Abstract
Catharanthus roseus is the sole commercial source of the monoterpenoid indole alkaloids (MIAs), vindoline and catharanthine, components of the commercially important anticancer dimers, vinblastine and vincristine. Carborundum abrasion technique was used to extract leaf epidermis–enriched mRNA, thus sampling the epidermome, or complement, of proteins expressed in the leaf epidermis. Random sequencing of the derived cDNA library established 3655 unique ESTs, composed of 1142 clusters and 2513 singletons. Virtually all known MIA pathway genes were found in this remarkable set of ESTs, while only four known genes were found in the publicly available Catharanthus EST data set. Several novel MIA pathway candidate genes were identified, as demonstrated by the cloning and functional characterization of loganic acid O-methyltransferase involved in secologanin biosynthesis. The pathways for triterpene biosynthesis were also identified, and metabolite analysis showed that oleanane-type triterpenes were localized exclusively to the cuticular wax layer. The pathways for flavonoid and very-long-chain fatty acid biosynthesis were also located in this cell type. The results illuminate the biochemical specialization of Catharanthus leaf epidermis for the production of multiple classes of metabolites. The value and versatility of this EST data set for biochemical and biological analysis of leaf epidermal cells is also discussed.
INTRODUCTION
Plant tissues are composed of various cell types with unique sizes, shapes, and biological functions that play different roles in normal plant growth, development, and reproduction. Each of these different roles calls for a different complement of proteins produced in each cell type. Among these cell types, the epidermis, composed of epidermal cells, trichomes, and guard cells, constitutes the surface layer of the plant that is directly exposed to the outside environment. The leaf epidermis usually constitutes a single layer of cells that serves as a protective barrier to environmental factors (i.e., UV light, water loss, herbivory, and pathogen attack). Similarly, the leaf epidermis also has specialized cells, such as glandular trichomes, that possess highly specialized systems for biosynthesis, secretion, and/or accumulation of toxic phytochemicals to defend plants against insects and pathogens. By contrast, guard cells are the important physical openings that control gas exchange between the plant tissue and the atmosphere. In underground plant parts, the root epidermis is involved in water uptake from soil, where these cells also encounter a variety of microorganisms in the rhizosphere.
Recently, the developmental regulation of epidermal layer cell patterning has been extensively studied to identify various genes that trigger the formation of guard cells and trichomes from parental epidermal cells (Martin and Glover, 2007). Unfortunately, the mechanisms involved in biochemical differentiation and global gene expression of epidermal cells remain largely unknown, partly due to perceived difficulties associated with selective isolation of leaf epidermal cells that are intimately associated with adjacent cells within the tissue. However, several protocols have been developed for the isolation of other leaf surface cells, like glandular trichomes that protrude on the leaf surface (Lange et al., 2000; Gang et al., 2002; Wagner et al., 2004).
Some biochemical features of epidermal cells have been well studied in the last few decades, mainly by chemical analysis and microscopy. Two key characteristics of epidermal cells include (1) the ubiquitous biosynthesis of cuticular wax for producing the protective barrier of the surface (Kunst and Samuels, 2003; Shepherd and Griffiths, 2006) and (2), with the exception of guard cells, the apparent lack of chlorophylls. The cuticle is composed of wax containing a mixture of very-long-chain fatty acids (VLFAs), primary and secondary alcohols, aldehydes, ketones and esters, and cutin polymer esters that are primarily composed of C16 and C18 unsaturated hydroxy fatty acid monomers. The extremely hydrophobic wax layer minimizes not only water loss from the plant body but also the growth of fungi or bacteria on the surface of the plant. By contrast, the chemistry of the cutin layer is slightly more hydrophilic than the wax layer, but its biological roles remain to be studied in detail. The chemical composition of wax and cutin layers as well as the morphology of wax crystalloids are quite variable among different plant species and therefore have been used for taxonomical analysis (Barthlott et al., 1998). Apart from guard cells, other epidermal cells do not usually contain significant levels of chlorophyll, but they have nongreen leucoplasts whose detailed biological functions have yet to be elucidated.
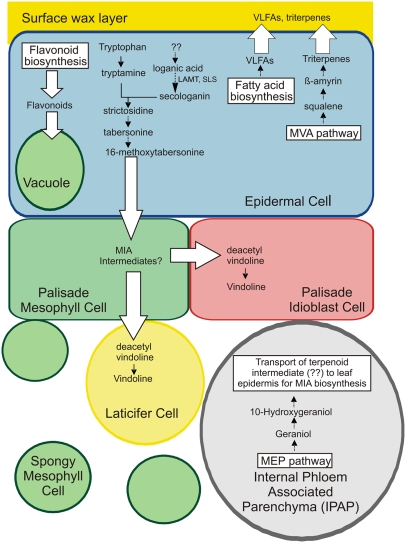
In most plant species, the epidermis also plays specialized roles in the biosynthesis and accumulation of a wide range of secondary metabolites, including flavonoids, terpenes, and alkaloids, as illustrated in recent localization studies (Dudareva et al., 2005; Kutchan, 2005; Murata and De Luca, 2005; Mahroug et al., 2006). In the case of Catharanthus roseus (Madagascar periwinkle), leaf epidermal cells appear to be specialized for monoterpenoid indole alkaloid (MIA) biosynthesis (St-Pierre et al., 1999; Murata and De Luca, 2005; Mahroug et al., 2006). This medicinal plant is the only commercial source of the valuable dimeric MIAs with anticancer activities vinblastine and vincristine, which are derived from combining vindoline and catharanthine monomers. Remarkably, studies using in situ RNA hybridization and immunolocalization techniques showed that vindoline biosynthesis occurred in at least three cell types (epidermal cells, idioblast cells, and laticifer cells) within the leaf (St-Pierre et al., 1999), with early steps occurring in leaf epidermal cells and terminal steps occurring in idioblast and laticifer cells found in the mesophyll leaf layer. More recently, laser capture microdissection (LCM) and carborundum abrasion technique (CA) (Murata and De Luca, 2005) were used to expand the importance of the epidermal cells as the primary site for biosynthesis of 16-methoxytabersonine from the secoiridoid, secologanin, and tryptamine. Further studies using in situ RNA hybridization showed that three genes involved in the plastid localized 2-C-methyl-d-erythritol-4-phosphate (MEP) pathway and geraniol-10-hydroxylase (G10H) that commits this pathway to the production of secoiridoids were preferentially expressed in the internal phloem-associated parenchyma (IPAP) of leaves. These latter studies implied that phloem parenchyma cells participated in MIA biosynthesis by supplying an unknown isoprenoid pathway intermediate that would be translocated to the leaf epidermis for elaboration into secologanin and into MIAs (Burlat et al., 2004; Mahroug et al., 2006).
To obtain novel genes involved in MIA biosynthesis, a useful approach for nonmodel systems like Catharanthus involves random sequencing and development of ESTs from cDNA libraries prepared from selected tissues. Random sequencing of Catharanthus cDNA libraries prepared from whole organs (Murata et al., 2006; Shukla et al., 2006) or from induced cell cultures (Rischer et al., 2006) have produced potentially valuable candidate genes for MIA biosynthesis; virtually no MIA pathway genes have been identified and functionally characterized using these approaches. These results suggest that the MIA pathway was poorly represented in the RNA populations derived from leaf, root, and induced cell cultures and that new approaches like LCM and CA technique (Murata and De Luca, 2005; Murata et al., 2006) were required to harvest cell types specialized in MIA biosynthesis.
This report applies CA to produce a leaf epidermis–enriched cDNA library that samples the Catharanthus epidermome that is the complement of genes expressed in the leaf epidermis. Because of the unique developmental and metabolic roles of the epidermis, the epidermome is predicted to contain genes for important pathways not expressed in other tissues of the plant. This work shows that the moderate population of 3655 ESTs (CROLF1NG data set) produced by random sequencing contains essentially all the known MIA pathway genes that are known to be preferentially expressed in Catharanthus leaf epidermis, including tabersonine-16-hydroxylase (T16H) (Schröder et al., 1999), but not deacetylvindoline-4-hydroxylase (D4H) nor deacetylvindoline-4-O-acetyltransferase (DAT), which are known to be preferentially expressed in Catharanthus idioblasts and laticifers (St. Pierre et al., 1999). Furthermore, the EST data set has been used to identify, clone, and functionally characterize loganic acid O-methyltransferase (LAMT) involved in secologanin biosynthesis described here, as well as 16-hydroxytabersonine 16-O-methyltransferase (16OMT) that catalyzes the fifth to last step in the tabersonine-to-vindoline pathway (Levac et al., 2008). The CROLF1NG data set also contains genes involved in the mevalonic acid (MVA) pathway, together with post-isopentenyl diphosphate (IPP) biosynthesis genes and oleanane triterpene biosynthesis genes. The exclusive localization of the triterpenes ursolic and oleanolic acid to the cuticular wax layer provided a basis for the leaf epidermis–localized expression of this pathway. These data suggest that epidermal cells are involved in the biosynthesis of triterpenes as well as the monoterpene component of MIAs. The identification the entire pathways for flavonoid and VLFA biosynthesis further demonstrated the high value of CA-based leaf epidermis–enriched cDNA libraries. The results clearly illustrate the biochemical specialization of Catharanthus leaf epidermis for the production of multiple classes of metabolites (i.e., MIAs, flavonoids, triterpenes, and VLFAs) and predict the putative mechanisms for directed transport of various metabolites from the epidermal cells to proper destinations: some MIAs to laticifer/idioblast cells, the triterpenes and VLFAs to the leaf surface, and the flavonoids to the plant vacuole. The value and versatility of this EST data set for biochemical and biological analysis of leaf epidermal cells also highlights the versatility of CA as a unique tool to identify the complex biochemical pathways associated with leaf epidermal cells.
RESULTS
Construction of the Leaf Epidermis–Enriched Catharanthus cDNA Library
To study the biochemical specialization of Catharanthus leaf epidermal cells, the CA technique was used to extract mRNA (Murata and De Luca, 2005) for producing a leaf epidermis–enriched cDNA library. Briefly, the upper and lower surface of young Catharanthus leaves (1.5 cm in length) were selectively abraded using a cotton swab coated with carborundum particles, and the leaf was dipped into Trizol reagent to release mRNA from leaf tissue. This RNA was then used to make a leaf epidermis–enriched cDNA library, and 8527 colonies were randomly chosen for sequencing to produce 3655 unique sequences composed of 1142 clusters and 2513 singletons. The average length of unique sequences from this library was 431.5 bp after removing low quality clones and vector sequences. Genes were annotated accord to their putative functions based on their similarity to sequences in the GenBank database using the BLASTX program coordinated by the FIESTA gene annotation system (http://bioinfo.pbi.nrc.ca/napgen.beta//login.html) developed at the Plant Biotechnology Institute (Saskatoon, Canada). This data set was named CROLF1NG according to rules developed by the Natural Products Genomic Resource (NAPGEN) consortium (http://pbi-ibp.nrc-cnrc.gc.ca/en/CEHH/napgen.htm) at the Plant Biotechnology Institute. The validity of putative annotations was checked manually through the FIESTA system.
Data Mining of the CROLF1NG Data Set and Categorizing the Genes of Interest
After annotations for each gene were verified manually, they were categorized into genes involved in the biosynthesis of (1) MIAs, (2) isoprenoid precursors, (3) other terpenoids, (4) flavonoids, and (5) lipids. Since many MIA biosynthetic genes remain to be cloned, additional genes encoding certain classes of putative enzymes were also annotated, including (6) methyltransferases, (7) acyltransferases, (8) cytochrome P450-dependent monooxygenases (CYPs), (9) glycosyltransferases, (10) dioxygenases, and (11) transcription factors.
The CROLF1NG Data Set Contains Virtually All Known MIA Biosynthetic Genes
Detailed analysis established that the CROLF1NG data set was a particularly rich source of genes for MIA biosynthesis, based on the representation of most functionally characterized genes, from the pathway for secoiridoid biosynthesis to the early steps in the conversion of tabersonine into vindoline. This included 10-hydroxygeraniol oxidoreductase (10HGO), secologanin synthase (SLS), cytochrome P450 reductase (CPR), tryptophan decarboxylase (TDC), strictosidine synthase (STR), strictosidine β-glucosidase (SGD), T16H, octadecanoid-derivative responsive Catharanthus AP2-domain3 (ORCA3), box P binding factor 1 (BPF1), and zinc finger Catharanthus transcription factor 2 (ZCT2) (Figure 1, Table 1). It was quite significant that SLS, TDC, SGD, and T16H, which have been shown by various localization studies to be expressed exclusively in Catharanthus leaf epidermal cells (St-Pierre et al., 1999; Irmler et al., 2000; Murata and De Luca, 2005), were also identified several times in the CROLF1NG data set. This is particularly striking with the SLS contig (CL19Contig4), which is represented by 25 ESTs and is considerably enriched in the leaf epidermis. These results dramatically show the effectiveness and the value of CA technique in constructing a leaf epidermis–enriched cDNA library and the specialization of this cell type for MIA biosynthesis.
