scott's Redi-Earth and grown under 24-h illumination.
Plant Transformation
T-DNA constructs were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation. Arabidopsis thaliana plants (ecotype Columbia) were transformed using a modified floral dip procedure (Clough and Bent, 1998). Transformed progeny were selected by germinating surface-sterilized T1 seeds on growth medium containing antibiotics. Resistant seedlings were transplanted to soil after 10 d of growth.
Cloning the AGL62 cDNA
To amplify a cDNA encompassing the entire open reading frame of AGL62, we performed RT-PCR on RNA extracted from pistils harvested from flowers at stage 13. RNA was extracted using the Qiagen RNeasy kit following the manufacturer's instructions. Aliquots of RNA (1 μg) were reverse transcribed using the RETROscript kit (Ambion) following the manufacturer's instructions. PCR was performed with primers AGL62-cDNA-F (5′-CAATGACAAGAAAGAAAGTG-3′) and AGL62-cDNA-R (5′-ACCTAATGGAACCATGTTT-3′). The cDNA was cloned into the pCRII-TOPO vector using the TOPO TA cloning kit (Invitrogen) resulting in plasmid pCRII-AGL62.
We identified the 5′ and 3′ untranslated sequences with RACE using the First Choice RLM-RACE kit (Ambion). For 5′ RACE, the gene-specific outer primer was 5′RACE-Outer#2 (5′-AAGAGCGAGAGTTCGTGTACCTTC-3′) and the gene-specific inner primer was 5′RACE-Inner#2 (5′-AAGAATCGTTGGAAATGTAAGCAAG-3′). For 3′ RACE, the gene-specific outer primer was AGLSEQp4 (5′-AGCGACAGAAACTTTGAGGAGA-3′) and the gene-specific inner primer was AGLSEQp6 (5′-TCGCAGGATTGAGATTTTAC-3′). This analysis showed that AGL62 contains 5′ and 3′ untranslated regions of 35 and 196 bp, respectively.
Sequence Analysis
We used PROSITE (http://ca.expasy.org/prosite) to identify predicted functional domains of the AGL62 protein. This prediction tool identified the MADS domain but no other domains. We used PSORT (http://psort.nibb.ac.jp/form.html), WoLF PSORT (http://wolfpsort.org/), and PredictNLS (http://cubic.bioc.columbia.edu/predictNLS) to identify a nuclear localization signal in AGL62 protein; no putative nuclear localization signal was identified.
Construction of AGL62-GFP
The AGL62-GFP construct includes a 3262-bp DNA fragment containing 2073 bp of sequence upstream of the translational start codon and 1189 bp of AGL62 genomic coding sequence, excluding the stop codon. This region was obtained by PCR amplification using the primers AGL62-ProtF (5′-TGCCTGCAGGTCGACTACTGCAAAAGTAGTTTGTCT-3′) (contains a SalI site) and AGL62-ProtR (5′-TGCTCACCATGGATCCATAGTAATCAGATCTAGACTG-3′) (contains a BamHI site). The resulting PCR product was cloned into pBI-GFP(S65T) (Yadegari et al., 2000) using the SalI and BamHI sites, resulting in plasmid pBI-AGL62-GFP. The PromAGL62:GFP construct includes a DNA fragment containing 2074 bp of sequence upstream of the translational start codon. This region was obtained by PCR amplification using the primers AGL62Prom-F (5′-TGATTACGCCAAGCTTACTGCAAAAGTAGTTTGTCTC-3′) (contains a HindIII site) and AGL62prom-R (5′-TGCTCACCATGGATCCTTTGCTTTTTTTCACCATTTTT-3′) (contains a BamHI site). The resulting PCR product was cloned into pBI-GFP(S65T) (Yadegari et al., 2000) using the HindIII and BamHI sites, resulting in plasmid pBI101-PromAGL62-GFP. These construct were introduced into Arabidopsis plants as described above, and transformed plants were selected by germinating seeds on growth medium containing 30 μg/mL of kanamycin. We analyzed AGL62-GFP expression from 10 transformants.
