Abstract
Background
Kidneys have an important function in blood pressure (BP) regulation and elevated BP may lead to kidney failure. Chr2p12-p13 region linked to BP traits in multiple studies harbours a potential candidate for BP and renal function, N-acetyltransferase 8 (NAT8) expressed in embryonic and adult kidney and associated with nephrotoxicity response.
Methods/Results
We report the first study exploring NAT8 as a potential candidate gene for blood pressure and kidney function. The resequencing (n = 42, random Estonian samples) identified 15 NAT8 polymorphisms, including 6 novel variants. The diversity of NAT8 5' upstream region (π/bp = 0.00320) exceeded up to 10 times the variation in the NAT8 genic region (π/bp = 0.00037) as well as the average variation (π/bp = 0.00040) for the promoters of 29 reference genes associated with hypertension. We suggest that a potential source for such high variation could be an active gene conversion process from NAT8B duplicate gene to NAT8. Similarly to NAT8, several reference genes with the most variable upstream regions have also duplicate copies. The NAT8 promoter SNPs were targeted with pilot quantitative association studies for blood pressure (n = 137, healthy unrelated individuals) and for the index of kidney function – estimated glomerular filtration rate (eGFR; n = 157 hypertensives with and without nephropathy). Minor alleles of these polymorphisms revealed a significant protective effect against elevated systolic BP as well as kidney failure in hypertension patients (p < 0.05; linear regression model, addictive effect).
Conclusion
The full resequencing and pilot association study of a novel positional candidate gene for blood pressure and renal function, human N-acetyltransferase 8, suggested a contribution of highly variable NAT8 promoter polymorphisms in determination of systolic blood pressure and eGFR. Based on in silico analysis, we raise the hypothesis that the alternative SNP alleles of the NAT8 upstream region may have differential effect on gene expression.
Background
Twin studies have shown that approximately half of the inter-individual variance in blood pressure (BP) level is heritable [1,2]. As distinctive for complex diseases, pathologically high BP diagnosed as essential hypertension (repeated systolic and diastolic BP readings > 140 and/or > 90 mmHg) evolves in association with genetic predisposition and personal life-style. Identification of the mutations behind Mendelian forms leading to hypo- or hypertension has highlighted the central role of the renal metabolic pathways in blood pressure regulation [3]. Consistently, depending on the severity and duration of the hypertension, the patients may develop morphological changes in vasculature of kidneys followed by the progression renal failure [4-6]. In the other way around, it has also been proposed that the defective embryonic development of the kidney or loss of existing nephrons may act as a predisposing factor for primary hypertension [7-10].
Genome-wide linkage studies [11] have identified the short-arm of human Chr2 as one of the four major genomic regions to be responsible for BP regulation and therefore hold a risk for essential hypertension. Several candidate genes (ADD2; SLC8A1) from this region have been proposed and investigated in detail to play a role in the aetiology of essential hypertension [12,13]. For this study we screened the reported linkage regions at Chr2 for novel BP regulating genes involved in kidney function and renal metabolic pathways. Within the window of 10 Mb from the four reported linkage peaks [11] we have identified at 2p13.1-p12 a novel functionally relevant candidate gene N-acetyltransferase 8 (NAT8) that is highly expressed in normal human embryonic kidney and liver. NAT8 protein has been suggested to play an important role in development and maintenance of the structure and function in adult kidney and liver [14]. Although exact function of the NAT8 is unknown, its expression profile is related to detoxification pathways in kidney [15].
In the present study, we have carried out the resequencing of human NAT8 gene followed by a pilot association study of the common NAT8 sequence variation with blood pressure measurements and the index of kidney function. We show that most of the NAT8 variation is harboured in the 5'UTR and upstream regions, which is associated with both, systolic blood pressure levels and the estimated glomerular filtration rate (eGFR).
Methods
Study Population
The study has been approved by the Ethics Committee on Human Research of University of Tartu, Estonia (permissions no 122/13,22.12.2003; 137/20, 25.04.2005). The initial study for resequencing (n = 42) and the association studies (detailed description found below) were carried out using samples of recruited individuals from the Estonian population, classified as a typical North-European population based on its genetic composition [16]. All participating individuals gave their informed consent prior to their inclusion into the study.