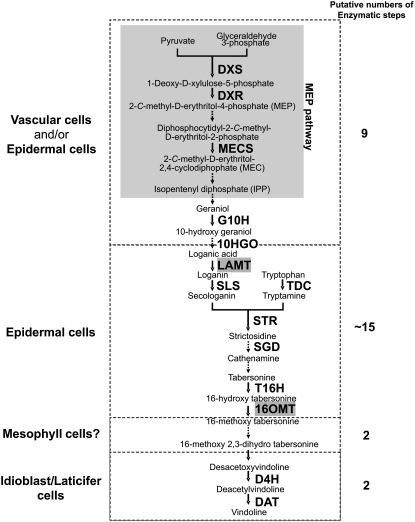
Figure 1.
Vindoline Biosynthesis and Its Proposed Localization within the Leaf in C. roseus.
Among ∼30 enzymatic steps involved in vindoline biosynthesis, 17 genes encoding known MIA biosynthesis enzymes and transcription factors and the recently cloned and characterized 16OMT (Levac et al., 2008) and LAMT described in this article are represented in the CROLF1NG data set. The leaf epidermal localization of 16OMT and LAMT (highlighted in gray) expands the list of genes and enzymes expressed in this tissue and highlights it as the major site of MIA biosynthesis up to 16-methoxytabersonine. Putative numbers of the enzymatic reactions in each cell type are shown. CPR is believed to be involved in all the reactions catalyzed by cytochrome P450 monooxygenases. DXS, 1-deoxy-d-xylulose 5-phosphate synthase; DXR, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; MECS, 2-C-methyl-d-erythritol-2,4-cyclodiphosphate synthase; G10H, geraniol 10-hydroxylase; LAMT, loganic acid methyltransferase; SGD, strictosidine β-glucosidase; T16H, tabersonine-16-hydroxylase; 16OMT, 16-hydroxytabersonine-O-methyltransferase; BPF1, box P binding factor 1; ZCT2, zinc finger Catharanthus transcription factor 2.
Table 1.
CROLF1NG Genes with Significant Similarity to Known MIA Biosynthetic Genes
| Gene | Category | Name | Description | Hit ID | E-Value | No. of ESTs |
|---|---|---|---|---|---|---|
| SLS (CYP72A1) | CYP | CL19Contig4 | Cytochrome P-450 protein (Catharanthus) | gi|167484| | 0 | 25 |
| G10H | CYP | 258-D02 | Geraniol-10-hydroxylase | gi|17065916| | 1.00E-121 | 1 |
| 10HGO-type | Dehydrogenase | CL24Contig1 | 10-Hydroxygeraniol oxidoreductase (Catharanthus) | gi|34013695| | 1.00E-114 | 24 |
| G10H-like | CYP | CL34Contig1 | Cytochrome P450 (Catharanthus) | gi|12657333| | 1.00E-138 | 18 |
| 10HGO-type | Dehydrogenase | CL36Contig1 | 10-Hydroxygeraniol oxidoreductase (Catharanthus) | gi|34013695| | 1.00E-103 | 17 |
| TDC | Decarboxylase | CL123Contig1 | TDC (Catharanthus) | gi|18226| | 1.00E-145 | 8 |
| CYP72B | CYP | CL19Contig3 | Cytochrome P450 | gi|404688| | 1.00E-101 | 5 |
| CPR | CYP | CL224Contig1 | NADPH–ferrihemoprotein reductase (Catharanthus) | gi|18139| | 1.00E-110 | 5 |
| SGD | β-Glucosidase | CL267Contig1 | Strictosidine β-glucosidase (Catharanthus) | gi|6840855| | 3.00E-99 | 4 |
| G10H-like | CYP | CL241Contig1 | Cytochrome P450 (Catharanthus) | gi|12657333| | 1.00E-102 | 4 |
| T16H | CYP | CL288Contig1 | Cytochrome P450 (Catharanthus) | gi|5921278| | 1.00E-109 | 4 |
| CYP72C | CYP | CL19Contig2 | Cytochrome P450 | gi|404690| | 1.00E-45 | 3 |
| CYP72C | CYP | CL19Contig1 | Cytochrome P450 | gi|404690| | 1.00E-102 | 2 |
| ZCT3-type | TF, ZF | CL972Contig1 | Zinc finger DNA binding protein (Catharanthus) | gi|55734108| | 2.00E-78 | 2 |
| PR | Reductase | CL578Contig1 | Perakine reductase (R. serpentina) | gi|59896631| | 1.00E-168 | 4 |
| G10H-like | CYP | 027-H04 | Cytochrome P450 (Catharanthus) | gi|12657333| | 3.00E-78 | 1 |
| CYP72C | CYP | 110-F10 | Cytochrome P450 | gi|404690| | 1.00E-130 | 1 |
| 10HGO-like | Dehydrogenase | 145-C09 | Geraniol dehydrogenase (Ocimum basilicum) | gi|62461968| | 9.00E-75 | 1 |
| 10HGO | Dehydrogenase | 056-A09 | 10-Hydroxygeraniol oxidoreductase (Catharanthus) | gi|34013695| | 4.00E-26 | 1 |
| 10HGO-like | Dehydrogenase | 165-G01 | 10-Hydroxygeraniol oxidoreductase (Camptotheca acuminata) | gi|33519154| | 2.00E-49 | 1 |
| STR | Others | 032-G06 | STR (Ophiorrhiza pumila) | gi|13928598| | 9.00E-31 | 1 |
| STR | Others | 164-E07 | STR precursor (Catharanthus) | gi|18222| | 1.00E-100 | 1 |
| ORCA3-type | TF, AP2 | 015-D04 | AP2-domain DNA binding protein (Catharanthus) | gi|8980315| | 6.00E-29 | 1 |
| ORCA3 | TF, AP2 | 118-A07 | AP2-domain DNA binding protein (Catharanthus) | gi|8980315| | 4.00E-75 | 1 |
| BPF1 | TF, MYB | 128-A08 | MYB-like DNA binding protein (Catharanthus) | gi|12043533| | 8.00E-78 | 1 |
| ZCT2 | TF, ZF | 158-E02 | Zinc finger DNA binding protein (Catharanthus) | gi|55734106| | 2.00E-19 | 1 |
Genes that are shown in bold are the clones with perfect identity at amino acid level with known and functionally characterized Catharanthus genes.
The representation of functionally characterized MIA pathway genes in the CROLF1NG data set also strongly suggests that other novel and uncharacterized pathway genes are very likely to be represented. For example, there are four different clones that belong to the CYP72 family of cytochrome P450 monooxygenases (Table 1). Since these genes have sequences that are extremely similar to (>95% identity at the amino acid level), but slightly different from SLS, they may encode 7-deoxyloganin hydroxylase that preceeds SLS (Yamamoto et al., 1999, 2000; Irmler et al., 2000). The functional characterization of these candidate genes will help to complete our understanding of the terminal two steps in secologanin biosynthesis in Catharanthus and in other species of plants that accumulate these secoiridoid compounds.
It is interesting that a single EST for G10H (CAC80883; Collu et al., 2001) was represented in the CROLF1NG data set as well as two additional CYP ESTs with significant similarity to a gene annotated as G10H (CAC27827; Meijer et al., 1993) but whose biochemical function has yet to be described. However, this clone (CAC27827) is not likely to catalyze the G10H reaction since it was more similar in sequence to other CYPs, including T16H, than to the functionally characterized G10H clone (CAC80883). In general, the ESTs that encoded MIA biosynthesis enzymes were represented several times compared with those of the three transcription factors (ORCA3, BPF1, and ZCT2) (van der Fits and Memelink, 2000; van der Fits et al., 2000; Pauw et al., 2004) that were only represented once. These relative differences implied that transcription factors were expressed at lower levels than those of the MIA pathway enzymes that they regulate (Table 1) and that the overall profile of mRNA expression in the leaf epidermal cells of Catharanthus was largely maintained in the CROLF1NG data set.
The CROLF1NG Data Set Contains the Genes for the MVA and Triterpenoid Biosynthesis Pathways
It is well known that plants possess two different biosynthetic pathways for making IPP, the key building block of all isoprenoids, including pigments (chlorophylls and carotenoids), phytohormones (gibberellins), sterols, and other terpenes (Rohmer et al., 1993; Lichtenthaler, 1999). It has been suggested that the cytosolic MVA pathway produces precursors for sterols, triterpenes, and polyterpenes, while the plastidic MEP pathway supplies IPP precursors for chlorophyll, carotenoid, phytol, monoterpene, and gibberellin (Lichtenthaler, 1999) biosynthesis. It is remarkable that one MEP pathway EST (4-diphosphocytidyl-2-C-methyl-d-erythritol synthase), four MVA pathway ESTs (HMGR, AACT1, AACT2, and HMGS), and three late isoprenoid pathway ESTs (GPS, GPPS, and IPPI) (see Supplemental Table 1 online) were represented in the CROLF1NG data set.
The enrichment in MVA pathway genes (13 ESTs in total) compared with the single MEP pathway EST identified strongly suggests that the leaf epidermis of Catharanthus expressed higher levels of the cytosolic MVA pathway. Since Catharanthus accumulates considerable amounts of the oleanane-type triterpenes, oleanolic and ursolic acid (Usia et al., 2005), it is possible that the MVA pathway in leaf epidermis is involved in their biosynthesis. The CROLF1NG data set also contained squalene monooxygenase (five ESTs), a key enzyme for sterol and triterpene biosynthesis and 13 β-amyrin synthase-like ESTs that may commit oxidosqualene to the formation of oleanane-type triterpenes (see Supplemental Table 2 online). The representation of these transcripts strongly implies that the leaf epidermis of Catharanthus may also be specialized for triterpene biosynthesis and accumulation.
The CROLF1NG Data Set Contains the Genes for Flavonoid Biosynthesis-Related Genes
Many genes involved in flavonoid biosynthesis were also found in the CROLF1NG data set (see Supplemental Table 3 online). Among the ESTs reported, only cinnamate 4-hydroxylase (C4H) had complete sequence identity to a functionally characterized C4H previously reported in Catharanthus (Hotze et al., 1995), whereas the other ESTs are novel with considerable similarity to known flavonoid pathway genes described in other plant species that include phenylalanine ammonia lyase (PAL), chalcone synthase, flavanone 3-hydroxylase, and 2′-hydroxy isoflavone/dihydroflavonol reductase (DFR). These data were not surprising since flavonoids appear to play vital roles in the control of epidermal cell fate (reviewed in Broun, 2005), have long been proposed to protect plants against UV-B radiation (Schmitz-Hoerner and Weissenböck, 2003), and have been shown to accumulate within the leaf epidermis of Catharanthus by microscopy (Mahroug et al., 2006). Together, these data suggest that Catharanthus epidermis is a primary site for flavonoid biosynthesis.
It is also interesting that another EST (CL314Contig1) is represented four times in the CROLF1NG data set and is annotated as a putative flavonoid O-methyltransferase (OMT) with 77% similarity to a previously functionally characterized flavonoid 4′-OMT (AAR02420; Schröder et al., 2004). By contrast, this EST was not represented in the CrUniGene set established from sequencing of cDNA libraries prepared from the base part of the leaf and from the root tip (Murata et al., 2006). The public database did contain one EST sequence (EG555799) from a C. roseus flower bud cDNA library identical to CL314Contig1, whereas the root tip library was only represented with OMT2 and OMT4 that had been previously cloned and functionally characterized as flavonoid OMTs. This novel OMT contig turned out to encode 16OMT (Levac et al., 2008).
The CROLF1NG Data Set Contains Many Genes for Lipid Biosynthesis
Leaf epidermal cells are important sites of biosynthesis of VLFA lipids that are secreted into cell walls to form the impermeable cuticular wax layer. Many fatty acid biosynthesis-related genes were represented in the CROLF1NG data set, including: fatty acyl-ACP thioesterase, fatty acyl-CoA synthases, fatty acid desaturase, carboxylic acid ester hydrolase, phospholipase D, and lipid transfer protein (LTP) (see Supplemental Table 4 online). Among these, the number of ESTs encoding LTP was quite high (121 ESTs), suggesting that this type of protein might be very abundant in the epidermal cells of young leaves. LTPs are composed of two subfamilies, LTP1 and LTP2, which are remotely related to each other by sequence similarity. LTPs bind to different types of fatty acids and hydrophobic substances in their hydrophobic groove in a noncovalent manner. Although there is much to be studied to assign in vivo functions of LTPs, this class of proteins is implicated in various biological processes, including cuticle biosynthesis. It has been claimed that some LTPs are localized preferentially to the cell wall of epidermal cells or in cuticular wax layer of various plants (Sterk et al., 1991; Pyee et al., 1994; Thoma et al., 1994). Moreover, barley (Hordeum vulgare) seedlings accumulate cuticular wax and express LTPs in response to the abiotic stress caused by heavy metals (Hollenbach et al., 1997). These previous observations support our result that LTPs are abundantly expressed in the leaf epidermis of Catharanthus. In addition, the putative ortholog (92% identity at the amino acid level) to Arabidopsis thaliana 3-ketoacyl CoA synthase (CUT1) was represented with seven ESTs in CL135Contig1. Previous studies showed that CUT1 was involved in VLFA biosynthesis and that it was expressed specifically in leaf epidermal cells (Millar et al., 1999).