Analysis of GFP Expression Patterns
Tissues from plants containing GFP constructs were initially analyzed using an Olympus SXZ12 compound UV dissecting microscope with epifluorescence. Samples were then analyzed using either a Zeiss Axioplan microscope or a Zeiss LSM510 confocal microscope to determine GFP expression patterns. Using the Zeiss Axioplan microscope, GFP was excited using a UV lamp and was detected using a 38 HE EGFP filter set, and images were captured using an AXIOCAM MRM REV2 camera with the AxioVision software package version 4.5 (Zeiss). Using the Zeiss LSM510 microscope, GFP was excited with an argon laser at a wavelength of 488 nm, and emission was detected between 500 and 530 nm.
For analysis of GFP expression in female gametophytes, we emasculated flowers at stage 12c, waited 24 h, and removed the flowers from the plants for analysis. For analysis of GFP expression in seeds, we emasculated flowers at stage 12c, waited 24 h, pollinated with self-pollen, waited the appropriate time, and removed the siliques from the plants for analysis. For analysis of expression of the maternal and paternal alleles of AGL62-GFP during endosperm development, we performed reciprocal crosses with the wild type and plants homozygous for the AGL62-GFP construct as described above and scored expression at 24 to 144 h after pollination. In all cases, we then removed the sepals, petals, and stamen using tweezers, dissected the carpel walls using a 30-gauge syringe needle, and mounted the exposed ovules/seeds on a slide in 10 mM phosphate buffer, pH 7.0.
Real-Time RT-PCR
For plant-wide real-time RT-PCR, we performed the experiments and analysis as described by Steffen et al. (2007). Tissue was harvested from plants and placed immediately into liquid nitrogen. Ovaries were harvested from ms1 at flower stages 12c (Christensen et al., 1997) and 13 (Smyth et al., 1990). Floral cluster tissue includes the inflorescence meristem and flowers at stages 1 to 10 (Smyth et al., 1990). Silique tissue includes siliques at 1 to 2 d after pollination. Leaf tissue includes leaves of sizes 5 to 12 mm. Roots were harvested from seedlings at 11 d after germination. Floral stem tissue includes internodes from 4-week-old plants. Anthers were collected from flowers at stages 11 to 13 (Smyth et al., 1990). RNA was extracted from two different biological samples for each tissue type.
RNA extractions, cDNA synthesis, and real-time RT-PCR were performed as described by Steffen et al. (2007). Each expression value is the result of three independent PCR reactions, including technical and biological replicates. The PCR primers used were IHM102-F (5′-TCATCTTACTCAGGTGTTGAGTCA-3′) and IHM102-R (5′-CGAGTTGAGATAACGCAAGTTCC-3′). We calculated relative expression levels as follows. We first normalized AGL62 transcript levels relative to a standard (ACTIN2) using the formula DCT = CT(AGL62) − CT(ACTIN2). We next calculated an average DCT value for each tissue. Silique tissue with the highest relative expression (lowest DCT value) was used as the standard for comparison of expression levels. We then calculated relative expression levels using the equation 100 × 2−(average DCT (tissue) – average DCT (silique).
Characterization of the agl62 Alleles
To characterize the left- and right-border T-DNA junctions, we performed PCR using T-DNA- and genomic-specific primers and determined the DNA sequences of the PCR products. The T-DNA in agl62-1 is inserted into the intron, 474 nucleotides downstream of the start codon, and is associated with a 75-bp deletion in the intron (nucleotides +474 to +548 deleted). The T-DNA in agl62-1 has a left border oriented toward the 3′ end of AGL62 and a right border oriented toward the 5′ end of AGL62. The right-border junction was determined using the T-DNA primer pBinProK2-RB1 (5′-TCAGTTCCAAACGTAAAACGGC-3′) combined with the genomic primer agl62-1rp (5′-AGTTGTGTTCTCACCTGGTCG-3′), and the left-border junction was determined using T-DNA primer LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) and genomic primer agl62rp (5′-CAAGAACAAGAAAAACAACAACAAC-3′).
