Abstract
Oxidation of amino acid residues in proteins can be caused by a variety of oxidizing agents normally produced by cells. The oxidation of methionine in proteins to methionine sulfoxide is implicated in aging as well as in pathological conditions, and it is a reversible reaction mediated by a ubiquitous enzyme, peptide methionine sulfoxide reductase. The reversibility of methionine oxidation suggests that it could act as a cellular regulatory mechanism although no such in vivo activity has been demonstrated. We show here that oxidation of a methionine residue in a voltage-dependent potassium channel modulates its inactivation. When this methionine residue is oxidized to methionine sulfoxide, the inactivation is disrupted, and it is reversed by coexpression with peptide methionine sulfoxide reductase. The results suggest that oxidation and reduction of methionine could play a dynamic role in the cellular signal transduction process in a variety of systems.
Keywords: inactivation, oocyte, methionine sulfoxide reductase
Oxidation of amino acid residues in proteins are known to drastically affect their functional properties. Although all amino acids can be oxidized, cysteine, methionine, histidine, tyrosine, and tryptophan are readily oxidized by biologically relevant oxidants such as H2O2, hydroxyl radicals, and hypochlorite (1). Oxidation is known to affect potassium channel proteins (2–5), which play crucial roles in the cellular signal transduction activities in excitable cells in the brain, heart, and muscle (6) as well as in nonexcitable cells such as lymphocytes (7). Although cysteine oxidation has been generally suspected, it is not clear which amino acid residues are oxidized to alter the channel properties. In addition to cysteine, methionine is also easily oxidized to form methionine sulfoxide [Met(O)]. Oxidation of methionine residues has been implicated in some pathological conditions, such as emphysema, adult respiratory distress syndrome, cataracts, and aging (1). Oxidation of methionine is unique in that the reaction is reversible and enzymatically controlled. Reduction of Met(O) to methionine is catalyzed by an enzyme, peptide Met(O) reductase (MsrA) (8), which has been recently cloned from Escherichia coli (9) and bovine adrenal medulla (10). MsrA is very specific for Met(O) residues and is found in a variety of tissues, especially in the kidney, retina, and neurons (10, 11). The reversible oxidation of methionine and reduction mediated by MsrA could provide an important biological regulatory mechanism like cysteine oxidation/reduction and phosphorylation/dephosphorylation. In particular, the high level of expression in neurons (11) hints that MsrA may have functional roles in regulating neuronal excitability.
Voltage-dependent K+ channels open in response to depolarization and then often undergo inactivation. Properties of the potassium channels, such as inactivation, can be modulated by a variety of means, potentially contributing to learning and memory. Inactivation kinetics of the K+ channels play critical roles in determining cellular excitability, including cardiac rhythmicity. Some genetic disorders, such as long QT syndrome and episodic ataxia, have been shown to involve defects in voltage-dependent K+ channels (12–14). In Shaker voltage-dependent K+ channels, fast inactivation is typically mediated by N-type inactivation in which approximately the first 20 amino acid residues form the inactivation ball structure, which physically occludes the channel pore to induce inactivation (15–17). The nonpolar nature of the first 10 amino acids is important in determining the rate constant of recovery from N-type inactivation, and the positive charges in the next 10 residues are important for the rate constant of entry into N-type inactivation (15, 18). NMR structures of the ball domain of two mammalian Shaker-like channels are now known (19). Because the biophysical and molecular properties of the Shaker channels have been studied extensively, these channels represent a good model system for which a real-time assay of the protein function is easily achieved. Thus, we examined the functional roles of methionine oxidation in regulation of the Shaker channel function. The results show that methionine oxidation and reduction facilitated by MsrA regulate the channel inactivation time course, suggesting that methionine oxidation could act as an important functional regulator of many proteins.
MATERIALS AND METHODS
Channel Expression and Electrophysiology.