The CROLF1NG Data Set Contains Genes for S-Adenosyl-l-Methionine–Dependent Methyltransferases
Several classes of genes were also identified that could be potential candidates for identifying novel MIA genes that remain to be characterized. In addition to 16OMT (CL314Contig1) described above, 20 putative OMTs were annotated, including three ESTs with identical sequences to known Catharanthus genes: methionine synthase (CL69Contig1) (Eichel et al., 1995), S-methyltransferase (169-E03) (Coiner et al., 2006), and caffeic acid OMT (101-B09) (Schröder et al., 2002). It is well known that the MIA pathway leading to vindoline biosynthesis involves three separate methyltransferases, loganic acid OMT (LAMT; Madyastha et al., 1973), 16-hydroxytabersonine-16-OMT (16OMT; Fahn et al., 1985; Cacace et al., 2003), and 16-methoxy-2,3-dihydro-3-hydroxytabersonine N-methyltransferase (NMT) (De Luca et al., 1987), that remain to be cloned and functionally characterized. Previous studies have suggested that LAMT might be expressed in the leaf epidermis, since SLS converts the LAMT reaction product into secologanin (Irmler et al., 2000; Murata and De Luca, 2005). Inspection of the CROLF1NG data set revealed a full-length cDNA (see Supplemental Table 5 online; CL57Contig1 composed of 12 ESTs) encoding a putative AdoMet-dependent carboxyl methyltransferase such as LAMT. However, the CROLF1NG data set is not expected to produce candidate genes for NMT, an activity associated with chloroplast thylakoid membranes within leaf mesophyl cells rather than with leaf epidermal cells (De Luca and Cutler, 1987; Murata and De Luca, 2005).
The CROLF1NG Data Set Contains Acyltransferases, Cytochrome P450 Monooxygenases, Glycosyltransferases, Dioxygenases, and Transcription Factors
The database contains a significant number of ESTs encoding acyltransferases (see Supplemental Table 6 online), Cytochrome P450 monooxygenases (see Supplemental Table 7 online), glycosyltransferases (see Supplemental Table 8 online), dioxygenases (see Supplemental Table 9 online), and transcription factors (see Supplemental Table 10 online). This rich source of genes promises to hold a number of MIA pathway and regulatory genes that need to be functionally characterized in future research.
Functional Characterization of a Novel LAMT Obtained from the CROLF1NG Data Set
The last step in loganin biosynthesis is catalyzed by LAMT, which has only been partially purified and characterized (Madyastha et al., 1973) (Figure 2). To demonstrate the value of the CROLF1NG data set, a full length candidate gene, obtained by overlapping 12 separate ESTs (C57Contig1; see Supplemental Table 5 online), was cloned and functionally characterized. This unique sequence was not found in the EST data sets obtained from cDNA libraries produced from young leaves or from root tips (CrUniGene database; Murata et al., 2006) nor from other Catharanthus tissues described in the public database. The clone was 1396 bp long and exhibited moderate similarities (44% amino acid sequence identity) to a putative carboxyl methyltransferase from Medicago truncatula (ABE92230) (see Supplemental Table 5 online). Since LAMT may catalyze a reaction preceding SLS shown to be expressed in the leaf epidermis (Irmler et al., 2000; Murata and De Luca, 2005), it was possible that C57Contig1 (Figure 3A) encoded LAMT.
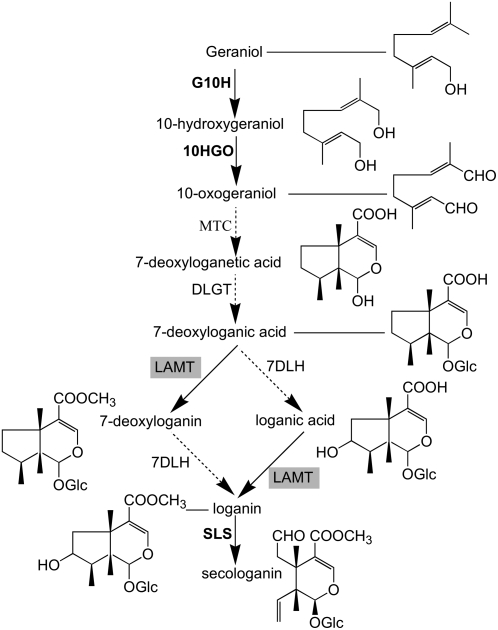
Figure 2.
The Proposed Pathway for Secologanin Biosynthesis in Catharanthus.
Enzymes whose corresponding genes have been cloned are shown in bold. LAMT, which is cloned and functionally characterized in this study, is shown in a gray box. Dotted arrows represent uncharacterized enzymatic steps. G10H, geraniol-10-hydroxylase; MTC, monoterpene cyclase; DLGT, deoxyloganetic acid-O-glucosyltransferase; LAMT, loganic acid-O-methyltransferase; 7DLH, 7-deoxyloganic acid hydroxylase.
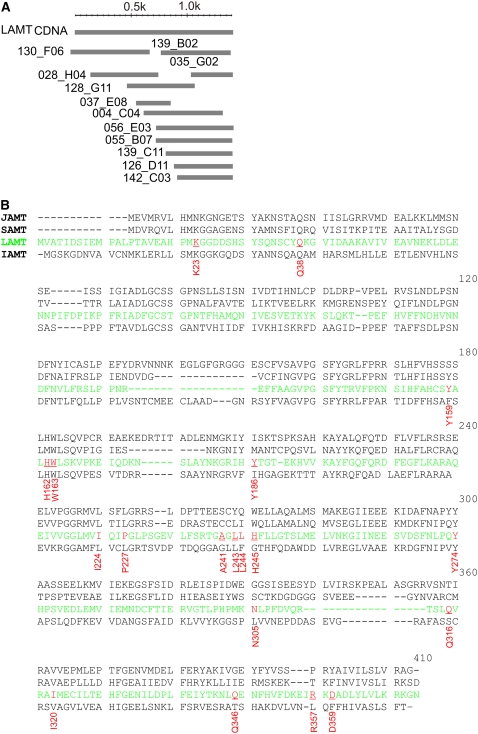
Figure 3.
Identification and Functional Characterization of Loganic Acid Methyltransferase from Catharanthus.
(A) Representation of CL57Contig1 composed of 13 ESTs that encode the full-length LAMT obtained from the CROLF1NG data set. The consensus sequence contains a putative open reading frame with significant similarity to known carboxyl methyltransferases.
(B) Protein alignment of JAMT from Arabidopsis (Q9AR07), SAMT from C. breweri (Q9SPV4), LAMT from Catharanthus (highlighted in green) (EU057974), and IAMT from Arabidopsis (NP20036). Highlighted in red are important active-site residues that form the hydrophobic core, based on previously reported active-site residues of the SAMT crystal structure (Zubieta et al., 2003). Interesting differences between LAMT and JAMT, and SAMT and IAMT are denoted by an acidic (Asp) and basic (His) amino acid substitution in amino acids 359 and 245, respectively.
The putative open reading frame of LAMT (Figure 3B) aligns to three (35, 36, and 36% amino acid sequence identity) previously characterized carboxyl methyltransferases from the SABATH family: Salicylic Acid Methyltransferase (SAMT) (Ross et al., 1999) from Clarkia breweri and Jasmonic Acid Methyltransferase (JAMT) (Seo et al., 2001) and Indole-3-Acetic Acid Methyltransferase (IAMT) (Qin et al., 2005) from Arabidopsis. Based on the crystal structure of SAMT (Zubieta et al., 2003), amino acid residues composing the putative hydrophobic active site of LAMT include Y159, W163, I224, P227, L244, H245, I320, and D359 (Figure 3B). Within these active-site residues, LAMT has two notable substitutions where the nonpolar aromatic residues W226 and F347 of SAMT are replaced with the polar-charged residues H245 and D359 (Figure 3B).
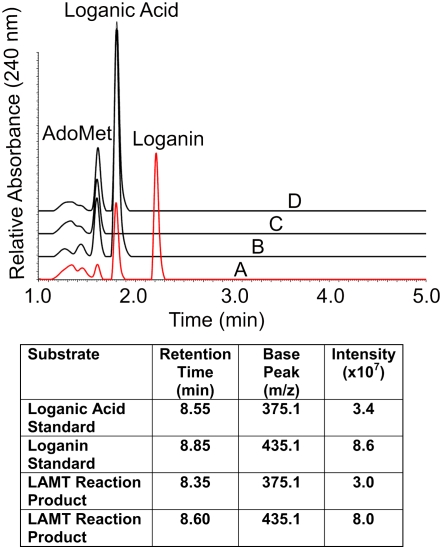
To test the activity of the C57Contig1 gene product, the corresponding cDNA clone was isolated by PCR amplification using specific primers based on the C57Contig1 sequence using the Catharanthus leaf base cDNA library as a template. The product was sequenced to obtain a 1113-bp product identical to C57Contig1 (Figure 3A) encoding a putative 371–amino acid open reading frame (Figure 3B). Recombinant LAMT protein was expressed in Escherichia coli cells to perform functional studies. Enzyme assays with crude recombinant protein, S-adenosyl-L-[methyl-14C]-methionine, and loganic acid produced a radioactive product corresponding to loganin (data not shown). These results were corroborated by repeating these assays at a larger scale and by isolating reaction products by ultraperformance liquid chromatography (UPLC) (Figure 4). Loganin production only occurred in the presence of recombinant enzyme, AdoMet and loganic acid (Figure 4, profile A), but not in reactions with boiled recombinant enzyme (Figure 4, profile B), in reactions lacking loganic acid (Figure 4, profile C), or in reactions with E. coli cell-free extracts expressing the empty vector (Figure 4, profile D). Mass spectra analysis (Figure 4, table) illustrated that the reaction product (Figure 4, profile A) is loganin based on its base peak at 435.1 (mass-to-charge ratio [m/z]), consistent with the mass obtained with loganin standard.
Figure 4.
Production of Loganin by E. coli Cell-Free Extracts Expressing Recombinant LAMT.
Loganin production only occurred in the presence of recombinant enzyme, AdoMet and loganic acid (A), but not in reactions with boiled recombinant enzyme (B), in reactions lacking loganic acid (C) nor in reactions with E. coli cell-free extracts expressing the empty vector (D). Note the loss of AdoMet and loganic acid with the appearance of loganin in (A). The table confirms that the reaction product in (A) is loganin based on its base peak at 435.1 (m/z). Analyses for A to D were performed by UPLC. The reaction products obtained in A were also analyzed by liquid chromatography–mass spectrometry (LC-MS) using a Bruker HCT+ System as described in Methods.
Recombinant LAMT Is a Highly Specific OMT
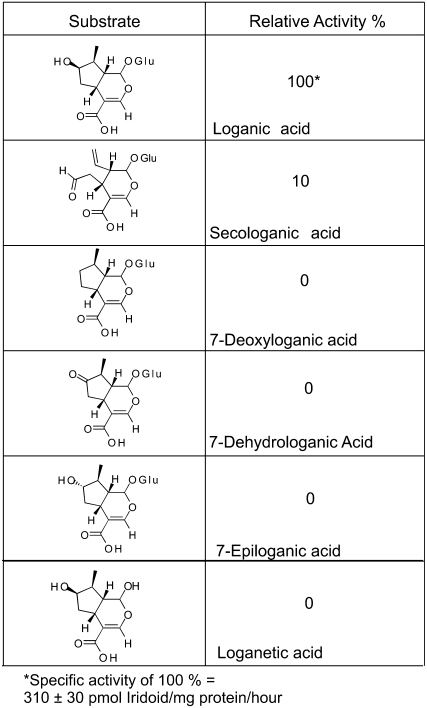
The recombinant LAMT (rLAMT) showed striking specificity for loganic acid since it was not active with other similar iridoids, including deoxyloganic, dehydrologanic, epiloganic, loganetic acid, or with the reaction product, loganin (Figure 5). Various benzoic acids (salicylic, benzoic, p-hydroxybenzoic, and anthranillic acids), phenylpropanoids (caffeic, ferulic, sinapic, gallic, chlorogenic, p-coumaric, and trans-cinammic acids), dicarboxylic acids (malic and tartaric), and hormones (indole acetic and abscisic acids) were not accepted as substrates by rLAMT nor was it active with MIAs (16-OH tabersonine and 3-OH tabersonine) (data not shown). Together, these results confirm that the 12 ESTs used to assemble C57Contig1 encoded authentic LAMT, that Catharanthus leaf epidermis is specialized for MIA biosynthesis, and that the CROLF1NG data set will be a valuable resource for cloning many more unknown and uncharacterized genes in this pathway.
Figure 5.
Substrate Specificity of rLAMT.
The LAMT only accepted loganic and secologanic acids as possible substrates but was not active with other similar compounds shown. Asterisk, the 100% value is related to the specific activity of the enzyme for loganic acid (310 pmol loganin/mg protein/h).
Biochemical and Kinetic Properties of rLAMT
The pH optimum of rLAMT was 7.5 with loganic acid as a substrate, and cofactors such as Mg2+ or K+ were not required for activity nor did they enhance enzyme activity (data not shown). The Km for loganic acid of rLAMT was 14.76 ± 1.7 mM (Table 2), similar to the results obtained for the partially purified enzyme isolated from cell suspension cultures of C. roseus (Km = 12.5 mM) (Madyastha et al., 1973). While the Km value for the cosubstrate AdoMet was not previously reported for LAMT, rLAMT showed a remarkably high Km (742.1 ± 37 μM) compared with JAMT (Seo et al., 2001) and SAMT (Ross et al., 1999) whose Km values were 6.3 and 9 μM, respectively.