The T-DNA in agl62-2 is inserted into the predicted second exon, 620 nucleotides downstream of the start codon, and is associated with a 9-bp deletion in the predicted second exon (nucleotides +620 to +628 deleted). The T-DNA in agl62-2 has a left border oriented toward the 3′ end of AGL62 and a right border oriented toward the 5′ end of AGL62. The right-border junction was determined using the T-DNA primer pBinProK2-RB1 (described above) combined with the genomic primer agl62-2lp (5′-TGGATCTTTCTGGCAGATTTG-3′), and the left-border junction was determined using T-DNA primer LBa1 (described above) and genomic primer agl62rp (described above).
The T-DNA in agl62-3 is inserted into the second exon, 1009 nucleotides downstream of the start codon, and is associated with a 12-bp deletion in the predicted second exon (nucleotides +1009 to +1020 deleted). The T-DNA in agl62-3 has two left borders. The left border toward the 5′ end of AGL62 was determined using the T-DNA primer LBa1 (described above) combined with the genomic primer agl62-3rp (5′-AGAAAAGACAAAAGCCCTTGG-3′), and the left border toward the 3′ end of AGL62 was determined using the T-DNA primer LBa1 (described above) combined with the genomic primer agl62-3lp (5′-TTTTGCTTGATTTTGAAAATTTC-3′).
Segregation Analysis
agl62/AGL62 females were either self-crossed or were crossed with wild-type males. In both cases, progeny seed was collected, and the genotypes of the progeny plants were assayed by both PCR and silique phenotype. For PCR genotyping, primers agl62lp (5′-ATTTGGTTTGTAATATTCTGCT-3′) and agl62rp (described above) were used to identify the wild-type allele, and primers LBa1 (described above) and agl62rp (described above) were used to identify the mutant alleles (both agl62-1 and agl62-2). For genotyping by silique phenotype, siliques were opened and seed set was scored; plants containing 25% defective seeds were scored as agl62/AGL62, and plants containing 100% green seeds were scored as AGL62/AGL62. Statistical analyses of these data were performed using the χ2 test.
Molecular Complementation
Molecular complementation was performed using pBI-AGL62-GFP (described above) as a rescue construct. Females homozygous for the pBI-AGL62-GFP construct were crossed with heterozygous agl62 males. Three AGL62-GFP transgenic lines (lines 1, 3, and 4) were used in these crosses. Line 1 expressed AGL62-GFP strongly, and lines 3 and 4 expressed AGL62-GFP relatively weakly. Line 1 was crossed with agl62-1, and lines 3 and 4 were crossed with agl62-2. In all cases, progeny seed was collected and the genotypes of the progeny plants were assayed by PCR to identify plants heterozygous for agl62 and hemizygous for pBI-AGL62-GFP. Primers LBa1 (described above) and agl62rp (described above) were used to identify the mutant alleles, and primers AGL62seq5 (5′-AAGCCCTTGGGAATTGGT-3′) and pBI101.gfp (5′-GCCGGTGGTGCAGATGAACT-3′) were used to identify the rescue construct. These F1 plants were allowed to self-pollinate, and the progeny were analyzed by PCR to identify plants homozygous/heterozygous for agl62-1 and containing the rescue construct. With the agl62-1 cross, these plants had full seed set. With the agl62-2 crosses, plants homozygous for both the mutant allele and the rescue construct were identified through analysis of progeny using PCR (identified lines in which 100% of the progeny contained agl62-2 and the rescue construct). These plants had ∼70% seed set. The partial rescue exhibited in the agl62-2 crosses is likely due to the weak expression of the AGL62-GFP transgenic lines (transgenic lines 3 and 4).
Confocal Analysis of the agl62 Phenotype
CLSM of seeds was performed as described previously (Christensen et al., 1997, 1998, 2002), with the modification that we used a Zeiss LSM510 microscope. The number of nuclei in wild-type and agl62-1 seeds was obtained by analyzing serial optical sections of seeds using images captured on the Zeiss LSM510 microscope with Zeiss LSM Image Browser software version 3.5.0.376. The average number of nuclei was calculated from nine seeds from at least three plants. To ensure seeds were at similar developmental stages, images were captured from the 10 seeds located proximal to the stigma.