The K+ channels are expressed in Xenopus oocytes essentially as described (15). The ShC/B DNA was linearized with Bgl2, and the RNA was synthesized using SP6 RNA polymerase using a commercially available kit (Ambion, Austin, TX). Whole oocyte currents were recorded using a Warner 725C amplifier (Warner Instruments, Hamden, CT). The electrodes typically had an initial input resistance of 0.5–0.8 Mohms when filled with 3 M KCl. The patch-clamp recordings were obtained using an AxoPatch 200A (Axon Instruments, Foster City, CA) with borosilicate pipettes. Unless otherwise indicated, the holding voltage was −90 mV. The signals were filtered through 8-pole Bessel filters (Frequency Devices, Haverhill, MA) and digitized with ITC-16 interfaces (Instrutech, Mineola, NY) attached to Apple Macintosh computers. The whole oocyte data presented were collected 1 day after RNA injection. The data were collected and analyzed using pulse/pulsefit (Heka Electronics, Lambrecht, Germany), igorpro (WaveMetrics, Lake Oswego, OR), and datadesk (DataDescriptions, Ithaca, NY). Linear leak and capacitative currents have been subtracted from the data presented. The external solution contained (in millimolars): 140 NaCl, 10 KCl, 2 CaCl2, and 10 Hepes, pH adjusted to 7.2 with N-methylglucamine. The internal/bath solution in the patch-clamp experiments contained (in millimolars): 140 KCl, 2 MgCl2, 10 EGTA, and 10 Hepes, pH adjusted to 7.2 with N-methylglucamine. Recordings were typically made 1 day after RNA injection. Statistical comparisons were made using the data collected from the same batches of oocytes. Chloramine-T (Sigma) solutions were prepared immediately before use. All procedures conformed to an animal use protocol approved by the University of Iowa Animal Care and Use Committee.
The ShC/B M3L channel was constructed from the ShB channel using the standard PCR-mediated cassette mutagenesis as described (20). The ShB·pSP72B was cut with PflM1 and Rsr2, and the DNA segment corresponding to the ShC/B amino terminus with the M3L mutation was ligated in. The sequence of the PCR-amplified DNA segment was verified (The University of Iowa DNA Core Facility). The ShC/B M3L DNA was linearized with NdeI and transcribed with T7 polymerase.
MsrA Expression.
The coding sequence of MsrA was PCR amplified (5′-GTGGAATTCCAACCAAAGATATTAAACTAACCATGCTCTCGGTCACCAGGAGGGCCCTCCAGCTCTTTCAC-3′, 5′-CAGATCAAGCTTCATGGGGAGAAATTACTTTTTAATACC-3′) from the plasmid described by Moskovitz et al. (10), digested with EcoRI and HindIII, and subcloned into an oocyte expression plasmid vector (pGEMHE) (21) cut with the same enzymes. The PCR-amplified DNA sequence was verified (The University of Iowa DNA Core Facility). The MsrA DNA was linearized with NheI and was transcribed using T7 RNA polymerase. The MsrA activity was assayed essentially as described (22). The oocytes were suspended in a Tris buffer solution, sonicated, and centrifuged. The cell-free extract below the lipid layer was removed and incubated with DTT (15 mM) and 3[H]N-acetylmethionine sulfoxide. After the incubation, the mixture was acidified, the 3[H]N-acetylmethionine formed was extracted with ethyl acetate, and the radioactivity in the ethyl acetate phase was determined. The MsrA activity in uninjected cells was typically <0.1 pmol/μg protein/hour and the activity increased to 5 pmol/μg protein/hour 1 day after MsrA RNA injection.
RESULTS
Inactivation Time Course Is Variable in ShC/B.
Outward K+ currents recorded from one alternatively spliced variant of the Shaker gene, ShC/B (23, 24), heterologously expressed in Xenopus oocytes, are shown in Fig. 1A. Although identical stimuli were applied, the currents recorded from different cells showed a marked variability in the inactivation time course. In some cells, the inactivation time course was fast and well described by a single exponential, consistent with the N-type inactivation mechanism being dominant. In others, however, the inactivation time course was slower and much more complex, requiring a sum of at least two exponential components. In the latter cells, the overall inactivation time course exhibited both the fast inactivation component and a slow inactivation component. The variability of the inactivation time course could be described as a variable presence of the slow inactivation component. The inactivation time course of K+ currents recorded from cells expressing another splicing variant, ShB (25), which differs from ShC/B only in the amino terminus, did not show a similar variability (data not shown).
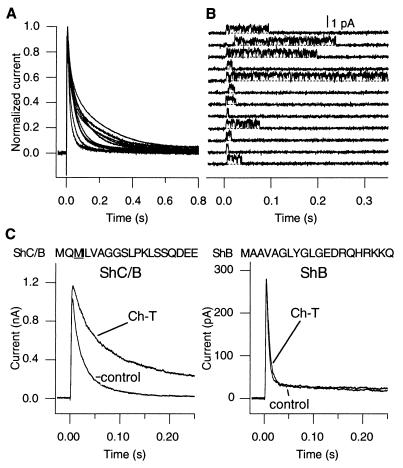
Figure 1.