Table 2.
CROLF1NG Is Enriched in ESTs for Enzymes in the MIA Biosynthetic Pathway
| LF (3655) | LB (1231) | RT (1546) | Pub (9850) | |
|---|---|---|---|---|
| G10H | 1 | 0 | 3 | 1 |
| 10HGO | 1 | 0 | 1 | 0 |
| LAMT | 12 | 0 | 0 | 1 |
| SLS | 25 | 2 | 3 | 0 |
| TDC | 8 | 0 | 0 | 0 |
| STR | 1 | 0 | 0 | 0 |
| SGD | 4 | 1 | 1 | 2 |
| T16H | 4 | 0 | 0 | 0 |
| 16OMT | 4 | 0 | 0 | 1 |
| D4H | 0 | 0 | 0 | 0 |
| DAT | 0 | 0 | 0 | 0 |
| CPR | 5 | 0 | 0 | 0 |
| ORCA3 | 1 | 0 | 0 | 0 |
| ZCT2 | 1 | 0 | 3 | 0 |
| BPF1 | 1 | 0 | 0 | 0 |
Comparison of the numbers of the MIA-related ESTs that were obtained by sequencing cDNA libraries produced from leaf epidermis–enriched mRNA (LF) in this study compared with previous studies involving random sequencing of cDNA libraries produced from mRNA extracted from the leaf base of young Catharanthus leaves (LB; Murata et al., 2006) from hairy root tips (RT; Murata et al., 2006) or from EST sequences found in the GenBank database (Pub). The numbers of unique EST sequences found in each case are shown in parentheses.
The high Km values for both substrate and cosubstrate suggest that the catalytic efficiency of rLAMT appears to be very low (Table 2) compared with other plant OMTs. The turnover number of rLAMT for loganic acid was quite low (0.327 s−1) compared with nonhistidine-tagged (2.8 s−1) and histidine-tagged (14 s−1) rSAMT (Ross et al., 1999) and to JAMT (25 s−1) (Seo et al., 2001). Remarkably, inhibition studies with the reaction products revealed that loganin is a strong inhibitor for rLAMT with a Ki of 215 ± 30 μM, since it has 50 times greater affinity for loganin compared with loganic acid. By contrast, the Ki (400 ± 47 μM) for S-adenosyl-L-homocysteine (AdoHys) was only twofold higher then that of AdoMet. Since the AdoHys inhibition constant was approximately twofold higher than that of AdoMet, inhibition of LAMT by AdoHys would depend on the ability of Catharanthus cells to recycle AdoHys into AdoMet through the AdoMet cycle.
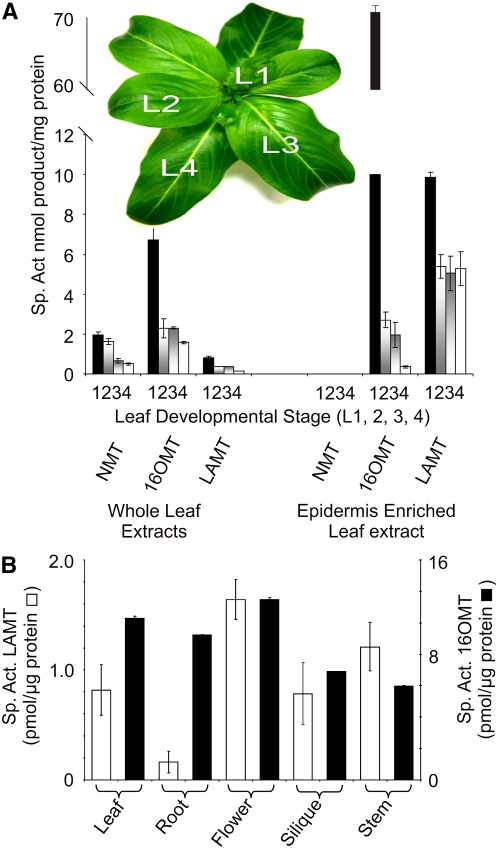
LAMT Enzyme Activity Is Preferentially Expressed in Catharanthus Leaf Epidermis
To localize LAMT in Catharanthus leaf, the enzyme activity of LAMT was measured using extracts either from whole leaves in various developmental stages or epidermis extracts of corresponding leaves obtained by CA. Previous studies with 16OMT (Levac et al., 2008) showed that this enzyme activity was highly enriched in the epidermis of young Catharanthus leaves, whereas the activity of the chloroplast thylakoid-associated NMT (De Luca and Cutler, 1987; Murata and De Luca, 2005) is found within cells inside the leaf. In accordance with these findings, NMT activity was only detected in whole leaves but not in epidermis-enriched extracts in all developmental stages tested (Figure 6A). By contrast, 16OMT-specific enzyme activity was 10 times higher in leaf epidermis–enriched extracts compared with the activity found in whole young leaves (Figure 6A). The 10-fold higher specific activity of LAMT in the leaf epidermis than in whole leaves strongly suggests that this enzyme is also preferentially localized to the leaf epidermis (Figure 6A). However, the activity of 16OMT was 35 times lower in epidermis-enriched leaf extracts of L2 compared with those of L1 leaves, whereas the activity of LAMT only decreased by ∼50% in epidermis-enriched leaf extracts of L2, L3, and L4 leaves compared with those of L1 leaves. These data suggest that unlike highly regulated 16OMT, significant LAMT activity is maintained as Catharanthus leaves age. The reasons for this difference in regulation remain to be determined. Leaf, hairy root, stem, silique, and flower extracts contained both 16OMT and LAMT activities in all these tissues (Figure 6B). However, the levels of LAMT (Figure 6B) in hairy roots were 4 to 8 times lower than the activities found in the other plant organs, respectively.
Figure 6.
Differential Distribution of LAMT Enzyme Activity in Catharanthus Leaf Epidermis.
(A) Catharanthus leaves of different ages (L1 to L4) were extracted in triplicate either as whole leaves or by CA treatment to obtain epidermis-enriched leaf extracts as described in Methods. The two sets of desalted extracts from each stage of leaf development were assayed for NMT, 16OMT, or LAMT to identify the differential distribution of these enzyme activities.
(B) Whole Catharanthus leaves, hairy roots, flowers, siliques, and stems were extracted in triplicate as described in Methods. Desalted extracts were assayed for LAMT and 16OMT to compare the distribution of these enzyme activities within different plant organs. Each point represents the mean of three assays ± sd.
Biosynthesis of Pentacyclic Triterpenes with an Oleanane Skeleton May Be Exclusively Localized to the Epidermal Cells of Catharanthus Leaf
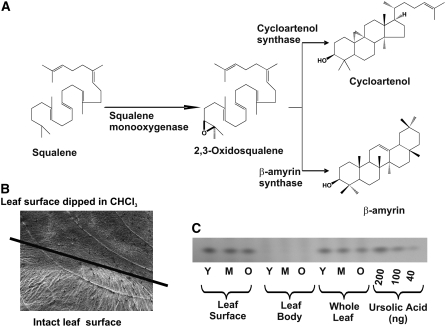
The identification in leaf epidermis of putative β-amyrin synthase (CL50Contig1; 13 ESTs) and squalene monooxygenase (CL247Contig1; 5 ESTs) genes (see Supplemental Table 2 online) suggested epidermal specialization for biosynthesis of pentacyclic triterpenes having an oleanane skeleton in Catharanthus (Figure 7A), since none of these genes were represented in the CrUniGene set (Murata and De Luca, 2005) or in the >10,000 Catharanthus ESTs found in the public database. Furthermore, cuticular wax components were selectively extracted by briefly dipping the leaf into chloroform (Figure 7B). The triterpene component of the extract was analyzed by thin layer chromatography (TLC) to show that triterpene alcohols, composed primarily of ursolic acid and small amounts of oleanolic acid (Usia et al., 2005), are exclusively localized to the cuticular wax layer (Figure 7C), suggesting that the leaf epidermal cells are the primary location of their biosynthesis. Together, ursolic and oleanolic acid compose ∼2.5% of the dry weight of the young Catharanthus leaf (Usia et al., 2005), illustrating the importance of this pathway in the leaf epidermis. Triterpene alcohols within the extract analyzed by capillary HPLC MS identified ursolic acid as a major triterpene component based on its elution with the same retention time (16 to 16.2 min) and the same molecular mass (455.3 m/z) as authentic ursolic acid standard.
Figure 7.
The Oleanane-Type Triterpenes Are Restricted to Catharanthus Leaf Epidermis.
(A) The common pathway for oleanane-type triterpene and phytosterol biosynthesis involves squalene monooxygenase, whereas β-amyrin synthase and cycloartenol synthase catalyze the key cyclization of 2,3-oxidosqualene to form β-amyrin and cycloartenol, respectively. These three genes were represented 5, 14, and 2 times in the CROLF1NG EST data base (see Supplemental Table 2 online).
(B) Scanning electron microscopy analysis of the surface of the leaf treated with chloroform. The line indicates the border between the areas that have been dipped into chloroform prior to the observation of the surface and the intact leaf. The surface of chloroform-dipped part of the leaf is clearly stripped of surface waxes compared with the untreated part of the leaf.
(C) Triterpenes are localized to the surface in Catharanthus leaves. Triterpene extracts were obtained by dipping leaves of various stages of the development into chloroform to obtain the leaf surface extract. The remainder of the chloroform-dipped leaves and whole leaves were extracted separately as described in Methods to obtain the leaf body and whole leaf extracts, respectively. Each extract was analyzed by TLC as described in Methods, and the levels of oleanolic triterpenes were compared with standard levels of ursolic acid (40, 100, and 200 ng) that were also applied to the TLC. The quantities of ursolic acid were also analyzed by UPLC and by MS analysis as described in Methods.
DISCUSSION
Toward a Complete Understanding of MIA Biosynthesis in Catharanthus
Significant efforts by various research groups have yielded >25,000 Catharanthus ESTs with >12,000 unique sequences that have partly been submitted to the GenBank database (Murata et al., 2006; Rischer et al., 2006; Shukla et al., 2006). While these efforts identified only four known MIA biosynthesis pathway genes (Table 2), this study, using a cDNA library prepared from leaf epidermis–enriched mRNA, identified ESTs for virtually all known MIA biosynthesis and regulatory genes up to and including T16H (Figure 1) and 16OMT (Levac et al., 2008). These results strongly support the biochemical specialization of young leaf epidermis as the primary site for expression of most of the pathway in vindoline biosynthesis from primary iridoid precursors and tryptamine to 16-methoxytabersonine (Murata and De Luca, 2005). Most significantly, this data set is very likely to contain the remaining uncharacterized biosynthesis and regulatory genes for the MIA pathway as shown by the CROLF1NG (3655 unique sequences) guided identification and functional characterization of LAMT (Figures 3 to 6) and 16OMT (Levac et al., 2008). In this respect, it is relevant that the LAMT and 16OMT sequences were represented 12 and four times, respectively, in the CROLF1NG data set, while only one EST for each sequence was found in published Catharanthus EST data sets (Table 2). These results highlight the remarkable value of random sequencing of leaf epidermis–enriched cDNA libraries for investigating the specialized chemistry of this cell type. The results also suggest that selection of candidate genes based on their relative abundance in this library is a useful approach to identify MIA biosynthesis enzyme candidates. In this context, further sequencing of this epidermis-enriched cDNA library should be considered to obtain more reliable data on the redundancies of the ESTs (Table 1) and to reach saturation with respect to the numbers of ESTs that encode enzymes that are related to MIA biosynthesis. In addition, gene families with particular biochemical functions could be selected for expression analysis by RT-PCR using RNA harvested from different organs or cell types (Murata and De Luca, 2005) to select for candidates to be screened directly for biochemical function.
Use of the CROLF1NG Data Set to Elucidate the Pathway for Secologanin Biosynthesis
Iridoids are cyclic monoterpenes, found in various plant species, where they play important defensive roles in plant–herbivore interactions. For example, aucubin and catalpol found in Plantago lanceolata deter and retard the growth rate of generalist insect herbivores (Harvey et al., 2005), whereas insect specialists are attracted to and consume host plants to sequester iridoids for defensive purposes or to use them as oviposition cues (Pereyra and Bowers, 1988; Peñuelas et al., 2006). In the case of MIA biosynthesis, the iridoid secologanin is important as the source for the terpene moiety of all the MIAs. While iridoids have also been used in chemotaxonomy because of their structural diversity in various plant species (Lopes et al., 2004), their localization within the plant as well as their biochemistry and molecular biology remain largely unknown.