Analysis of AGL62-GFP Expression in fis Seeds
We crossed plants homozygous for AGL62-GFP as males with fie-1/FIE (Ohad et al., 1999), fis2-8/FIS2 (Wang et al., 2006), and mea-3/MEA (Kiyosue et al., 1999) females and scored the number of seeds expressing AGL62-GFP at 3 to 7 d after pollination. Tissue was dissected and expression was scored as described above. For each time point, we scored >105 seeds from each of three siliques from at least two different plants. Plants were scored as described above.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EU493093 (AGL62).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Yeast Two-Hybrid Analysis of AGL62–AGL80 Interaction.
Supplemental Figure 2. Expression of the Paternally Derived Allele of AGL62-GFP during Early Endosperm Development.
Supplemental Figure 3. Silique Phenotypes of agl62-1 and agl62-2 Mutants at 10 d after Pollination.
Supplemental Figure 4. Silique Phenotypes of Self-Pollinated agl62-1/AGL62 and agl62-2/AGL62 Plants at 2 to 10 d after Pollination.
Supplemental Table 1. Expression of the Maternal and Paternal Alleles of AGL62-GFP during Endosperm Development.
Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Ramin Yadegari, Anna Koltunow, and members of the Drews lab for critical review of this manuscript. We thank Ramin Yadegari for providing the fie-1, fis2-8, and mea-3 seeds and the pBI-GFP(S65T) vector. We thank Ed King and the Department of Biology Microscopy Facility for guidance with the microscopy analysis. This work was supported by a Department of Energy grant (DE-FG02-04ER15620) to G.N.D and a National Institutes of Health Developmental Biology Training grant appointment (5T32HD007491-12) to J.G.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gary N. Drews (drews@bioscience.utah.edu).
Online version contains Web-only data.
References
- Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H.G., and Scott, R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127 2493–2502. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J., and Berger, F. (2001). Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocklehurst, P.A. (1977). Factors controlling grain weight in wheat. Nature 266 348–349. [Google Scholar]
- Brown, R.C., Lemmon, B.E., and Nguyen, H. (2003). Events during the first four rounds of mitosis establish three developmental domains in the syncytial endosperm of Arabidopsis thaliana. Protoplasma 222 167–174. [DOI] [PubMed] [Google Scholar]
- Brown, R.C., Lemmon, B.E., Nguyen, H., and Olsen, O.-A. (1999). Development of endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 12 32–42. [Google Scholar]
- Bushell, C., Spielman, M., and Scott, R.J. (2003). The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojecki, A.J.S., Bayliss, M.W., and Gale, M.D. (1986). Cell production and DNA accumulation in the wheat endosperm, and their association with grain weight. Ann. Bot. (Lond.) 58 809–817. [Google Scholar]
- Christensen, C.A., Gorsich, S.W., Brown, R.H., Jones, L.G., Brown, J., Shaw, J.M., and Drews, G.N. (2002). Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14 2215–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10 49–64. [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202 136–151. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cooper, D.C. (1951). Caryopsis development following matings between diploid and tetraploid strains of Zea mays. Am. J. Bot. 38 702–708. [Google Scholar]
- de Folter, S., and Angenent, G.C. (2006). trans meets cis in MADS science. Trends Plant Sci. 11 224–231. [DOI] [PubMed] [Google Scholar]
- de Folter, S., Busscher, J., Colombo, L., Losa, A., and Angenent, G.C. (2004). Transcript profiling of transcription factor genes during silique development in Arabidopsis. Plant Mol. Biol. 56 351–366. [DOI] [PubMed] [Google Scholar]
- de Folter, S., Immink, R.G., Kieffer, M., Parenicova, L., Henz, S.R., Weigel, D., Busscher, M., Kooiker, M., Colombo, L., Kater, M.M., Davies, B., and Angenent, G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D., Fitz Gerald, J.N., and Berger, F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D., Saingery, V., Chambrier, P., Mayer, U., Jurgens, G., and Berger, F. (2003). Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Guitton, A.E., Page, D.R., Chambrier, P., Lionnet, C., Faure, J.E., Grossniklaus, U., and Berger, F. (2004). Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131 2971–2981. [DOI] [PubMed] [Google Scholar]