Resequencing
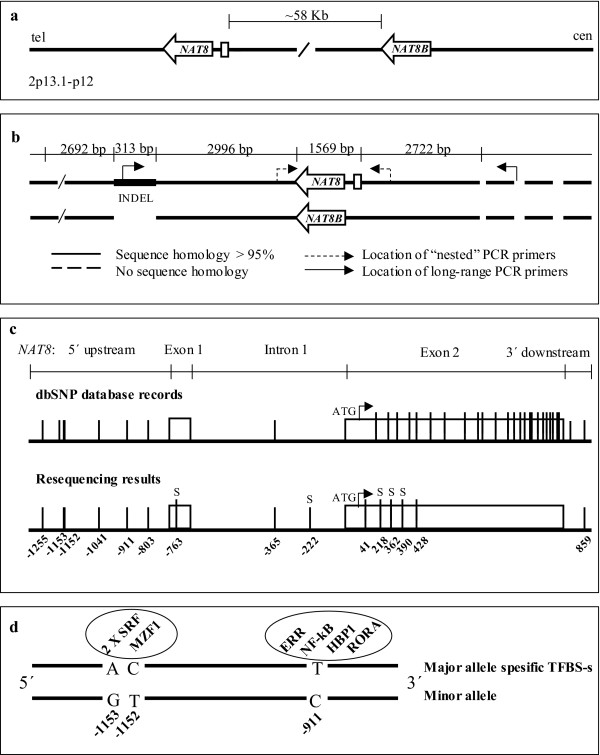
The sequence of the human NAT8 genomic region has been obtained from NCBI GeneBank database. As NAT8 possesses a duplicate gene NAT8B located about 58 Kb downstream from NAT8, the design of the amplification of human NAT8 gene involved two steps (long-range PCR and nested-PCR) (Figure 1a). Alignments between NAT8/NAT8B and their flanking areas (web-based global alignment tool CLUSTALW [17]) revealed a high nucleotide homology between extended genomic regions. The forward primer for NAT8 was positioned within a 313 bp NAT8-specific insertion (Figure 1b). The Primer3 primer designing software [18] was used to search for the matching reverse primer from the region upstream of NAT8, where the sequence homology with NAT8B region did not continue (> 2722 bp upstream; Figure 1b). The uniqueness of long-range PCR primers was checked using BLAST [19]. PCR reactions were performed using Long PCR Enzyme Mix (MBI Fermentas, Vilnius, Lithuania) and Smart-Taq Hot DNA polymerase (Naxo, Estonia) according to the standard protocol recommended by the manufacturers. Detailed conditions for long-range (10341 bp) and nested (2322 bp) PCR, product purification, sequencing, sequence contig assembly and polymorphism identification are described in detail elsewhere [20]. The resequenced region covered 2185 bp including the entire NAT8 (1569 bp) as well as 541 bp of the 5' upstream and 75 bp of the 3' downstream regions. All primers used for the NAT8 PCR amplification and resequencing are listed in Table 1.
Figure 1.
(a) The structure of NAT8/NAT8B cluster: NAT8/NAT8B location in chromosome 2p13.1-p12; (b) Schematic presentation of the alignment of NAT8/NAT8B and their flanking regions and the positioning of NAT8-specific PCR primers; (c) NAT8 resequencing results and SNP positions compared to dbSNP database records; (d) Missing Binding Sites for Transcription Factors (TFBS) on chromosomes carrying the minor alleles of SNPs -911, -1151 and -1153. "S" – singleton polymorphisms.
Table 1.