Recent studies in Catharanthus have shown that while G10H is preferentially expressed in IPAP cells (Burlat et al., 2004), SLS (Figure 2) is preferentially expressed in leaf epidermal cells (Irmler et al., 2000; Burlat et al., 2004; Murata and De Luca, 2005) and in roots. This preferential expression of G10H and several steps in the methyl erythritol phosphate (MEP) pathway has led to the suggestion that the precursors for secoridoid biosynthesis may be produced in IPAP cells and that an unknown intermediate is transported to the leaf epidermis for elaboration into secologanin (Burlat et al., 2004). Further investigations into this question have been limited since other enzymes in this pathway remain to be cloned. This report has identified and functionally characterized LAMT, which catalyzes O-methylation of loganic acid (Figure 3) and is strongly expressed in (Figure 6) but not restricted to leaf epidermal cells (data not shown). In addition, the representation of a single MEP pathway gene (4-diphosphocytidyl-2-C-methyl-d-erythritol synthase, three ESTs; see Supplemental Table 1 online) and the G10H (one EST; Table 1) gene in the CROLF1NG data set, together with the abundance of 10-hydroxygeraniol oxidoreductase–like transcripts and SLS transcripts (Table 1) in epidermal cells, may mean that the complete secologanin biosynthesis pathway is expressed in the leaf epidermis (Murata and De Luca, 2005). These results remain ambiguous and further detailed studies are required to elucidate if an isoprenoid intermediate produced in IPAP cells is required for the biosynthesis of secologanin in Catharanthus leaf epidermal cells.
The epidermal localization of secologanin biosynthesis in Catharanthus leaf fits their possible antiherbivory role since the leaf surface is the first contact point between herbivores and the plant. It would be interesting to know if other iridoid-producing plants, including Plantago and Lonicera species (Pereyra and Bowers, 1988; Harvey et al., 2005; Peñuelas et al., 2006), also preferentially produce their iridoids in leaf epidermal cells.
Two additional candidate ESTs in the CROLF1NG data set could encode 7-deoxyloganetic acid 1-O-glucosyltransferase (Yamamoto et al., 2002) (see Supplemental Table 8 online) and 7-deoxyloganin 7-hydroxylase (Yamamoto et al., 1999) (CYP72B and CYP72C in Supplemental Table 9 online) involved in earlier steps in the secologanin pathway (Figure 2). While these two enzymes were partially purified and characterized from Lonicera japonica cell cultures, their corresponding genes remain to be cloned. Further experiments with these two candidate genes will be performed to identify their possible role in these biochemical functions.
Features of the Active Site of LAMT Based on the Crystal Structure of SAMT
The SABATH family of carboxyl methyltransferases has distinct characteristics that separates them from other OMTs. These enzymes do not require any catalytic residues but rather provide a hydrophobic pocket or reaction center where the desolvated carboxylate moiety can attack the AdoMet (Zubieta et al., 2003). SABATH family OMTs, such as JAMT, FAMT, and SAMT (Ross et al., 1999; Seo et al., 2001; Yang et al., 2006), that O-methylate fairly hydrophobic substrates require that the carboxyl moiety be devoid of water to increase its reactivity. By contrast, loganic acid with its glucose moiety and its higher water solubility may require special conditions for eliminating the glucose-associated water as loganic acid enters the active site of LAMT. The protein alignment of LAMT to IAMT, SAMT, and JAMT in Figure 3B reveals that active-site residues required for the hydrophobic core (Y159, W163, I224, P227, L244, and I320) are conserved in LAMT when compared with those based on the crystal structure of SAMT (Zubieta et al., 2003). Two important differences involve the substitution of SAMT amino acids W226 and F347 with H245 and D359, respectively, in LAMT.
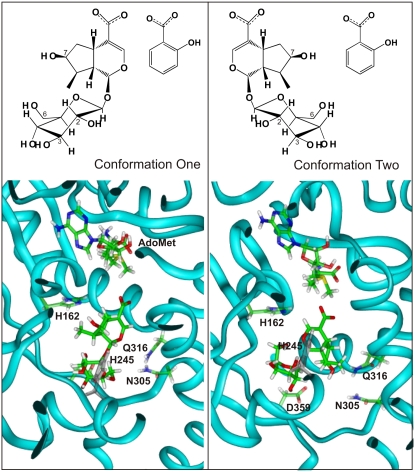
Preliminary modeling studies of LAMT with loganic acid superimposed onto the salicylic acid binding site of SAMT (Figure 8) showed that two different orientations of loganic acid had favorable van der Waals and electrostatic interactions with the protein and could be accommodated comfortably. The average CHARMM interaction energy, >100 minimum energy poses, for the first conformation within the LAMT comparative model was −100.57 ± 5.2 kcal/mol with an overall range of −113.32 kcal/mol to −86.78 kcal/mol. For the second conformation, it was −87.98 ± 14 kcal/mol with an overall range of −139.29 kcal/mol to −70.37 kcal/mol. Given that both conformers have similar interaction energies with the protein, neither model can be excluded from consideration without further experimental evidence.
Figure 8.
Computational Model for Accommodating Two Conformations of Loganic Acid in the Active Site of LAMT.
The top panel shows the two orientations of loganic acid with respect to salicylic acid that were used to initially position the substrate in the binding site of the comparative model of LAMT. The bottom panel shows an image of the lowest energy pose obtained for conformers 1 and 2 from the 100 minimum energy conformations obtained from the modeling of loganic acid and AdoMet within the active site of the LAMT model. The LAMT peptide backbone is described by a ribbon structure, and the polar/charged amino acids putatively involved in substrate binding are labeled. H245 is colored in gray to differentiate it from the sugar moiety of loganic acid.
We examined five amino acids in the putative binding site of LAMT, H162, H245, Q316, N305, and D359, which are distinct from those of the SAMT for putative hydrogen bonding with loganic acid. The 100 minimum energy poses of conformer 1 (Figure 8, conformer 1), consistently made good hydrogen bonds (i.e., those having lengths <3.0 Å) with both D359 and H162. The carboxylate of D359 forms hydrogen bonds to the C3 and C6 hydroxyls of the glucose moiety, and the 7-hydroxyl group of the five-membered ring of loganic acid displays hydrogen bonding to the Nɛ of H162.
Analysis of the 100 minimum energy poses of the second conformer of loganic acid in the putative binding site (Figure 8, conformer 2) revealed that in this orientation, the carboxylate side chain of D359 consistently forms a hydrogen bond with the C2 hydroxyl of the glucose moiety and occasionally with the 7-hydroxyl substituent of the five-membered ring. Concomitantly, the N305 amide side chain was sufficiently close to the oxygen of the 7-hydroxyl group to make good hydrogen bonds in 65% of the minimum energy conformations. The Nɛ of H245 also showed putative hydrogen bonds with the C3 hydroxyl of the glucose moiety in just under a quarter of the 100 minimum energy poses of conformer 2.
The putative association of the 7-hydroxyl group of loganic acid with H162 (conformer 1) or N305/D359 (conformer 2) may explain why 7-deoxyloganic acid and 7-epiloganic acid are not accepted as substrates, since hydrogen bonding may be required for substrate binding and specificity. However, without a crystal structure for LAMT, this remains a hypothesis. Regardless, it is evident that the high Km (Table 2) value for loganic acid may be due to the tendency of water to associate with the glucose moiety, and this could affect its binding affinity to LAMT. Although beyond the immediate scope of this work, the above modeling study suggests that appropriate mutational studies of LAMT might support conformer 1 versus 2 as the actual binding site orientation of loganic acid.
Kinetic Analyses and Substrate Specificity Studies with rLAMT Suggest a Preferred Route of Secologanin Biosynthesis in Catharanthus
It has been shown that LAMT from crude Catharanthus leaf extracts O-methylated loganic acid but not 7-deoxyloganic acid, suggesting that hydroxylation preceeds O-methylation (Madyastha et al., 1973). Studies with L. japonica cell cultures suggested that O-methylation could precede hydroxylation since 7-deoxyloganin could be converted into loganin (Figure 2) (Yamamoto et al., 1999). The substrate specificity studies (Figure 5) with cloned rLAMT show that the preferred iridoid substrate is loganic acid and that O-methylation does appear to proceed after hydroxylation of 7-deoxyloganic acid (Figure 2) (Madyastha et al., 1973) in Catharanthus rather than before it (Yamamoto et al., 1999). Further studies with the LAMTs of other secologanin-producing species may also show that the substrate specificity of this reaction rather than that of the hydroxylase could define the order of secologanin biosynthesis.
The Km for loganic acid (12.5 mM) found for partially purified C. roseus LAMT (Madyastha et al., 1973) was very similar to that determined for the rLAMT (14.76 ± 1.7 mM) (Table 2). Remarkably, rLAMT had a relatively high Km for AdoMet (742.1 ± 37 μM) compared with other carboxyl methyltransferases from the SABATH family (JAMT, Km 6.3 μM; SAMT, Km 9 μM) (Ross et al., 1999; Seo et al., 2001). The high Km values for both loganic acid and AdoMet suggest that the catalytic activity of LAMT may be limiting the production of terpenoid component of MIA biosynthesis in C. roseus (Peebles et al., 2006).
Inhibition studies with the reaction products revealed that loganin was a very good inhibitor (Ki of 215 ± 30 μM) for LAMT, with its 50 × greater affinity for loganin than for loganic acid. These results suggest that for LAMT to be active, loganin concentrations would need to be kept very low in the cell, either by rapid turnover of loganin into secologanin or by subcellular compartmentation. Substrate specificity studies have suggested that only loganin can serve as a substrate for SLS (Yamamoto et al., 2000). The association of this cytochrome P450 with endoplasmic reticulum membranes or related vesicles could rapidly convert loganin into secologanin followed by its transfer to the vacuole (Contin et al., 1999) together with tryptamine for conversion into strictosidine by vacuole-associated STR (Stevens et al., 1993). In this way, the SLS-mediated conversion of loganin into secologanin, together with its subcellular compartmentalization for MIA biosynthesis, could prevent loganin from inhibiting LAMT.
Catharanthus Leaf Epidermis as the Exclusive Site of Oleanane Triterpene Biosynthesis
The plant kingdom produces many thousands of triterpenes exhibiting a broad range of pharmacological activities based on >80 different carbon skeletons produced from (3S)-oxidosqualene (Ebizuka et al., 2003). While the accumulation of high levels of ursolic acid in C. roseus has been documented (Usia et al., 2005), the sites of accumulation of these products have not been elucidated. Studies with other plants showed that ursolic and oleanolic acid accumulate on the surfaces of apple (Malus domestica) (Bringe et al., 2005) and of grape berry (Vitis vinifera) exocarp (Grncarevic and Radler, 1971) as well as in the epidermis of grape leaves. In other studies with Avena strigosa, the expression of β-amyrin synthase was localized to epidermal cells of root tips that accumulate triterpene avenacins (Qi et al., 2006). The CROLF1NG data set suggests that the MVA pathway (see Supplemental Table 1 online) and two key triterpene biosynthesis genes (squalene monooxygenase [five ESTs] and β-amyrin synthase [14 ESTs]) (see Supplemental Table 2 online) were preferentially expressed in Catharanthus leaf epidermis. The selective and quantitative extraction of surface leaf triterpenes with chloroform (Figures 7B and 7C) suggests that once they are made inside Catharanthus leaf epidermal cells, they are secreted where they accumulate on the surface of leaf epidermal cells (Figure 9).
Figure 9.
Catharanthus Leaf Epidermal Cells Express the MIA, MVA/Triterpene, VLFA, and Flavonoid Biosynthetic Pathways.
The EST sequencing analysis described in this study suggests that the complete MVA/triterpene pathway is expressed in leaf epidermal cells and that the triterpenes being produced are exported outside the cell into the surface of the leaves. Similarly the entire pathway for VLFA biosynthesis is also expressed in this cell type and is exported to produce the surface wax layer of Catharanthus leaves. Flavonoid biosynthesis also appears within these same epidermal cells where flavonoid products accumulate within epidermal cell vacuoles as suggested by Mahroug et al. (2006). The model also proposes that most of the vindoline pathway up to and including 16-methoxytabersonine is expressed in the leaf epidermis and an undetermined intermediate is then transported to adjacent mesophyll cells, specialized idioblasts, and/or laticifers, for final elaboration into vindoline. To permit this complex transport, strict control mechanisms appear to be operating and these have yet to be characterized. In contrast with leaf epidermal cells that are enriched in the MVA pathway that supplies IPP for the biosynthesis of triterpenes, there is strong evidence that the IPAP cells preferentially express the MEP pathway and G10H. This raises questions about the roles played by these two cell types in supplying the necessary IPP for the biosynthesis of secologanin within the leaf epidermis.
It is interesting that Catharanthus leaf epidermal cells are the site of biosynthesis of triterpenes that are secreted into epicuticular wax as well as of secologanin that is then incorporated into MIAs. The more frequent representation of five MVA pathway–specific genes compared with the single representation of a MEP pathway gene (see Supplemental Table 1 online; 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase) does suggest that MVA pathway transcripts are selectively expressed in leaf epidermal cells compared with those of the MEP pathway. These differences might be attributed to the greater demand for biosynthetic precursors for triterpene biosynthesis, since triterpenes accumulate at much higher levels in Catharanthus than do MIAs (Usia et al., 2005). Further biochemical and chemo-ecological analyses will be required to elucidate the mechanism of triterpene accumulate in the leaf epicuticular waxes, their in vivo roles, and the involvement of the MEP and MVA pathways in Catharanthus leaf epidermal cells in relation to MIA and triterpene biosynthesis.