(A) ShC/B expressed in Xenopus oocytes shows marked variability in the inactivation time course. The whole-oocyte K+ currents recorded at 0 mV from 12 different cells 1 day after RNA injection were scaled and shown superimposed. The mean peak current amplitude was 10.5 μA (SEM = 1.6 μA). (B) One ShC/B channel displays two patterns of inactivation. Openings of one ShC/B channel were elicited by repeated depolarizing pulses to 0 mV every 15 sec in the cell-attached configuration of the patch clamp. The channel occasionally failed to inactivate during the pulse. The mean open times in the sweeps during which the channel failed to inactivate were not greater than those in the sweeps during which the channel inactivated rapidly. There was only a very modest positive correlation between the mean open and the open probability in each depolarizing epoch (r = 0.66, Spearman rank correlation). This patch contained only one active channel as judged by the absence of overlapping openings in more than 150 sweeps. The data were filtered through an eight-pole Bessel filter at 1.2 kHz and digitized at 20 kHz. Essentially similar inactivation patterns were observed in two other single-channel patches. (C) Ch-T slows down the inactivation time course in ShC/B but not in ShB. Macroscopic currents from the ShC/B and ShB channels were recorded in response to depolarizing pulses to +50 mV in the inside-out configuration of the patch clamp. Ch-T (50 μM), prepared fresh immediately before use, was applied to the cytoplasmic side. The first 20 deduced amino acid residues in ShB and ShC/B are also shown. Intracellular Ch-T, up to 0.6 mM, did not markedly affect the ShB inactivation time course.
The single-channel recording from a single ShC/B channel showed that one channel protein is capable of showing two patterns of inactivation (Fig. 1B; also see ref. 24). In response to depolarizing pulses, the ShC/B channel normally entered a long-lived, inactivated state after a few openings. Intermittently, however, the channel inactivated much more slowly and continued to open and close throughout out the pulse. These results indicate that the reversible and intermittent loss of fast inactivation leaving only slow inactivation is responsible for the observed variability in inactivation.
Consistent with the observation that two inactivation patterns were observed from one channel protein, the overall inactivation time course of the macroscopic ShC/B currents sometimes spontaneously slowed down, especially in the cell-free configuration. This slowing was accompanied by an increase in the slow component of inactivation. Spontaneous changes in the inactivation properties upon patch excision have been noted in other K+ channels (2, 26). In one case, cysteine oxidation was shown to account for the loss of N-type inactivation (2). Reducing agents such as DTT and glutathione were able to restore the loss of inactivation caused by cysteine oxidation. To see if oxidation of amino acid residues in the ShC/B protein is responsible for the variability in the inactivation time course (Fig. 1A) and the intermittent loss of inactivation (Fig. 1B), we examined the effects of an oxidant, chloramine-T (Ch-T), on the inactivation time course of ShC/B and ShB. Application of Ch-T, which preferentially oxidizes cysteine and methionine residues, to the cytoplasmic side in the inside-out patch configuration slowed the inactivation time course of ShC/B but not of ShB at the concentrations examined (up to 600 μM) (Fig. 1C). Consistent with the removal of fast N-type inactivation, the slowing of inactivation by low concentrations of Ch-T (<300 μM) was accompanied by a small increase in the peak current amplitude. At greater concentrations (1 mM), however, Ch-T slowed the apparent activation time course and reduced the peak amplitudes of the ShC/B currents. These effects of Ch-T at the high concentrations are probably mediated by the effects on other gating processes (27). Internal DTT (5 mM) in the inside-out patches did not have any consistent effects on the ShC/B currents (data not shown). The observations that ShB does not show the inactivation variability or intermittent loss of inactivation and that Ch-T has differential effects on ShC/B and ShB suggest that the oxidation of a sensitive amino acid unique to ShC/B may be responsible for the inactivation variability of ShC/B.
Coexpression of ShC/B with MsrA Accelerates Inactivation.