- Hayes, T.E., Sengupta, P., and Cochran, B.H. (1988). The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 2 1713–1722. [DOI] [PubMed] [Google Scholar]
- Ingouff, M., Haseloff, J., and Berger, F. (2005. a). Polycomb group genes control developmental timing of endosperm. Plant J. 42 663–674. [DOI] [PubMed] [Google Scholar]
- Ingouff, M., Fitz Gerald, J.N., Guerin, C., Robert, H., Sorensen, M.B., Van Damme, D., Geelen, D., Blanchoin, L., and Berger, F. (2005. b). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7 374–380. [DOI] [PubMed] [Google Scholar]
- Irish, V.F. (2003). The evolution of floral homeotic gene function. Bioessays 25 637–646. [DOI] [PubMed] [Google Scholar]
- Jones, R.J., Schreiber, B.M.N., and Roessler, J.A. (1996). Kernel sink capacity in maize: Genotypic and maternal regulation. Crop Sci. 36 301–306. [Google Scholar]
- Kaufmann, K., Melzer, R., and Theissen, G. (2005). MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 347 183–198. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Yadegari, R., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., Katz, A., Margossian, L., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji, R., Sumikawa, N., Yamasaki, M., Kondo, K., Ueda, K., Ito, M., and Hasebe, M. (2003). Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 20 1963–1977. [DOI] [PubMed] [Google Scholar]
- Kohler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., and Gruissem, W. (2003. b). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003. a). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, C., Page, D.R., Gagliardini, V., and Grossniklaus, U. (2005). The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37 28–30. [DOI] [PubMed] [Google Scholar]
- Lauber, M., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jurgerns, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, J., Zhang, J., and Cao, X. (2001). Grain sink strength may be related to the poor grain filling of indica-japonica rice (Oryza sativa) hybrids. Physiol. Plant. 112 470–477. [DOI] [PubMed] [Google Scholar]
- Lopes, M.A., and Larkins, B.A. (1993). Endosperm origin, development, and function. Plant Cell 5 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Dennis, E.S., Berger, F., Peacock, W.J., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich, G., Leroy, O., Akinci, U., Schubert, D., Clarenz, O., Goodrich, J., Grossniklaus, U., and Kohler, C. (2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1990). Endosperm cellularization in Arabidopsis thaliana L. Arab. Inf. Serv. 27 65–72. [Google Scholar]
- Martinez-Castilla, L.P., and Alvarez-Buylla, E.R. (2003). Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc. Natl. Acad. Sci. USA 100 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C., and Robaglia, C. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99 6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy, F., and Dubois, E. (2003). Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316 1–21. [DOI] [PubMed] [Google Scholar]
- Muller, S., Fuchs, E., Ovecka, M., Wysocka-Diller, J., Benfy, P., and Hauser, M. (2002). Two new loci, PLEIADE and HYADE, implicate organ specific regulation of cytokinesis in Arabidopsis. Plant Physiol. 130 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer, M.G., and Sheridan, W.F. (1980). Defective kernel mutants of maize. I. Genetic and lethality studies. Genetics 95 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H., Brown, R.C., and Lemmon, B.E. (2000). The specialized chalazal endosperm in Arabidopsis thaliana and Lepidium virginicum (Brassicaceae). Protoplasma 212 99–110. [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, O.-A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 S214–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, O.A. (2001). Endosperm development: Cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 233–267. [DOI] [PubMed] [Google Scholar]