The Possible Roles of the Plastidic MEP and Cytosolic MVA Pathways in Secologanin Biosynthesis
Previous extensive analyses of the MEP pathway in MIA-producing plants have suggested that the terpenoid moiety of MIAs is predominantly, but not exclusively, derived from MEP pathway in cell suspension and hairy root culture systems of C. roseus and Rauvolfia serpentina cell cultures (Contin et al., 1998; Eichinger et al., 1999; Hong et al., 2003). These results are supported by studies in Arabidopsis that monoterpenes are produced principally via the plastidic MEP pathway (Lichtenthaler, 1999). In the case of C. roseus, recent in situ RNA hybridization studies showed that the MEP pathway genes DXS, DXR, MECS, and G10H, the first committed enzyme in iridoid biosynthesis, were expressed predominantly in IPAP cells (Burlat et al., 2004) of the leaf vasculature. Recent immunocytolabeling studies provided more quantitative information to corroborate the high enrichment of the MEP pathway in IPAP cells, but it was also found at low levels in other cell types, including epidermal and mesophyll cells (Oudin et al., 2007). The expression of G10H was used to further suggest that MEP pathway–derived isopentenyl pyrophosphate was converted to 10-hydroxygeraniol or another derivative for mobilization to the leaf epidermis for further elaboration into secologanin and MIAs. While the baseline representation of MEP pathway genes compared with those of the MVA pathway in the CROLF1NG data set (see Supplemental Table 1 online) provides support for the MEP pathway enrichment of phloem parenchyma (Burlat et al., 2004), it also establishes the differential enrichment of the MEP and MVA pathways in each cell type.
Since only a single G10H EST was detected in the CROLF1NG data set, it does support in situ hybridization studies that show preferential expression of G10H in phloem parenchyma (Burlat, et al., 2004). Remarkably, it has been implied that the biosynthesis of the similar iridoids in leaf beetles belonging to the Chrysomelidae family is divided between the fat body and a specialized glandular reservoir that also accumulates these defensive secretory compounds (Burse et al., 2007). The fat body expresses the MVA pathway and a geraniol hydroxylase and glucosyltransferase to produce 10-hydroxygeraniol-10-O-β-glucoside. This glucoside is then transported to the glandular reservoir where a β-glucosidase releases 10-hydroxygeraniol for enzyme-mediated oxidation and cyclization/isomerization for the formation of toxic iridoids (Burse et al., 2007). In the case of Catharanthus MIA biosynthesis, the identification of a similar pathway step or intermediate in the IPAP cells could be very helpful to completely prove that MEP pathway–derived precursors are transported to the leaf epidermis from the internal phloem parenchyma for the biosynthesis of secologanin (Burlat et al., 2004). Similarly, the hypothesis that leaf epidermal cells alone are competent to supply sufficient MEP pathway intermediates for secologanin biosynthesis and MIA accumulation in Catharanthus leaves also needs to be fully tested (Murata and De Luca, 2005).
The unique biochemical properties of distinct cells (i.e., epidermal cells, mesophyll cells, and phloem parenchyma cells) with respect to IPP biosynthesis are clearly related to their complex roles to produce different metabolites. For example, neither leaf epidermal cells nor IPAP cells, in contrast with those of the palisade and spongy mesophyll, require IPP for biosynthesis of chlorophyll and yet these two cell types require increased expression of the MVA/MEP pathways for their unique metabolic needs. The data presented here highlight the complexity involved and present novel approaches to study these questions.
The Versatility of the CA Technique and Its Possible Use for Epidermome Analysis
Unlike LCM, CA can be used to harvest epidermis-enriched materials from intact leaves, making the analysis of mRNA, active protein, and metabolite analyses possible without significant technical difficulties. The technical simplicity of CA technique makes it valuable for (1) localizing enzyme activities to the leaf epidermis (Murata and De Luca, 2005), (2) large-scale purification of leaf epidermal proteins (Levac et al., 2008), and (3) targeted random sequencing of cDNA libraries produced from leaf epidermis–enriched mRNA, as shown in this study.
Our effort to establish leaf epidermis–specific ESTs not only revealed the capability of the epidermis to manufacture monoterpenes and triterpenes but indicated the value of the CROLF1NG data set as a gene discovery tool with respect to various pathways in the leaf epidermis. For example, numerous putative candidate genes involved in fatty acid biosynthesis were identified (see Supplemental Table 4 online) presumably since fatty acid biosynthesis is elevated for the production of cuticular waxes that are secreted to the surface of the epidermis. Similarly, Catharanthus leaf epidermis may express the entire flavonoid pathway (Figure 9), as suggested previously (Mahroug et al., 2006), since the majority of the genes encoding the putative pathway for flavonoid biosynthesis, from PAL to DFR, were also represented (see Supplemental Table 5 online). It is noteworthy that the whole pathway for flavonoid biosynthesis appears within the same cell type, in clear contrast with the vindoline pathway that appears to require the involvement of three or more leaf cell types (St. Pierre et al., 1999; Burlat et al., 2004; Murata and De Luca, 2005). VLFA and triterpene alcohols, on the other hand, are synthesized in the leaf epidermis and are secreted out to the cuticular wax layer (Figure 9). The wide range of biochemical pathways described here require the movement of different end products within the cell, outside to leaf epidermis or toward the mesophyll, and suggests that the leaf epidermis also expresses the complex transport mechanisms and controls that permit the proper distribution of metabolites to appropriate destinations.
The discovery of entire pathways for the biosynthesis of triterpenes, VLFAs, and flavonoids also promote the usefulness of the CA technique as a very promising tool for global profiling of gene expression, enzyme activities, and metabolites within the leaf epidermal cells. The CA technique should in fact be considered to conduct analyses for any plant where the specialized biological role of the leaf epidermis is of interest.
METHODS
Plant Material
The Catharanthus roseus (cv Little Delicata) plants were grown in a greenhouse under a long-day photoperiod at 30°C. Young leaves (1.5 cm in length) were harvested for cDNA library construction, and more mature leaves were also used for other experiments.
CA for cDNA Library Construction
CA was performed primarily as described previously (Murata and De Luca, 2005), but with some modifications. The upper and lower epidermis from 5 g of young Catharanthus leaves were selectively abraded with carborundum (SiC) (Fisher Scientific) using a cotton swab to apply even pressure. The epidermis was rubbed eight times per side (upper and lower surface), and the leaf was then dipped in 4.0 mL of Trizol (Invitrogen) at 4°C and gently agitated for 5 s to release the epidermal cell content into the solution to obtain 2.5 mL of extract from 5 g of abraded leaves. The extract was then used for RNA isolation according to the manufacturer's protocol.
Construction and Sequencing of Leaf Epidermis–Enriched cDNA Library
The Catharanthus leaf epidermis–specific cDNA library was constructed using the SMART cDNA library construction kit (Clontech Laboratories) according to the manufacturer's instructions. The cDNA was then amplified by PCR prior to the packaging to Gigapack III gold packaging extract (Stratagene). The primary library (1.0 × 106 plaque-forming units) was directly converted into plasmids by in vivo excision, and the Escherichia coli colonies obtained were randomly picked for single-path sequencing using primers from the 5′ end of the inserts. The sequencing reactions were performed using the Templiphi DNA sequencing template preparation kit (GE Healthcare), and the resulting DNA templates were sequenced using ABI Prism Big Dye terminator sequencing kits and an ABI 3730 genetic analyzer (Applied Biosystems)
Sequence Analysis
The sequence files in ABI format were analyzed using the BLASTX algorithm (Altschul et al., 1997). Multiple clones with the overlapping areas of identical sequences were clustered and classified as “Clustered,” while sequences that appeared only once in the ESTs were classified as “Singletons.” The threshold of the sequence similarity was set as E-values at 10−6 and lower, while sequences that did not show significant homology were named “No hits.” The sequences were archived in the FIESTA software package (http://bioinfo.pbi.nrc.ca/napgen.beta//login.html) at the Plant Biotechnology Institute of the National Research Council of Canada. The functional categorization was first done automatically to produce putative annotations, followed by the manual inspection to verify the reasons for the annotation. All the sequences described in this report, including those found in Tables 1 and 2, those found in Supplemental Tables 1 to 10 online, those produced from the leaf base (Murata et al., 2006), and those produced from the root tip cDNA (Murata et al., 2006) libraries, are available in the GenBank (dbST) database.
Extraction and Analysis of Triterpenes in Catharanthus Leaves
Fresh leaves from different developmental stages (120 mg of young 1.5 cm long [Y], mid-aged 3.0 cm long [M], and older 4.5 cm long [O]) were harvested. To obtain surface extracts, leaves were dipped in 3 mL of chloroform (Gniwotta et al., 2005), vortexed for 2 min, and incubated for 5 min at room temperature. Surface-stripped leaves were then rinsed with water to remove excess chloroform. To extract intact leaves or chloroform surface-stripped leaves, they were separately frozen in liquid nitrogen and homogenized with 3 mL of chloroform with a mortar and pestle. The extracts were filtered through one layer of miracloth (VWR Canlab), and the filtrates were transferred to 15-mL conical tubes together with 3 mL of water. The samples were mixed by vortex treatment (Genie 2 vortex set at 10; Fisher Scientific) and then centrifuged at 5000g for 10 min to separate the phases. The chloroform component was harvested and evaporated to dryness by vacuum centrifugation in the SPD SpeedVac (Fisher Scientific). The dry residues were resuspended in acetone (300 μL); 2 μL were spotted on Machery Nagel Silica Gel G thin layer chromatograms (Fisher Scientific) and submitted to TLC using ethyl ether:petroleum ether (9:1) as solvent (Wagner et al., 1984). Chromatograms were sprayed with vanillin-phosphoric acid reagent (500 mg vanillin dissolved in 50 mL of 50% phosphoric acid) and developed over a hot plate for 5 min until triterpenes could be visualized. Ursolic acid levels could be determined using an ursolic acid standard curve.
Capillary HPLC MS Analysis of Triterpenes
Ursolic acid standard (Mr 456) (Sigma Aldrich) and C. roseus triterpenoids (crude extracts or samples partially purified by TLC) were submitted to analysis using a Bruker HCT+ LC-MS system (Mode:ESI-Negative Inlet Agilent 1100 LC; solvent system, acetonitrile/water; column, 50-mm C18 with a precolumn guard). Both crude and partially TLC purified triterpenoid samples displayed similar retention times (16.0 to 16.2 and 16.4 to 16.6 min) and identical masses (455.3 m/z) as authentic ursolic acid standard. This verified that the major triterpene of Catharanthus surface extracts was ursolic acid, as suggested in previous studies with whole plants (Usia et al., 2005).
Scanning Electron Microscopy
The images were obtained using an AMRAY 1600 Turbo scanning electron microscope as described previously (Murata and De Luca, 2005).
LCM for the Isolation of RNA from Single Cells
C. roseus leaf was fixed and embedded in paraffin as described previously (Murata and De Luca, 2005). LCM, mRNA isolation, and amplification were performed as described previously (Murata and De Luca, 2005).
LAMT Full-Length cDNA Cloning and Protein Expression in E. coli
The putative LAMT full-length cDNA sequence (accession number EUO57974) was assembled in silico from 12 different ESTs to produce CL57Contig1. PCR primers (LAMT 5′ ATG, 5′-CACCATGGTTGCCACAATTGATTCC-3′; LAMT 3′ UTR, 5′-TTAATTTCCCTTGCGTTTCAAG-3′) were designed to amplify the putative open reading frame based on its similarity to known carboxylic acid methyltransferases. The PCR conditions used were as follows: 20 μL reaction volume containing Takara- Ex Taq Polymerase and a template composed of SMART cDNA from the leaf base of C. roseus; denature, 15 s at 94°C; re-anneal, 10 s at 55°C; elongation, 40 s at 72°C; final extension of 5 min at 72°C. Since the coding sequence of LaMT had five restriction sites (EcoRI at 320 bp, EcoRI at 549 bp, SacI at 640 bp, XmnI at 969 bp, and BamHI at 1003 bp), the open reading frame was inserted in the expression vector pET 30b (Novagen) using NcoI and SacI. For this reason, the original PCR primers also contained NcoI and SacI sites that were used for the subcloning.
The amplified PCR product was ligated into pGEM-TEasy (Fisher Scientific), and the ligation mixture was used to transform heat shock competent DE3-MRF' E. coli cells. Cells were grown at 37°C for 1 h and then streaked onto Luria-Bertani agar plates with ampicillin as the selectable marker. Plates were incubated overnight at 37°C. Positive inserts were confirmed with colony PCR using single colonies as a template and the primers LAMT 5′ ATG and LAMT 3′ UTR. The LAMT insert was excised from the pGEM-T EASY vector using the restriction endonuclease SalI and NcoI (Fisher Scientific). The protein expression vector was digested with the restriction endonuclease SalI, EcoRV, and NcoI (Fisher Scientific). The vector and LAMT insert were ligated with Takara DNA Ligase (Fisher Scientific). The ligation mixture was used to transform heat shock–competent DE3-MRF' E. coli cells. Cells were grown at 37°C for 1 h and then streaked onto Luria-Bertani agar plates with kanamycin as the selectable marker. Positive inserts were verified with colony PCR using individual colonies as a template and the primers LAMT 5′ ATG and LAMT 3′ UTR. The plasmids with positive inserts were sequenced (Robarts Research Institute, London, Ontario, Canada). The original sequence from CL57Contig1 contained a stop codon in the middle of the open reading frame that turned out to be derived from sequencing error (data not shown). The cloned 1116-bp LAMT coding sequence encoded a 371–amino acid ORF with calculated molecular mass of 42,018 D.