ShB and ShC/B differ only in the distal amino terminus in their deduced amino acid sequences (see Fig. 1C). Although cysteine residues are present at the equivalent positions in both channels, ShC/B contains an additional methionine residue in the distal amino terminus, which forms the inactivation ball domain. Methionine residues in proteins are readily oxidized to Met(O), and they are physiologically reduced back to methionine by MsrA using thioredoxin as the reducing system (8, 10). If methionine oxidation is responsible for the inactivation variability and intermittent loss of fast inactivation, coexpression of ShC/B with MsrA should accelerate the overall inactivation time course by reducing the slow component in inactivation. The coexpression should also eliminate the large variability in inactivation. Thus, we examined the effects of MsrA expression on the ShC/B K+ currents. The uninjected oocytes showed a low endogenous level of MsrA activity, and injection of MsrA RNA increased the enzyme activity by >30-fold as measured by the conversion of 3[H]N-acetylmethionine sulfoxide to 3[H]N-acetylmethionine (Fig. 2A). Scaled K+ currents recorded from cells expressing both ShC/B and MsrA are compared with those expressing ShC/B alone in Fig. 2B. MsrA coexpression consistently accelerated the inactivation time course. Furthermore, coexpression with MsrA dramatically reduced the variability in inactivation time course. The inactivation time course was fitted with a sum of two exponentials, and the inactivation parameters are shown in Fig. 2C using boxplots. The MsrA coexpression specifically reduced the slow inactivation component from ≈60 to 10% without markedly affecting the time constant of the fast component or slow component. These results strongly support the hypothesis that methionine oxidation is responsible for the inactivation variability in ShC/B. In addition to the inactivation time course, the MsrA coexpression also affected the time course of recovery from inactivation. As with the inactivation time course during depolarizing pulses, the time course of recovery from inactivation in ShC/B, as measured by a double pulse protocol, was quite variable (Fig. 3B). As predicted, MsrA coexpression reduced the variability and slowed the recovery from inactivation (Fig. 3B).
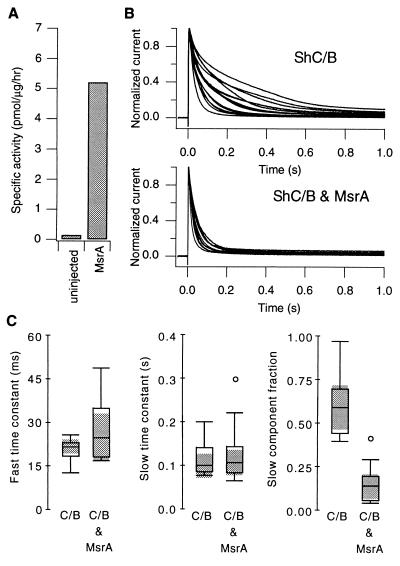
Figure 2.
(A) The MsrA activities measured from uninjected oocytes and the oocytes injected with MsrA RNA (50% dilution, 1 day) are compared using boxplots (Left). Specific activity is defined as the picomol of N-acetylmethionine sulfoxide reduced to N-acetylmethionine per microgram of protein per hour at 37°C. (B) MsrA coexpression accelerates the inactivation time course of ShC/B and reduces the inactivation variability. Scaled whole oocyte K+ currents at +30 mV recorded from the cells expressing ShC/B alone (15 cells, Upper) and ShC/B and MsrA together (12 cells, Lower) are shown. Each oocyte in the control group was injected with 40 nl of 30% dilution ShC/B RNA. In the coexpression group, each oocyte was injected with 40 nl of ShC/B (50%) and MsrA (50%) RNAs. The mean peak K+ current amplitudes were 27.8 μA in the control group (SEM = 3.1 μA) and 15.1 μA in the coexpression group (SEM = 2.1 μA). Similar effects of MsrA coexpression were observed in nine separate batches of oocytes involving ≈230 cells with different expression levels. (C) MsrA specifically decreases the slow inactivation component. The whole oocyte currents elicited at +30 mV were fitted with a sum of two exponentials, and the parameters are compared using boxplots. From left to right, the fast inactivation time constant, slow inactivation time constant, and the fractional amplitude of the slow inactivation component are shown. The shaded areas represent the 95% confidence intervals of the median. Pearson product-moment correlation between the peak current amplitude and the slow component fraction was low (r = −0.14 for the control group and r =−0.23 for the coexpression group). The difference between the two medians of the slow component fraction was statistically significant at P = 0.0006 using the Mann–Whitney U test.
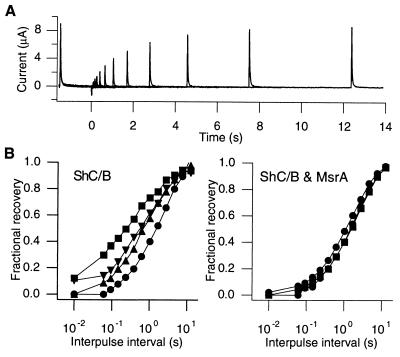
Figure 3.