- Otegui, M., and Staehelin, L.A. (2000). Syncytial-type cell plates: A novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12 933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M.S., Mastronarde, D.N., Kang, B.H., Bednarek, S.Y., and Staehelin, L.A. (2001). Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell 13 2033–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicova, L., de Folter, S., Kieffer, M., Horner, D.S., Favalli, C., Busscher, J., Cook, H.E., Ingram, R.M., Kater, M.M., Davies, B., Angenent, G.C., and Colombo, L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko, M.F., Lloyd, A., Steffen, J.G., Punwani, J.A., Otsuga, D., and Drews, G.N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley, M. (1978). Factors affecting grain enlargement in wheat. J. Exp. Bot. 29 919–934. [Google Scholar]
- Ray, S., Park, S.-S., and Ray, A. (1997). Pollen tube guidance by the female gametophyte. Development 124 2489–2498. [DOI] [PubMed] [Google Scholar]
- Reddy, V.M., and Daynard, T.B. (1983). Endopserm characteristics associated with rate of grain filling and kernel size in corn. Maydica 28 339–355. [Google Scholar]
- Robles, P., and Pelaz, S. (2005). Flower and fruit development in Arabidopsis thaliana. Int. J. Dev. Biol. 49 633–643. [DOI] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J., and Dickinson, H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 3329–3341. [DOI] [PubMed] [Google Scholar]
- Singh, B.K., and Jenner, C.F. (1982). Association between concentrations of organic nutrients in the grain, endosperm cell number and grain dry weight within the ear of wheat. Aust. J. Plant Physiol. 9 83–95. [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, M.B., Chaudhury, A.M., Robert, H., Bancharel, E., and Berger, F. (2001). Polycomb group genes control pattern formation in plant seed. Curr. Biol. 11 277–281. [DOI] [PubMed] [Google Scholar]
- Sorensen, M.B., Mayer, U., Lukowitz, W., Robert, H., Chambrier, P., Jurgens, G., Somerville, C., Lepiniec, L., and Berger, F. (2002). Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129 5567–5576. [DOI] [PubMed] [Google Scholar]
- Steffen, J.G., Kang, I.-H., Macfarlane, J., and Drews, G.N. (2007). Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51 281–292. [DOI] [PubMed] [Google Scholar]
- Strompen, G., Kasmi, F., Richter, S., Lukowitz, W., Assaad, F., Jurgens, G., and Mayer, U. (2002). The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr. Biol. 12 153–158. [DOI] [PubMed] [Google Scholar]
- Tilly, J.J., Allen, D.W., and Jack, T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125 1647–1657. [DOI] [PubMed] [Google Scholar]
- Treisman, R. (1992). The serum response element. Trends Biochem. Sci. 17 423–426. [DOI] [PubMed] [Google Scholar]
- Verelst, W., Saedler, H., and Munster, T. (2007). MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol. 143 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., and Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenoog, R., Spielman, M., Adams, S., Fischer, R.L., Dickinson, H.G., and Scott, R.J. (2000). Hypomethylation promotes autonomous endosperm development and rescues postfertilization lethality in fie mutants. Plant Cell 12 2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim, K.H., and Peterson, H.P. (2004). Aberrant endosperm development in interploidy crosses reveals a timer of differentiation. Dev. Biol. 270 277–289. [DOI] [PubMed] [Google Scholar]
- Wang, D., Tyson, M.D., Jackson, S.S., and Yadegari, R. (2006). Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 13244–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., Brown, R.C., Lemmon, B.E., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2006). Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 142 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y., Nakashima, K., Harada, J.J., Goldberg, R.B., Fischer, R.L., and Ohad, N. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12 2367–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Zhang, J., Huang, Z., Wang, Z., Zhu, Q., and Liu, L. (2002). Correlation of cytokinin levels in the endosperms and roots with cell number and cell division activity during endosperm development in rice. Ann. Bot. (Lond.) 90 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.K., Lee, J.S., and Ahn, J.H. (2006). Overexpression of AGAMOUS-LIKE 28 (AGL28) promotes flowering by upregulating expression of floral promoters within the autonomous pathway. Biochem. Biophys. Res. Commun. 348 929–936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.