PCR and resequencing primers for human NAT8
| Primer ID | Primer sequence 5'-3' |
| NAT8 resequencing | |
| Long-range PCR primers | |
| NAT8_10kb_F | TGTAATCCCACACTTTTAGAGGCCAAG |
| NAT8_10kb_R | GAGTAGGTAAACAAAGCAGCCAGGAA |
| Nested PCR primers | |
| NAT8_2kb_F | GCAAAATTGCTGACTTCAGACTC |
| NAT8_2kb_R | CAACAGCCAGAACACACACAAG |
| Sequencing primers | |
| NAT8_SEQ_1F | TGCAACATTATGCTGGCACT |
| NAT8_SEQ_2F | CAGACTCAGCCACCCAGAAG |
| NAT8_SEQ_3F | AGGGTCAGAAAGACCTGCATT |
| NAT8_SEQ_4F | GCATCCAGGAGATACCGTCT |
| NAT8_SEQ_1R | ATTGCTGAAGCTGCCTCGAAC |
| NAT8_SEQ_2R | AGGTAGGTGCCCCCTCAAT |
| NAT8_SEQ_3R | CCTGAACAGGCAAGAGAGGT |
| NAT8_SEQ_4R | GTCAAGCCGAATGTCAACCT |
In the extended association study of the 5'upstream SNPs, only the sequencing primers 4F and 4R were used. For the comparison of sequence diversity parameters, we resequenced (n = 14) the promoter regions (+100 bp up to -500 bp relative to mRNA transcription initiation site) for 29 previously established blood pressure associated genes from literature: ACE, ADD1, ADD2, AGT, AGTR1, AGTR2, KCNJ1, CYP17A1, NPR1, CLCNKA, HSD11B1, CYP11B2, CLCNKB, KLK1, NPR2, BSND, NR3C2, CYP21A2, SAH, SGK, SLC12A3, SCNN1A, IL1A, SLC14A2, SLC22A2, REN, ATP1A, SCNN1B and SLC8A1 (primers used for PCR and resequencing, see additional file 1: Primers).
Subjects for Association Studies
Association study for blood pressure
Long-term blood donors (n = 137; 76 males, 61 females; age 40 ± 7.8 years; BMI 25 ± 2.6) were recruited in the framework of HYPEST study (participation rate 51.5%; Org et al., unpublished data) from the North Estonian Blood Center [21] coordinating the blood donation in northern, eastern, and western Estonia, and the Blood Centre of the Tartu University Clinics [22] coordinating the blood donation activity in central and southern Estonia. The exclusion criteria for donors included cardiovascular diseases, diabetes and antihypertensive treatment ascertained by confidential self-reported signed questionnaires and medical examination. Both Blood Centres have standardized protocols for measuring blood pressure by responsible trained clinicians. After a rest in a sitting position, the blood pressure reading of a donor was obtained during each visit to the Blood Donation Centre. All recruited individuals possess a documented history of multiple systolic and diastolic blood pressure (SBP, DBP) readings (on average 4 readings per individual, range 2–14) during 3 ± 2.5 years (range 1–7 years). Most of the BP measurements per subject have been obtained using a standard mercury column sphygmomanometer and size-adjusted cuffs. Occasionally, outside of blood donation centres BP of the participants was measured by oscillometric method using NAIS DIAGNOSTEC blood pressure watch (Matsushita Electronic Works) after a rest in a sitting position and with the arm placed at the heart level. No systematic deviation was observed between the two devices. The BP parameters across the whole cohort study group were as following: SBP 134.2 ± 11.5 mmHg within a range of 113.5 – 171 mmHg; and DBP 84.6 ± 7.6 mmHg within a range of 65.5 – 110 mmHg. Quantitative analysis was performed with a calculated median of a subject's BP readings adjusted to age and sex as co-variables.
Association study for estimated glomerular filtration rate (eGFR)
The study group of long-term essential hypertension (EH) patients (n = 157; 78 males, 79 females; age at the time of EH diagnosis 47 ± 9.7 years; age at recruitment 59 ± 9 years; BMI 28 ± 3.8) without and with hypertensive nephropathy were recruited (participation rate 42.3%) by blood pressure specialists during the patients' ambulatory visit or hospitalization at the North Estonia Medical Center, Centre of Cardiology (cardiologist M.V.) or at the Cardiology (coordinated by G.V.) and Internal Medicine Clinics (nephrologist M.R.), Tartu University Hospital, Estonia. The selection criteria for the study group were as following:
- Long term (mean duration 12.5 y) essential hypertension diagnosed by a clinical specialist (SBP > 140 mmHg, DBP > 90 mmHg).