(A) Representative currents obtained from a cell expressing both the ShC/B channels and MsrA in response to a double-pulse protocol to measure recovery from inactivation. Two 1.5-s pulses to +30 mV separated by the variable intervals were given every 25 s. The interpulse voltage was −100 mV. (B) MsrA coexpression slows the time course of recovery from inactivation and reduces its variability. The left plot shows the recovery curves from four cells expressing ShC/B only, and the right plot shows the recovery curves from three cells expressing both ShC/B and MsrA. The fractional recovery is defined as the ratio of the peak current during the second pulse over that during the first pulse. The channels were expressed in Xenopus oocytes and the whole-oocyte currents were recorded as in A.
Methionine in the Amino-Terminal Ball Domain Is Oxidized to Slow Down Inactivation.
Because ShC/B contains an additional methionine residue near the amino terminus at position 3 (M3), which is not present in ShB, we hypothesized that intermittent oxidation of this amino-terminal methionine to Met(O) is responsible for the intermittent loss of inactivation and the inactivation variability. Previous results have shown that the nonpolar nature of the first 10 amino acid residues is important in N-type inactivation (15, 18). Presence of the polar oxygen group at this location would be expected to disrupt N-type inactivation. If so, the channels with the methionine residue at position three oxidized to Met(O) could be responsible for the slow component of inactivation whereas the intact channels are responsible for the fast component. According to this hypothesis, mutation of the methionine residue (M3) to the less readily oxidized leucine should eliminate the variability in inactivation time course by reducing the slow component, and there should be no effect of coexpression of MsrA on the inactivation time course. K+ currents recorded from cells expressing the mutant ShC/B M3L channels are shown in Fig. 4A. Consistent with the hypothesis, the overall inactivation time course of the K+ currents from the mutant M3L channels was faster and did not show a marked slow component. Furthermore, the inactivation time course was very consistent among the cells examined. The effect of this mutation was specific in that it did not affect the time constant of the fast inactivation phase (Fig. 4B). Internal Ch-T (up to 300 μM) affected neither the peak current amplitudes nor the inactivation kinetics of the ShC/B M3L current (data not shown) although at greater concentrations (>600 μM) it reduced the peak current and slowed the inactivation kinetics. Thus, the results strongly indicate that oxidation of methionine to Met(O) is responsible for the inactivation variability.
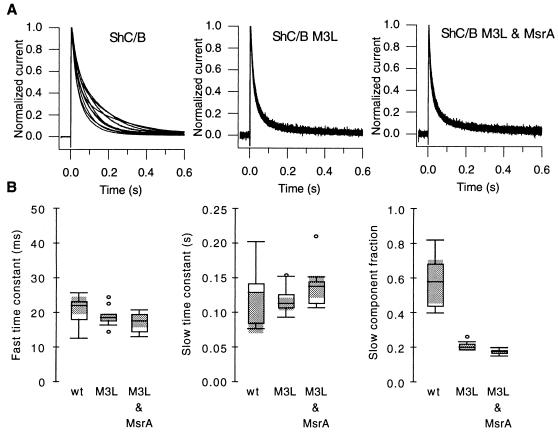
Figure 4.
(A) The M3L mutation accelerates the inactivation time course and eliminates the regulation by MsrA. Currents recorded at +30 mV from cells expressing ShC/B alone (Left, 10 cells), ShC/B M3L alone (Center, 9 cells) and ShC/B M3L and MsrA together (Right, 10 cells) are shown. The mean peak currents for these three groups were 34 μA (SEM = 5.4 μA), 5.1 μA (SEM = 0.74 μA), 2.6 μA (SEM = 0.19 μA), respectively. (B) The M3L mutation specifically decreases the slow inactivation component. The inactivation time course of the currents shown in A was fitted with a sum of two exponentials and the parameters are compared using boxplots. From left to right, the fast inactivation time constant, slow inactivation time constant, and the fractional amplitude of the slow inactivation component are shown. The shaded areas represent the 95% confidence intervals of the median.