Enzyme Assays for rLAMT
The standard radioactive enzyme assay (100 μL) contained 1 μg of crude desalted rLAMT protein, 2.5 nCi S-Adenosyl-L-[14C]-methionine (specific activity 58 mCi/mmol; GE Healthcare Canada), 0.5 mM loganic acid, and LAMT enzyme assay buffer (100 mM Tris-HCl, pH 7.5, 14 mM β-mercaptoethanol, and 25 mM KCl). Enzyme assays were incubated at 37°C for 30 min. Assays were immediately frozen in liquid nitrogen and lyophilized overnight. The dried material was resuspended into 15 μL methanol and centrifuged at 10,400 rpm for 10 min in a ThermoIEC, Micromass RF microcentrifuge (Fisher Scientific). The supernatant was directly added to a 0.20-mm Polygram Sil G/UV254 Macherey-Nagel TLC plate (Fisher Scientific), and the reaction products were developed with chloroform:methanol (7:3). The radioactivity was detected by exposure of the TLC to a storage phosphor screen (GE Healthcare Canada) for 16 h, and the emissions were detected using the FLA-3000 PhosphorImager (Fujifilm) with Multi Gauge version 3.0 software (Fujifilm).
The standard nonradioactive enzyme assay (100 μL) contained 4 μg of crude desalted bacterial protein with rLAMT, 7.5 mM AdoMet, 1.3 mM loganic acid, and LAMT enzyme assay buffer (100 mM Tris-HCl, pH 7.5, 14 mM β-mercaptoethanol, and 25 mM KCl). Enzyme assays were incubated at 37°C for 30 min. Assays were immediately frozen in liquid nitrogen and lyophilized overnight. The dried material was resuspended into 200 μL of 0.5% formic acid and 50 μL of acetonitrile. Samples were centrifuged at 10,000g for 10 min. An aliquot of 200 μL was then filtered through (0.22 μm) PALL filter (VWR Canada).
Reaction products were analyzed with UPLC (Waters). The analytes were separated using an Aquity UPLC BEH C18 with a particle size of 1.7 μm and column dimensions of 1.0 × 50 mm. The solvent systems containing A (0.5% formic acid) and B (100% acetonitrile) were used to form a linear gradient (time 0 to 2.99 min [0.1 to 30% B], 2.99 to 3.50 [30% B], 3.51 to 4.00 [30 to 50% B], and 4.01 to 5.00 [50 to 0.1% B]) at a flow rate of 0.150 mL/min. The reaction product detected at 240 nm eluted with an identical elution time and UV spectrum as authentic loganin (retention time 2.2 min).
This reaction product and the loganin standard were analyzed using a Bruker HCT+ LC-MS system (Mode:ESI-Negative Inlet Agilent 1100 LC) (solvent system, acetonitrile/water [0.1% formic acid]; column, 50 mm C18 with a precolumn guard). The reaction product displayed similar retention times (8.6 to 8.8 min) and identical masses (445.1 m/z) as authentic loganin standard.
Computational Methods for LAMT Sequence Alignment and Modeling of the Active Site
ClustalW (http://align.genome.jp) was used to produce an alignment of JAMT, SAMT, LAMT, and JAMT using a gap penalty of 10 and a gap extension of 0.1. The protein alignment was then submitted to SWISS-MODELLER using the alignment interface (http://swissmodel.expasy.org//SWISS-MODEL.html) (Guex and Peitsch, 1997; Schwede et al., 2003; Arnold et al., 2006). The software predicted the crystal structure of SAMT (Protein Databank number 1M6E).
Using QUANTA 2005 (Accelrys), three-dimensional all-atom models of both S-adenosyl-l-methionine and loganic acid were built. Partial charges on the atoms were assigned using charge templates of QUANTA, and the charge was smoothed over all atoms to give a total charge of +1 for SAM and −1 for loganic acid. The initial conformation of SAM in the model binding site of LAMT was obtained by superimposing atoms onto the corresponding coordinates of the S-adenosylhomocysteine in the crystal structure of salicylic acid carboxyl methyltransferase (1M6E) (Zubieta et al., 2003). Two different orientations of loganic acid within the putative binding pocket were obtained by superposition of the carboxylate, Cα and Cβ of loganic acid, onto the coordinates of the substrate salicylic acid in the 1M6E structure. The hydrogen atoms on the LAMT protein were generated using the H-build algorithm of CHARMM 31.1. (Accelrys; Brooks et al., 1983)
The CHARMM 31.1 algorithm was also used for all computer simulations employed to locate low-energy conformations of loganic acid in the putative binding site of the model for LAMT. Nonbonded interactions were smoothed to zero beginning for atoms whose center-to-center distance was ≥11.0 Å, with a final cutoff radius of 14.0 Å. Nonbonded neighbor lists were employed, for which the radius was set to 15.0 Å; the neighbor lists were updated using heuristic methods (Fincham and Ralston, 1981). All energies were computed in vacuo. Covalent bonds to hydrogen atoms were constrained using the SHAKE algorithm. (Ryckaert et al., 1977). During molecular dynamics simulations, a time-step of 1 fs was used.
An initial energy minimization was done of both AdoMet and loganic acid in the binding site. The atoms of the protein were fixed, and unfavorable interactions between the protein and the cofactor and substrate were relaxed by a sufficient number of steps of adopted-basis Newton-Raphson (ABNR) energy minimization so that the final root mean square gradient (RMSG) of the potential fell below 0.5 kcal/(mol Å). Subsequently, constraints on the protein side chains were released and another round of ABNR energy minimization was done until the RMSG was reduced to below 0.4 kcal/(mol Å). To allow the system to overcome energy barriers and explore nearby low-energy conformations, quenched molecular dynamics runs were done starting from these energy-relaxed structures. The backbone atoms of the protein remained fixed during these simulations, and amino acid side chains in the active site as well as AdoMet and loganic acid were allowed to move freely. During the initial 5 ps of computer simulation, the system was heated to 500K by rescaling the velocities of unconstrained atoms and increasing the temperature by 5K every 0.05 ps. Subsequently, the system was allowed to equilibrate at 500K for 100 ps, during which time period the atomic velocities of unconstrained atoms were rescaled only if the average temperature fell outside a temperature window of 500 ± 10K. Conformers were saved every 1 ps. Subsequent energy minimization of these 100 conformations comprised the quenching stage: the ABNR minimizations converged with all RMSG of 0.001 kcal/(mol Å) satisfied. This resulted in 100 low-energy conformations that were analyzed for both interaction energies and hydrogen bonds between loganic acid with the protein.
Accession Numbers
The EST sequences in Tables 1 and 2 as well as in Supplemental Tables 1 to 10 can be found in the GenBank (dbEST) database under accession numbers FD660726 to FD661300.
Supplemental Data
The following materials are available in the online version of this article
Supplemental Table 1. CROLF1NG Genes Related to Isoprenoid Biosynthesis.
Supplemental Table 2. CROLF1NG Genes Related to Downstream Terpenoid Biosynthesis.
Supplemental Table 3. CROLF1NG Genes Related to Flavonoid Biosynthesis.
Supplemental Table 4. CROLF1NG Genes Related to Lipid Biosynthesis.
Supplemental Table 5. CROLF1NG Genes Related to Methyltransferases.
Supplemental Table 6. CROLF1NG Genes Related to Acyltransferases.
Supplemental Table 7. CROLF1NG Genes Related Cytochrome P450–Dependent Monooxygenases.
Supplemental Table 8. CROLF1NG Genes Related to Glycosyltransferases
Supplemental Table 9. CROLF1NG Genes Related to 2-Oxoglutarate–Dependent Dioxygenases.
Supplemental Table 10. CROLF1NG Genes Related to Transcription Factors.
Supplementary Material
Acknowledgments
We thank Larry Pelcher and his team in the DNA Technologies Unit for DNA sequencing and Jacek Nowak and Dustin Cram in the Bioinformatics Unit (NAPGEN at the Plant Biotechnology Institute, National Research Council of Canada, Saskatoon) for their help in sequencing and in annotating the ESTs. We also thank Glenda Hooper (Brock University) for technical assistance in scanning electron microscopy and Tim Jones (Brock University) for mass spectrometric analysis. V.D.L. holds a Canada Research Chair in Plant Biotechnology. This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to V.D.L and by a joint NSERC Strategic grant held together with Peter Facchini (University of Calgary, Canada).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Vincenzo De Luca (vdeluca@brocku.ca).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSIBLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modeling. Bioinformatics 22 195–201. [DOI] [PubMed] [Google Scholar]
- Barthlott, W., Neinhuis, C., Cutler, D., Ditsch, F., Meusel, I., Theisen, I., and Wilhelmi, H. (1998). Classification and terminology of plant epicuticular waxes. Bot. J. Linn. Soc. 126 237–260. [Google Scholar]
- Bringe, K., Schumacher, C.F.A., Schmitz-Eiberger, M., Steiner, U., and Oerke, E.C. (2005). Ontogenetic variation in chemical and physical characteristics of adaxial apple leaf surfaces. Phytochemistry 67 161–170. [DOI] [PubMed] [Google Scholar]
- Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S., and Karplus, M. (1983). CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4 187–217. [Google Scholar]
- Broun, P. (2005). Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8 272–279. [DOI] [PubMed] [Google Scholar]
- Burlat, V., Oudin, A., Courtois, M., Rideau, M., and St-Pierre, B. (2004). Co-expression of three MEP pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J. 38 131–141. [DOI] [PubMed] [Google Scholar]
- Burse, A., Schmidt, A., Frick, S., Kuhn, J., Gershenzon, J., and Boland, W. (2007). Iridoid biosynthesis in Chrysomelina larvae: Fat body produces early terpenoid precursors. Insect Biochem. Mol. Biol. 37 255–265. [DOI] [PubMed] [Google Scholar]
- Cacace, S., Schröder, G., Wehinger, E., Strack, D., Schmidt, J., and Schröder, J. (2003). A flavonol O-methyltransferase from Catharanthus roseus performing two sequential methylations. Phytochemistry 62 127–137. [DOI] [PubMed] [Google Scholar]
- Coiner, H., Schröder, G., Wehinger, E., Liu, C.J., Noel, J.P., Schwab, W., and Schröder, J. (2006). Methylation of sulfhydryl groups: A new function in a family of plant O-methyltransferases. Plant J. 46 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collu, G., Unver, N., Peltenburg-Looman, A.M., van der Heijden, R., Verpoorte, R., and Memelink, J. (2001). Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett. 508 215–220. [DOI] [PubMed] [Google Scholar]
- Contin, A., van der Heijden, R., Lefeber, A.W.M., and Verpoorte, R. (1998). The iridoid glucoside secologanin is derived from the novel triose phosphate/pyruvate pathway in a Catharanthus roseus cell culture. FEBS Lett. 434 413–416. [DOI] [PubMed] [Google Scholar]
- Contin, A., van der Heijden, R., and Verpoorte, R. (1999). Accumulation of loganin and secologanin in vacuoles from suspension cultured Catharanthus roseus cells. Plant Sci. 147 177–183. [Google Scholar]
- De Luca, V., Balsevich, J., Tyler, R.T., and Kurz, W.G.W. (1987). Characterization of a novel N-methyltransferase (NMT) from Catharanthus roseus plants. Plant Cell Rep. 6 458–461. [DOI] [PubMed] [Google Scholar]
- De Luca, V., and Cutler, A.J. (1987). Subcellular localization of enzymes involved in indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 85 1099–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva, N., Andersson, S., Orlova, I., Gatto, N., Reichelt, M., Rhodes, D., Boland, W., and Gershenzon, J. (2005). The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 102 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebizuka, Y., Katsube, Y., Tsutsumi, T., Kushiro, T., and Shibuya, M. (2003). Functional genomics approach to the study of triterpene biosynthesis. Pure Appl. Chem. 75 369–374. [Google Scholar]
- Eichel, J., Gonzalez, J.C., Hotze, M., Matthews, R.G., and Schröder, J. (1995). Vitamin-B12-independent methionine synthase from a higher plant (Catharanthus roseus). Molecular characterization, regulation, heterologous expression, and enzyme properties. Eur. J. Biochem. 230 1053–1058. [DOI] [PubMed] [Google Scholar]
- Eichinger, D., Bacher, A., Zenk, M.H., and Eisenreich, W. (1999). Analysis of metabolic pathways via quantitative prediction of isotope labeling patterns: a retrobiosynthetic 13C NMR study on the monoterpene loganin. Phytochemistry 51 223–226. [Google Scholar]
- Fincham, D., and Ralston, B.J. (1981). Molecular dynamics simulation using the CRAY-1 vector processing computer. Comput. Phys. Commun. 23 127–134. [Google Scholar]
- Fahn, W., Laussermair, E., Deus-Neumann, B., and Stöckigt, J. (1985). Late enzymes of vindoline biosynthesis. S-Adenosyl-L-methionine: 11-O-demethyl-17-O-deacetylvindoline-11-O-methyltransferase and unspecific acetylesterase. Plant Cell Rep. 4 337–340. [DOI] [PubMed] [Google Scholar]