- Exclusion of patients with diabetes and primary renal failure by a clinical specialist
- All recruited hypertension patients received antihypertensive treatment
Diagnose of hypertensive nephropathy was assigned by nephrologists (M.R.) based on longstanding blood pressure elevation together with proteinuria and presence of progressive chronic kidney disease. Differential diagnosis was established using methods per exclusionem i.e. the patients possessed no symptoms of glomerulonephritis, systemic diseases, amyloidosis or other glomerulopathies, confirmed by diagnostic labororatory tests.
The venous blood for serum biomarker analysis was drawn in the morning after an overnight fast. Urea (measurement uncertainty 9.7%), creatinine (m.u. 17.2%) and albumine (m.u. 6.9%) in the serum were measured by standardized assays (Cobas Integra 800® analytical platform, Roche Diagnostics, Inc.) at the United Laboratories, Tartu University Clinics [22] or at the Diagnostics Division Laboratory, the North Estonia Medical Centre [23]. The tests of for the serum biomarkers have been accredited by the Estonian Accreditation Centre [24] according to the standards of the European co-operation for Accreditation [25]. Measurement uncertainty was estimated using EURACHEM guidelines [26]. It is defined as the parameter associated with the result of a measurement that characterizes the dispersion of the values that could reasonably be attributed to the measurand (e.g. the concentration of a biomarker).
Kidney function was evaluated using estimated glomerular filtration rate (eGFR), the most recommended overall index of kidney function in health and disease [27]. The formula [28] includes both, demographic (age, sex, race) and blood serum (urea – SUN [mg/dl], creatinine – PCr [mg/dl], albumin – Alb [g/dl]) variables.
eGRF (ml/min/1,73 m2) = 170× (PCr)-0,999 × (Age)-0,176 × (0,762 if patient is female) × (SUN)-0,17 × (Alb)+0,318
A decrease of eGFR below 60 ml/min/1.73 m2 precedes the onset of kidney failure (K/DOQI 2002). The distribution of eGFR values for our study population was 72 ± 28.5 (range 5 – 143).
Statistical analysis
Methods used for Hardy-Weinberg equilibrium estimations, sequence diversity calculations and in silico prediction of transcription factor binding sites are described in detail elsewhere [29,30]. Quantitative genetic association analyses for blood pressure and serum biomarkers were performed with association analysis toolset PLINK [31]. Association was tested using linear regression analysis with additive effect model and likelihood-ratio test (LRT) with age, sex and BMI as co-variables. Statistical significance for the different distribution of the quantitative measurements (BP, GFR) in two subgroups based on the genotype was verified using non-parametric Mann-Whitney two-sided U-test performed with a web-based interface [32]. In all computed statistical tests a p-value < 0.05 was considered statistically significant and p < 0.1 as suggestive.
Results
Detailed diversity patterns of NAT8 by resequencing
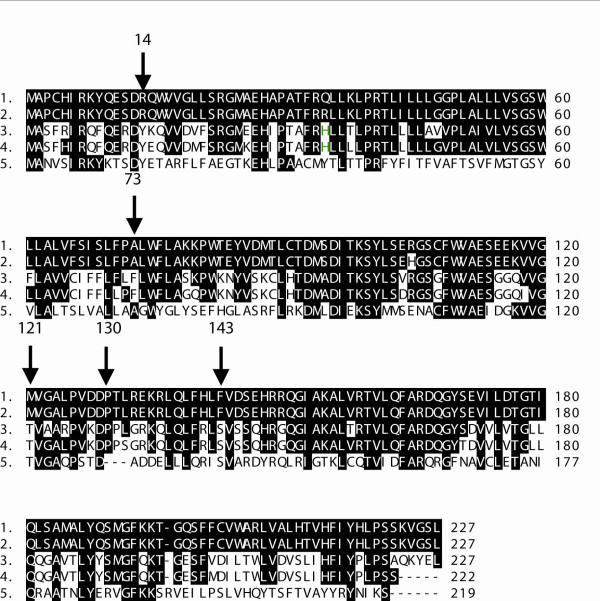
To uncover the detailed genetic variation of a putative human blood pressure candidate gene NAT8 (N-acetyltransferase 8; 2p13.1-p12) we resequenced the gene in a random Estonian population sample (n = 42). The analyzed region (1569 bp of the entire NAT8; 541 bp of 5' upstream and 75 bp of 3' downstream) harboured 15 SNPs including 10 rare (Minor allele frequency, MAF < 10%) and five common (MAF > 10%) polymorphisms (Table 2). Nine SNPs (n = 6 in 5'upstream region; n = 1 in intron 1, exon 2 and 3'downstream region) were previously characterized in dbSNP database, including five HAPMAP SNPs (rs482951, rs2280506, rs2280507, rs6744273, rs13538). Six rare variants are novel, neither represented in dbSNP nor reported elsewhere: one SNP in untranslated exon 1, one in intron 1 and four nonsynonymous substitutions in exon 2. The alignment of NAT8 protein sequences (Figure 2) of several vertebrate species (Pan troglodytes, Mus musculus, Rattus norvegicus, Xenopus tropicalis) indicated that the human polymorphic amino acid positions are not conserved in evolution. Thus we did not include these SNPs for further association study.