If MsrA is accelerating the inactivation time course by enhancing the reduction of the Met(O) at position 3 in ShC/B, it is expected that the inactivation time course of ShC/B M3L should be unaffected by MsrA. Fig. 4A shows the currents recorded from the cells expressing both the ShC/B M3L channels and MsrA. The inactivation time course of the ShC/B M3L channel was not affected by MsrA coexpression, confirming that MsrA exerts its effects by acting on the Met(O) at the third position in the channel protein
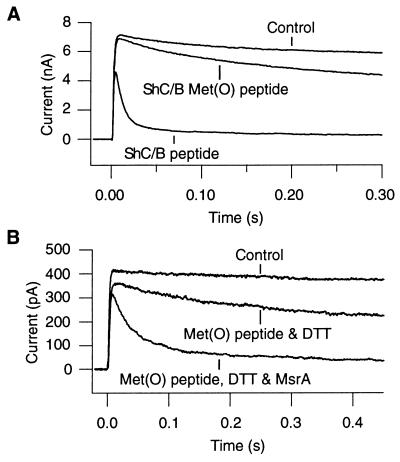
Synthetic peptides corresponding to the amino-terminal ball domains of the Shaker channels are known to restore inactivation (16, 18). Fig. 5 shows that intracellular application of a synthetic peptide corresponding to the first 16 amino acid residues of the ShC/B channel induced inactivation in the ShBΔ6–46:T449V channel, which shows neither N-type nor C-type inactivation in oocytes (15, 20). A mutant ShC/B synthetic peptide, where methionine at position 3 is replaced with Met(O), was much less effective in inducing inactivation, confirming that oxidation of this methionine to Met(O) interferes with N-type inactivation. Furthermore, the Met(O)-containing ShC/B peptide, preincubated with purified recombinant MsrA (0.06 μg/μl) (10) and DTT (1 mM), was more effective in inducing inactivation than the Met(O) peptide treated with DTT alone (Fig. 5B). The result suggests that MsrA was able to catalyze reduction of Met(O) at position 3 to methionine, thus enhancing the inactivation process.
Figure 5.
(A) The synthetic wild-type ShC/B peptide induces inactivation whereas the Met(O)-containing ShC/B peptide is much less effective. The ShBΔ6–46:T449V macroscopic currents at 0 mV were recorded in the inside-out configuration. Intracellular application of the wild-type ShC/B peptide (200 μM) rapidly induced inactivation whereas the Met(O)-containing ShC/B peptide (200 μM) was much less effective. The ShC/B peptide (Met-Glu-Met-ILe-Leu-Val-Ala-Gly-Gly-Ser-Leu-Pro-Lys-Leu-Ser-Ser) and the Met(O)-containing ShC/B peptide [Met-Glu-Met(O)-ILe-Leu-Val-Ala-Gly-Gly-Ser-Leu-Pro-Lys-Leu-Ser-Ser] were obtained from Genemed Biotechnologies, South San Francisco, CA (>85% pure). (B) Preincubation of the Met(O)-containing ShC/B peptide with MsrA and DTT (1 mM) restores its ability to induce inactivation. The ShBΔ6–46:T449V macroscopic currents were recorded as in A. The Met(O)-containing peptide incubated with DTT (1 mM) but not with MsrA is not markedly effective in inducing inactivation (125 μM). The Met(O)-containing ShC/B peptide (125 μM) was incubated with purified recombinant MsrA (0.03 μg/μl) (10) in the presence of DTT (1 mM) for 1.5 h at 37°C and applied to the patch.
DISCUSSION
The results presented above argue that oxidation of the methionine at position 3 in ShC/B (M3) has an important functional role in regulating the N-type inactivation mechanism. When methionine is present, the amino terminus is able to form the inactivation ball structure and stabilize N-type inactivation. However, when the methionine is oxidized, the presence of the polar oxygen alters the properties of the ball domain and allows the channel to recover very rapidly from the inactivated state, thus effectively eliminating the fast inactivation process. The single-channel data shown in Fig. 1B suggest that oxidation and reduction of this residue can take place rapidly, within several seconds. This conversion process is dramatically enhanced by heterologously expressed MsrA as its expression accelerates the inactivation time course and delays the recovery time course.
Oxidation of proteins as a result of reactions with various biological oxidants is important in many physiological and pathological conditions (28–32). Oxidation is considered to play major roles in the immune system, cardiovascular function/dysfunction (33, 34), Alzheimer disease (35), and other aging-related phenomena (29–32, 36). Oxidation of amino acid residues may also be a part of many signal transduction pathways. For example, some effects of nitric oxide (NO), implicated in many physiological phenomena, including learning and memory, could be mediated by oxidation (1, 37). Although oxidation of cysteine residues is well known, methionine residues can also be readily oxidized to form Met(O). Oxidation of methionine to Met(O) causes a marked difference in polarity, essentially converting a nonpolar amino acid into a polar hydrophilic one. Methionine oxidation in proteins was considered to be one of the consequences of oxidative damage to cells, which in many cases leads to the loss of biological activity. More recently, it was suggested that reversible oxidation of methionine residues may act as an oxidant scavenger in the cellular antioxidant defense mechanism because many methionine residues in a protein might be oxidized without affecting its activity (38). The results presented here demonstrate that methionine oxidation/reduction has important functional consequences and could act as a functional regulatory mechanism. In the ShC/B channel, oxidation of a methionine residue in the inactivation ball slows down the inactivation process, and the effect is efficiently reversed by MsrA. It will be important to determine what induces oxidation of this methionine residue. One tantalizing possibility is that the membrane depolarization itself may induce oxidation (39). Electrolysis of intracellular/extracellular fluids in heart has been suggested to produce reactive oxygen species (40).