- Gang, D.R., Beuerle, T., Ullmann, P., Werck-Reichhart, D., and Pichersky, E. (2002). Differential production of meta-hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases. Plant Physiol. 130 1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gniwotta, F., Vogg, G., Gartmann, V., Carver, T.L.W., Riederer, M., and Jetter, R. (2005). What do microbes encounter at the plant surface? Chemical composition of pea leaf cuticular waxes. Plant Physiol. 139 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grncarevic, M., and Radler, F. (1971). A review of the surface lipids of grapes and their importance in the drying process. Am. J. Enol. Vitic. 22 80–86. [Google Scholar]
- Guex, N., and Peitsch, M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modelling. Electrophoresis 18 2714–2723. [DOI] [PubMed] [Google Scholar]
- Harvey, J.A., van Nouhuys, S., and Biere, A. (2005). Effects of quantitative variation in allelochemicals in Plantago lanceolata on development of a generalist and a specialist herbivore and their endoparasitoids. J. Chem. Ecol. 31 287–302. [DOI] [PubMed] [Google Scholar]
- Hollenbach, B., Schreiber, L., Hartung, W., and Dietz, K.J. (1997). Cadmium leads to stimulated expression of the lipid transfer protein genes in barley: Implications for the involvement of lipid transfer proteins in wax assembly. Planta 203 9–19. [DOI] [PubMed] [Google Scholar]
- Hong, S.B., Hughes, E.H., Shanks, J.V., San, K.Y., and Gibson, S.I. (2003). Role of the non-mevalonate pathway in indole alkaloid production by Catharanthus roseus hairy roots. Biotechnol. Prog. 19 1105–1108. [DOI] [PubMed] [Google Scholar]
- Hotze, M., Schröder, G., and Schröder, J. (1995). Cinnamate-4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in Escherichia coli. FEBS Lett. 374 345–350. [DOI] [PubMed] [Google Scholar]
- Irmler, S., Schröder, G., St-Pierre, B., Crouch, N.P., Hotze, M., Schmidt, J., Strack, D., Matern, U., and Schröder, J. (2000). Indole alkaloid biosynthesis in Catharanthus roseus: New activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J. 24 797–804. [DOI] [PubMed] [Google Scholar]
- Kunst, L., and Samuels, A.L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42 51–80. [DOI] [PubMed] [Google Scholar]
- Kutchan, T.M. (2005). A role for intra- and intercellular translocation in natural product biosynthesis. Curr. Opin. Plant Biol. 8 292–300. [DOI] [PubMed] [Google Scholar]
- Lange, B.M., Wildung, M.R., Stauber, E.J., Sanchez, C., Pouchnik, D., and Croteau, R. (2000). Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc. Natl. Acad. Sci. USA 97 2934–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levac, D., Murata, J., Kim, W.S., and De Luca, V. (2008). Application of carborundum abrasion for investigating leaf epidermis: Molecular cloning of Catharanthus roseus 16-hydroxytabersonine-16-O-methyltransferase. Plant J. 53 225–236. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1999). The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 47–65. [DOI] [PubMed] [Google Scholar]
- Lopes, S., von Poser, G.L., Kerber, V.A., Farias, F.M., Konrath, E.L., Moreno, P., Sobral, M.E., Zuanazzi, J.A.S., and Henriques, A.T. (2004). Taxonomic significance of alkaloids and iridoids glucosides in the tribe Psychotrieae (Rubiaceae). Biochem. Syst. Ecol. 32 1187–1195. [Google Scholar]
- Madyastha, K.M., Guarnaccia, R., Baxter, C., and Coscia, C.J. (1973). S-Adenosyl-L-methionine: Loganic acid methyltransferase. J. Biol. Chem. 248 2497–2501. [PubMed] [Google Scholar]
- Mahroug, S., Courdavault, V., Thiersault, M., St-Pierre, B., and Burlat, V. (2006). Epidermis is a pivotal site of at least four secondary metabolic pathways in Catharanthus roseus aerial organs. Planta 223 1191–1200. [DOI] [PubMed] [Google Scholar]
- Martin, C., and Glover, B.J. (2007). Functional aspects of cell patterning in aerial epidermis. Curr. Opin. Plant Biol. 10 70–82. [DOI] [PubMed] [Google Scholar]
- Meijer, A.H., Souer, E., Verpoorte, R., and Hoge, J.H. (1993). Isolation of cytochrome P-450 cDNA clones from the higher plant Catharanthus roseus by a PCR strategy. Plant Mol. Biol. 22 379–383. [DOI] [PubMed] [Google Scholar]
- Millar, A.A., Clemens, S., Zachgo, S., Giblin, E.M., Taylor, D.C., and Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, J., Bienzle, D., Brandle, J.E., Sensen, C.W., and De Luca, V. (2006). Expressed sequence tags from Madagascar periwinkle (Catharanthus roseus). FEBS Lett. 580 4501–4507. [DOI] [PubMed] [Google Scholar]
- Murata, J., and De Luca, V. (2005). Localization of tabersonine 16-hydroxylase and 16-OH tabersonine-16-O-methyltransferase to leaf epidermal cells defines them as a major site of precursor biosynthesis in the vindoline pathway in Catharanthus roseus. Plant J. 44 581–594. [DOI] [PubMed] [Google Scholar]
- Oudin, A., Mahroug, S., Courdavault, V., Hervouet, N., Zelwer, C., Rodríguez-Concepción, M., St-Pierre, B., and Burlat, V. (2007). Spatial distribution and hormonal regulation of gene products from methyl erythritol phosphate and monoterpene-secoiridoid pathways in Catharanthus roseus. Plant Mol. Biol. 65 13–30. [DOI] [PubMed] [Google Scholar]
- Pauw, B., Hilliou, F.A.O., Sandonis, V., Chatel, M.G., de Wolf, C.J.F., Champion, A., Pré, M., van Duijn, B., Kijne, J.W., van der Fits, L., and Memelink, J. (2004). Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J. Biol. Chem. 279 52940–52948. [DOI] [PubMed] [Google Scholar]
- Peebles, C.A.M., Hong, S.B., Gibson, S.I., Shanks, J.V., and San, K.Y. (2006). Effects of terpenoid precursor feeding on Catharanthus roseus hairy roots over-expressing the alpha or the alpha and beta subunits of anthranilate synthase. Biotechnol. Bioeng. 93 534–540. [DOI] [PubMed] [Google Scholar]
- Peñuelas, J., Sardans, J., Stefanescu, C., Parella, T., and Filella, I. (2006). Lonicera implexa leaves bearing naturally laid eggs of the specialist herbivore Euphydryas aurinia have dramatically greater concentrations of iridoid glycosides than other leaves. J. Chem. Ecol. 32 1925–1933. [DOI] [PubMed] [Google Scholar]
- Pereyra, P.C., and Bowers, M.D. (1988). Iridoid glycosides as oviposition stimulants for the buckeye butterfly, Junonia coenia (Nymphalidae). J. Chem. Ecol. 14 917–928. [DOI] [PubMed] [Google Scholar]
- Pyee, J., Yu, H., and Kolattukudy, P.E. (1994). Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Arch. Biochem. Biophys. 311 460–468. [DOI] [PubMed] [Google Scholar]
- Qi, X., Bakht, S., Qin, B., Leggett, M., Hemmings, A., Mellon, F., Eagles, J., Werck-Reichhart, D., Schaller, H., Lesot, A., Melton, R., and Osbourn, A. (2006). A different function for a member of an ancient and highly conserved cytochrome P450 family from essential sterols to plant defense. Proc. Natl. Acad. Sci. USA 103 18848–18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, G., Gu, H., Zhao, Y., Ma, Z., Shi, G., Yang, Y., Pichersky, E., Chen, H., Liu, M., Chen, Z., and Qu, L.J. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rischer, H., Oresic, M., Seppanen-Laakso, T., Katajamaa, M., Lammertyn, F., Ardiles-Diaz, W., Montagu, M.C.E., Inzé, D., Oksman-Caldentey, K.M., and Goossens, A. (2006). Gene-to-metabolite networks for terpenoid indole alkaloid biosynthesis in Catharanthus roseus cells. Proc. Natl. Acad. Sci. USA 103 5614–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer, M., Knani, M., Simonin, P., Sutter, B., and Sahm, H. (1993). Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 295 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J., Nam, K., D'Auria, J., and Pichersky, E. (1999). S-Adenosyl-L-methionine: Salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 367 9–16. [DOI] [PubMed] [Google Scholar]
- Ryckaert, J.P., Ciccotti, G., and Berendsen, H.J.C. (1977). Numerical-integration of cartesian equations of motion of a system with constraints – Molecular dynamics of N-alkanes. J. Comput. Phys. 23 327–341. [Google Scholar]
- Schmitz-Hoerner, R., and Weissenböck, G. (2003). Contribution of phenolic compounds to the UV-B screening capacity of developing barley primary leaves in relation to DNA damage and repair under elevated UV-B levels. Phytochemistry 64 243–255. [DOI] [PubMed] [Google Scholar]
- Schröder, G., Unterbusch, E., Kaltenbach, M., Schmidt, J., Strack, D., De Luca, V., and Schröder, J. (1999). Light-induced cytochrome P450-dependent enzyme in indole alkaloid biosynthesis: Tabersonine 16-hydroxylase. FEBS Lett. 458 97–102. [DOI] [PubMed] [Google Scholar]
- Schröder, G., Wehinger, E., and Schröder, J. (2002). Predicting the substrates of cloned plant O-methyltransferases. Phytochemistry 59 1–8. [DOI] [PubMed] [Google Scholar]
- Schröder, G., Wehinger, E., Lukacin, R., Wellmann, F., Seefelder, W., Schwab, W., and Schröder, J. (2004). Flavonoid methylation: A novel 4′-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases. Phytochemistry 65 1085–1094. [DOI] [PubMed] [Google Scholar]
- Schwede, T., Kopp, J., Guex, N., and Peitsch, M.C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Song, J.T., Cheong, J.J., Lee, Y.H., Lee, Y.W., Hwang, I., Lee, J.S., and Choi, Y.D. (2001). Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98 4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, T., and Griffiths, D.W. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171 469–499. [DOI] [PubMed] [Google Scholar]
- Shukla, A.K., Shasany, A.K., Gupta, M.M., and Khanuja, S.P.S. (2006). Transcriptome analysis in Catharanthus roseus leaves and roots for comparative terpenoid indole alkaloid profiles. J. Exp. Bot. 57 3921–3932. [DOI] [PubMed] [Google Scholar]
- Sterk, P., Booij, H., Schellekens, G.A., Van Kammen, A., De Vries, S.C. (1991). Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell 3 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, L.H., Blom, T.J.M., and Verpoorte, R. (1993). Subcellular localization of tryptophan decarboxylase, strictosidine synthase and strictosidine glucosidase in suspension cultured cells of Catharanthus roseus and Tabernaemontana divaricata. Plant Cell Rep. 12 573–576. [DOI] [PubMed] [Google Scholar]
- St-Pierre, B., Vazquez-Flota, F.A., and De Luca, V. (1999). Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 11 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma, S., Hecht, U., Kippers, A., Botella, J., de Vries, S., and Somerville, C. (1994). Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol. 105 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usia, T., Watabe, T., Kadota, S., and Tezuka, Y. (2005). Cytochrome P450 2D6 (CYP2D6) inhibitory constituents of Catharanthus roseus. Biol. Pharm. Bull. 28 1021–1024. [DOI] [PubMed] [Google Scholar]
- van der Fits, L., and Memelink, J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289 295–297. [DOI] [PubMed] [Google Scholar]
- van der Fits, L., Zhang, H., Menke, F.L.H., Deneka, M., and Memelink, J. (2000). Catharanthus roseus BPF-1 homologue interacts with an elicitor-responsive region of the secondary metabolite biosynthetic gene Str and is induced by elicitor via a JA-independent signal transduction pathway. Plant Mol. Biol. 44 675–685. [DOI] [PubMed] [Google Scholar]
- Wagner, G.J., Wang, E., and Shepherd, R.W. (2004). New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. (Lond.) 93 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, H., Bladt, S., and Zgainski, E. (1984). Plant Drug Analysis: A Thin Layer Chromatography Atlas. (New York: Springer-Verlag).
- Yamamoto, H., Katano, N., Ooi, A., and Inoue, K. (1999). Transformation of loganin and 7-deoxyloganin into secologanin by Lonicera japonica cell suspension cultures. Phytochemistry 50 417–422. [Google Scholar]
- Yamamoto, H., Katano, N., Ooi, A., and Inoue, K. (2000). Secologanin synthase which catalyzes the oxidative cleavage of loganin into secologanin is a cytochrome P450. Phytochemistry 53 7–12. [DOI] [PubMed] [Google Scholar]
- Yamamoto, H., Sha, M., Kitamura, Y., Yamaguchi, M., Kitano, N., and Inoue, K. (2002). Iridoid biosynthesis: 7-Deoxyloganetic acid 1-O-glucosyltransferase in cultured Lonicera japonica cells. Plant Biotechnol. 19 295–301. [Google Scholar]
- Yang, Y., Yuan, J.S., Ross, J., Noel, J.P., Pichersky, E., and Chen, F. (2006). An Arabidopsis thaliana methyltransferase capable of methylating farnesoic acid. Arch. Biochem. Biophys. 448 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta, C., Ross, J.R., Koscheski, P., Yang, Y., Pichersky, E., and Noel, J.P. (2003). Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.