Table 2.
NAT8 genomic region SNPs identified by resequencing in random Estonian population samples (n = 42)
| Region | Locationa | dbSNP rs# | SNPb | Codon change | MAF |
| 5' upstream | -1255 | rs4852954 | T/C | - | 0.25 |
| -1153 | rs4852953 | A/G | - | 0.226 | |
| -1152 | rs4852952 | C/T | - | 0.226 | |
| -1041 | rs4852951 | G/C | - | 0.238 | |
| -911 | rs2280506 | T/C | - | 0.083 | |
| -803 | rs2280507 | C/T | - | 0.071 | |
| Exon1 | -763 | - | T/C | - | 0.012 (S) |
| Intron1 | -365 | rs6744273 | A/G | - | 0.071 |
| -222 | - | T/G | - | 0.012 (S) | |
| Exon2 | 41 | - | C/T | Arg-His | 0.024 |
| 218 | - | G/A | Ala-Val | 0.012 (S) | |
| 362 | - | A/G | Met-Thr | 0.012 (S) | |
| 390 | - | G/T | Pro-Pro | 0.012 (S) | |
| 428 | rs13538 | A/G | Phe-Ser | 0.167 | |
| 3'downstream | 859 | rs1879662 | G/A | - | 0.071 |
Abbreviations: MAF, minor allele frequency; S, singletons
aLocation relative to ATG
bMajor/minor allele
Figure 2.
The alignment of NAT8 protein sequences: 1- Homo sapiens, 2- Pan troglodytes, 3- Mus musculus, 4- Rattus norvegicus, 5-Xenopus tropicalis. Arrows pointing at polymorphic amino acid positions (14 Arg-His, 73 Ala-Val, 121 Met-Thr, 130 Pro-Pro, 143 Phe-Ser).
The NAT8 resequencing data deviated strongly from the polymorphism pattern in public dbSNP database [33] with 39 SNP entries for NAT8 region (Figure 1c). The higher number of SNPs in dbSNP might represent paralogous sequence variants (PSVs) between NAT8 and its duplicate gene NAT8B as all the unidentified dbSNP entries co-located with the sequence positions where NAT8 and NAT8B diverge (see additional file 2: Alignment).
Direct estimation of per-site heterozygosity (π) was derived from the average pairwise sequence difference. The NAT8 gene (1569 bp; π/bp = 0.00037) and mRNA (953 bp; (π/bp = 0.00045) exhibit two times lower sequence diversity compared to a data set of 74 genes [34] (π/bp = 0.00080 for European Americans; Table 3). Tajima's D (DT) statistic was used to estimate the difference between the parameters π and θ, representing the expected per-site heterozygosity based on the number of segregating sites. In case of neutrality, π equals θ and thus DT statistic equals zero. For NAT8 coding region, the calculated negative DT values refer to the excess of rare SNPs suggesting a purifying selection pressure. In contrast, the NAT8 5'upstream region exhibits approximately eight times higher diversity (π/bp = 0.00320; Table 3) and a positive DT value indicating an excess of common SNPs. Accordingly, four of the five NAT8 SNPs with minor allele frequency > 10% are located in the upstream region. Notably, this high diversity is uncommon in the dataset of promoter regions (+100 bp to -500 bp relative to mRNA transcription start) of 29 previously studied blood pressure genes resequenced to compare with NAT8 (Table 3). The diversity level of NAT8 upstream area (6 SNPs) exceeded about eight times the average of 29 reference genes (π/bp = 0.00320 versus π/bp = 0.0004). Of the 29 studied gene upstream regions, 17 (59%) had no SNPs, in the rest of the genes (n = 12) the number of SNPs ranged from one to four with the heterozygosity parameter (π) being 0.00023-0.0017. Interestingly, similarly to NAT8, several genes with the most variable upstream regions (CYP11B2, IL1A, SLC14A2) have also duplicate copies.