The bovine MsrA contains several consensus sites for phosphorylation by protein kinase C, suggesting that the reductase activity could be modulated as part of a transduction pathway controlling cellular activity. If the membrane depolarization itself could induce oxidation as suggested by previous experiments (39, 40), membrane depolarization could have a profound impact on the channel function, which might outlast the original length of depolarization. The duration of the channel modification is limited by the action of MsrA, which in turn could be regulated by protein kinase C. Thus, oxidation of methionine residues in ion channel proteins and its reversal by MsrA may play roles in an activity-dependent modulation of the cellular function, as in learning and memory. MsrA may be important not only as a repair enzyme but also as a key player in the cellular signal transduction cascade. Dynamic functional roles of methionine oxidation and its reversal by MsrA in a variety of systems need to be explored.
Acknowledgments
We thank Dr. J. Thommandru and Mina Masrorpour for technical assistance and L. W. Polsfuss for instrument design. This work was in part supported by the Human Frontier Science Program (T.H., S.H.H.). M.A.C. was supported by the American Heart Association. T.H. also was supported in part by the Klingenstein Foundation and McKnight Foundation.
ABBREVIATIONS
- MsrA
methionine sulfoxide reductase
- Met(O)
methionine sulfoxide, Ch-T, chloramine-T
References
- 1.Vogt W. Free Radical Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 2.Ruppersberg J P, Stocker M, Pongs O, Heinemann S H, Frank R, Koenen M. Nature (London) 1991;352:711–714. doi: 10.1038/352711a0. [DOI] [PubMed] [Google Scholar]
- 3.Duprat F, Guillemare E, Romey G, Fink M, Lesage F, Lazdunski M, Honore E. Proc Natl Acad Sci USA. 1995;92:11796–11800. doi: 10.1073/pnas.92.25.11796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vega-Saenz de Miera E, Rudy B. Biochem Biophys Res Commun. 1992;186:1681–1687. doi: 10.1016/s0006-291x(05)81602-x. [DOI] [PubMed] [Google Scholar]
- 5.Stephens G J, Owen D G, Robertson B. Pflügers Arch Eur J Physiol. 1996;431:435–442. doi: 10.1007/BF02207283. [DOI] [PubMed] [Google Scholar]
- 6.Pallotta B S, Wagoner P K. Physiol Rev. 1992;72:S49–67. doi: 10.1152/physrev.1992.72.suppl_4.S49. [DOI] [PubMed] [Google Scholar]
- 7.Lewis R S, Cahalan M D. Annu Rev Immunol. 1995;13:623–653. doi: 10.1146/annurev.iy.13.040195.003203. [DOI] [PubMed] [Google Scholar]
- 8.Brot N, Weissbach L, Werth J, Weissbach H. Proc Natl Acad Sci USA. 1981;78:2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman M A, Nelson H, Weissbach H, Brot N. J Biol Chem. 1992;267:15549–15551. [PubMed] [Google Scholar]
- 10.Moskovitz J, Weissbach H, Brot N. Proc Natl Acad Sci USA. 1996;93:2095–2099. doi: 10.1073/pnas.93.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moskovitz J, Jenkins N A, Gilbert D J, Copeland N G, Jursky F, Weissbach H, Brot N. Proc Natl Acad Sci USA. 1996;93:3205–3208. doi: 10.1073/pnas.93.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curran M E, Splawski I, Timothy K W, Vincent G M, Green E D, Keating M T. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 13.Sanguinetti M C, Curran M E, Zou A, Shen J, Spector P S, Atkinson D L, Keating M T. Nature (London) 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 14.Adelman J P, Bond C T, Pessia M, Maylie J. Neuron. 1995;15:1449–1454. doi: 10.1016/0896-6273(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 15.Hoshi T, Zagotta W N, Aldrich R W. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 16.Zagotta W N, Hoshi T, Aldrich R W. Science. 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- 17.Demo S D, Yellen G. Neuron. 1991;7:743–753. doi: 10.1016/0896-6273(91)90277-7. [DOI] [PubMed] [Google Scholar]
- 18.Murrell-Lagnado R D, Aldrich R W. J Gen Physiol. 1993;102:949–975. doi: 10.1085/jgp.102.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antz C, Geyer M, Fakler B, Schott M K, Guy H R, Frank R, Ruppersberg J P, Kalbitzer H R. Nature (London) 1997;385:272–275. doi: 10.1038/385272a0. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Barneo J, Hoshi T, Heinemann S H, Aldrich R W. Receptors Channels. 1993;1:61–71. [PubMed] [Google Scholar]
- 21.Liman E R, Tytgat J, Hess P. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 22.Brot N, Werth J, Koster D, Weissbach H. Anal Biochem. 1982;122:291–294. doi: 10.1016/0003-2697(82)90283-4. [DOI] [PubMed] [Google Scholar]
- 23.Zagotta W N, Hoshi T, Aldrich R W. Proc Natl Acad Sci USA. 1989;86:7243–7247. doi: 10.1073/pnas.86.18.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aldrich R W, Hoshi T, Zagotta W N. Cold Spring Harbor Symp Quant Biol. 1990;55:19–27. doi: 10.1101/sqb.1990.055.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz T L, Tempel B L, Papazian D M, Jan Y N, Jan L Y. Nature (London) 1988;331:137–142. doi: 10.1038/331137a0. [DOI] [PubMed] [Google Scholar]
- 26.Marom S, Goldstein S A N, Kupper J, Levitan I B. Receptors Channels. 1993;1:81–88. [PubMed] [Google Scholar]
- 27.Schlief T, Schonherr R, Heinemann S H. Pflugers Arch. 1996;431:483–493. doi: 10.1007/BF02191894. [DOI] [PubMed] [Google Scholar]
- 28.Dean R T, Gieseg S, Davies M J. Trends Biochem Sci. 1993;18:437–441. doi: 10.1016/0968-0004(93)90145-d. [DOI] [PubMed] [Google Scholar]
- 29.Stadtman E R. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 30.Rattan S I, Derventzi A, Clark B F. Ann NY Acad Sci. 1992;663:48–62. doi: 10.1111/j.1749-6632.1992.tb38648.x. [DOI] [PubMed] [Google Scholar]
- 31.Gibson G, Martins R, Blass J, Gandy S. Life Sci. 1996;59:477–489. doi: 10.1016/0024-3205(96)00327-x. [DOI] [PubMed] [Google Scholar]
- 32.Yu B P. Physiol Rev. 1994;74:139–162. doi: 10.1152/physrev.1994.74.1.139. [DOI] [PubMed] [Google Scholar]
- 33.Jackson C V, Mickelson J K, Pope T K, Rao P S, Lucchesi B R. Am J Physiol. 1986;251:H1225–H1231. doi: 10.1152/ajpheart.1986.251.6.H1225. [DOI] [PubMed] [Google Scholar]
- 34.Kukreja R C, Hess M L. Cardiovasc Res. 1992;26:641–655. doi: 10.1093/cvr/26.7.641. [DOI] [PubMed] [Google Scholar]
- 35.Good P F, Werner P, Hsu A, Olanow C W, Perl D P. Am J Pathol. 1996;149:21–28. [PMC free article] [PubMed] [Google Scholar]
- 36.Yu B P, Yang R. Ann NY Acad Sci. 1996;685:1–11. doi: 10.1111/j.1749-6632.1996.tb39047.x. [DOI] [PubMed] [Google Scholar]
- 37.Brune B, Mohr S, Messmer U K. Rev Physiol Biochem Pharmacol. 1996;127:1–30. doi: 10.1007/BFb0048263. [DOI] [PubMed] [Google Scholar]
- 38.Levine R L, Mosoni L, Berlett B S, Stadtman E R. Proc Natl Acad Sci USA. 1996;93:15306–15340. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.al-Mehdi A B, Ischiropoulos H, Fisher A B. J Cell Physiol. 1996;166:274–280. doi: 10.1002/(SICI)1097-4652(199602)166:2<274::AID-JCP4>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 40.Jackson C V, Mickelson J K, Stringer K, Rao P S, Lucchesi B R. J Pharmacol Methods. 1986;15:305–320. doi: 10.1016/0160-5402(86)90010-0. [DOI] [PubMed] [Google Scholar]