Table 3.
Diversity parameters for resequenced NAT8 gene and promoter regions of 29 genes associated with blood pressure determination
| Region | SNPs | π | θ | DT |
| A. NAT8 region | ||||
| Gene (1569 bp) | 9 | 0.00037 | 0.00102 | -1.58 |
| mRNA (953 bp) | 6 | 0.00045 | 0.00127 | -1.49 |
| Promoter (600 bp)a | 6 | 0.00320 | 0.00221 | 1.05 |
| B. Promoter regions (600 bp)a of 29 blood pressure associated genesb | ||||
| AGTR1 | 1 | 0.00082 | 0.00043 | 1.47 |
| CYP17A1 | 2 | 0.00058 | 0.00043 | 0.57 |
| CLCNKA | 2 | 0.00081 | 0.00086 | -0.11 |
| CYP11B2 | 3 | 0.00168 | 0.00085 | 2.02 |
| NPR2 | 1 | 0.00042 | 0.00043 | -0.018 |
| BSND | 2 | 0.00023 | 0.00043 | -0.74 |
| SAH | 2 | 0.00071 | 0.00087 | -0.38 |
| SGK | 1 | 0.00079 | 0.00043 | 1.36 |
| SLC12A3 | 1 | 0.00075 | 0.00043 | 1.216 |
| SCNN1A | 3 | 0.00167 | 0.00128 | 0.721 |
| IL1A | 2 | 0.0017 | 0.00086 | 2.02 |
| SLC14A2 | 4 | 0.00133 | 0.00085 | 1.17 |
Abbreviations: π, estimate of nucleotide diversity per site from average pairwise difference among individuals; θ, estimate of nucleotide diversity per site from number of segregating sites; DT, value of Tajima's D statistics expressing the difference between π and θ estimates
aRegion between -500 bp and +100 bp relative to mRNA transcription start point
bNo SNPs were detected in Estonian sample by resequencing the promoter regions for ACE, ADD1, ADD2, AGT, AGTR2, KCNJ1, NPR1, HSD11B1, CLCNKB, KLK1, NR3C2, CYP21A2, SLC22A2, REN, ATP1A1, SCNN1B, SLC8A1 genes
Association study of NAT8 5' upstream SNPs with blood pressure and eGFR
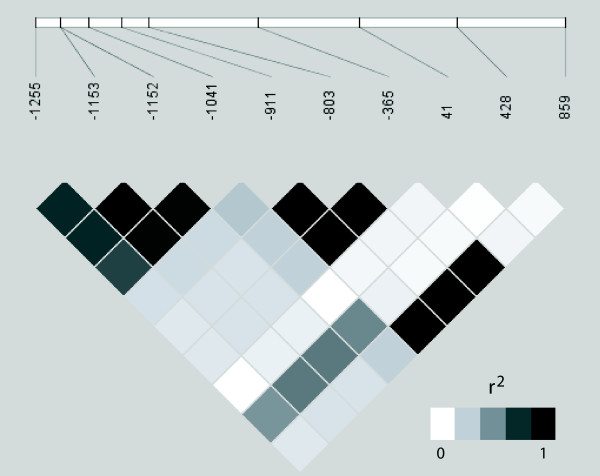
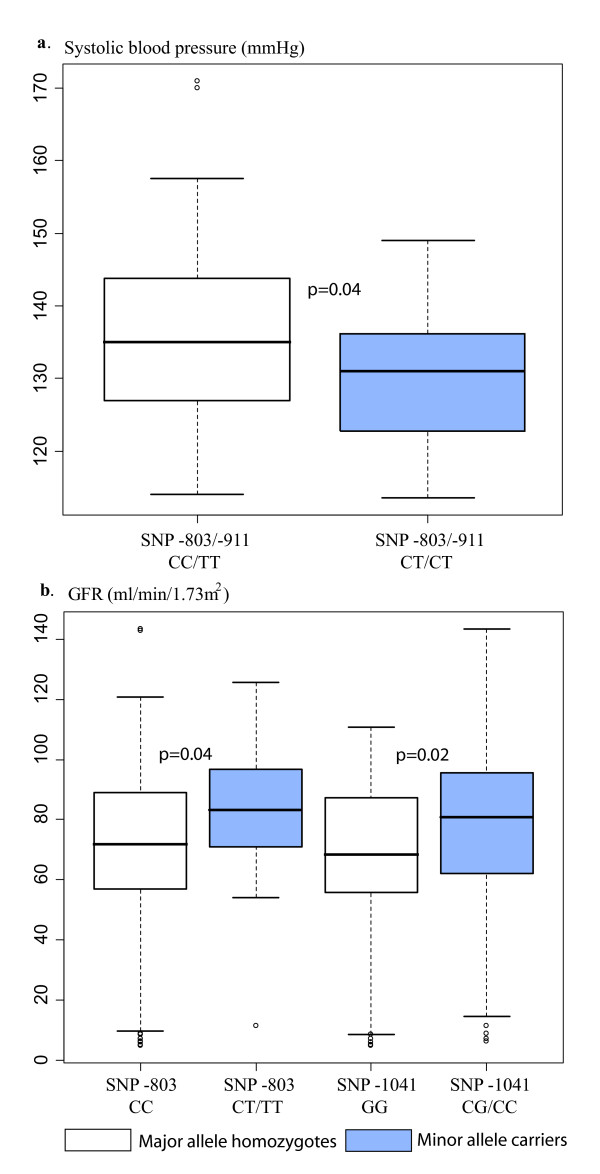
A NAT8 5'upstream putative promoter region [SNPs -1255 (T/C), rs4852954; -1153 (A/G), rs4852953; -1152 (C/T), rs4852952; -1041 (G/C), rs4852951; -911 (T/C), rs2280506 and -803 (C/T), rs2280507] was targeted in association studies for BP and estimated glomerular filtration rate (eGFR). Despite close vicinity, these SNPs form two distinct allelic association blocks: from -1225 to -1041 and from -911 to -365 (Figure 3). Notably, the latter region is also in LD with the 3' downstream SNP (+859 G/A; rs1879662). We used median values of systolic and diastolic blood pressure (SBP, DBP) readings in a cohort of long-term healthy Estonian donors (n = 137) with no history of cardiovascular diseases and respective treatments. SBP was significantly associated with NAT8 SNPs -803 (C/T) and -911 (T/C) using LRT (p = 0.028) and linear regression model (additive effects of allele dosage; p = 0.015) tests (Table 4). The significant negative regression coefficient (-5.034) referred to the association of the minor alleles of SNP -803/-911 with lower SBP values and indicated that these variants may possess a potential protective effect towards hypertension. Indeed SBP was significantly (p = 0.04; Mann-Whitney U test) lower by average 5 mmHg for the carriers of the minor alleles (median 131 mmHg; mean 130.5 ± 10.0; n = 33) compared to the major allele homozygotes (median 135 mmHg; mean 135.5 ± 11.7; n = 104) for SNPs -803/-911 (Figure 4a), supporting a protective effect of these alleles against blood pressure elevation. Association analysis of SBP with other upstream SNPs and of DBP with any NAT8 SNPs did not reveal any statistical significance (p = 0.1).
Figure 3.
LD structure of the resequenced NAT8 genomic region shown as r2-blot. Upper white bar depicts the positions of identified SNPs (Table 2) relative to the location of NAT8 gene ATG (A denoted as +1). Singleton polymorphisms were excluded from calculations.
Table 4.
P-values from association tests of NAT8 upstream SNPs: linear regression (additive model) adjusted for age, gender and BMI
| Blood pressure | |||
| SNP ID | SBP | DBP | eGFR |
| -803 (C/T) | 0.015 | 0.157 | 0.042 |
| -911 (T/C) | 0.015 | 0.100 | 0.060 |
| -1041 (G/C) | 0.519 | 0.943 | 0.050 |
| -1152 (C/T) | 0.373 | 0.828 | 0.081 |
| -1153 (A/G) | 0.373 | 0.828 | 0.081 |
| -1255 (T/C) | 0.659 | 0.870 | 0.070 |
Abbreviations: SBP, systolic blood pressure; DBP, diastolic blood pressure; e GFR, estimated glomerular filtration rate; p < 0.05; p ≤ 0.1.
Figure 4.
Notched box plots for the distribution of (a) systolic blood pressure (SBP) readings; and (b) estimated glomerular filtration rate (GFR) values for the major allele homozygotes and the minor allele carriers of NAT8 promoter SNPs. The boxes represent the 25th and 75th percentiles. The median is denoted as the line that bisects the boxes. The whiskers are lines extending from each end of the box covering the extent of the data on 1.5 × interquartile range. Circles represent the outlier values. P-values from Mann-Whitney U-test are shown, applied to test the statistical difference in the distributions of SBP or GFR estimates between two groups.
We further tested the association of NAT8 upstream SNPs with the reduction of kidney function as a consequence of essential hypertension using an Estonian cohort of long-term hypertension patients without and with hypertensive nephropathy. Linear regression analysis (additive model) and LRT test detected significant (SNP-803 and SNP-1041; p < 0.05) and borderline (p ≤ 0.08) associations of NAT8 upstream SNPs with eGFR estimates reflecting the efficiency of kidney function (Table 4). Significant positive linear regression coefficients indicated the association of SNP-803 (10.7) and SNP-1041 (6.4) with higher GFR values. The distribution of eGFR values for heterozygotes of SNP -803 (median 83.3; mean 82 ± 21.5; n = 33) and SNP -1041 (median 80.5; mean 77 ± 28; n = 90) was significantly higher (Mann-Whitney U test, p = 0.04 and p = 0.02) than for wild-type homozygotes (SNP -803: median 71.9; mean 70 ± 29.7, n = 124; SNP -1041: median 68.3; mean 66 ± 28; n = 67) on average 12 and 11 GFR units, respectively (Figure 4b).
Functional significance of NAT8 5'upstream SNP alleles
Possible functional significance of NAT8 upstream SNP alternative alleles was targeted with the in silico transcription factor binding site (TFBS) prediction using MatInspector [35] software. Several potential TFBSs within predicted classical promoter region (+100 to -500 bp relative to mRNA transcription start) are missing on the chromosomes that carry the minor alleles for NAT8 upstream SNPs -911 (major allele T/minor allele C), -1152 (C/T) and -1153 (A/G) (Figure 1d). Compared to the common T variant, the C allele of SNP -911 lacks potential binding sites for seven competing or co-activating TFs, including factors involved in inflammatory, cardiovascular or renal function: oestrogen related receptor (ERR) [36], RAR-related orphan receptor alpha1 (RORA) [37], HMG box-containing protein 1 (HBP1) [38], nuclear factor kB (NF-kB)/c-rel [39]. Allele- specific binding of ERR was confirmed by another TFBS prediction program AliBaba 2.1. [40]. Minor combinatory variant (T-G) for neighbouring SNPs -1152/-1153 miss two potential TFBSs, one for serum response factor (SRF) and the other for myeloid zinc finger protein (MZF1) located on the opposite strands. SRF is a transcription factor conserved from flies to humans and shown to regulate coordinated gene expression in cardiac and vascular smooth muscle cell differentiation [41].
Discussion
We report the first study targeting human N-acetyltransferase 8 (NAT8) as a potential blood pressure (BP) and kidney function associated gene. Although the knowledge about NAT8 is scarce, it was shown to be highly expressed in developing as well as adult human kidney and liver [14]. A recent study associated NAT8 expressional profile with nephrotoxicity response [15]. Kidney function has been associated with BP from two complimentary viewpoints: (1) link between elevated blood pressure and risk for kidney failure [4,6]; and (2) link between renal structural/functinonal abnormalities developed during intrauterine life as susceptibility to hypertension in adult life [7,10]. In listing the candidate genes for association studies of human blood pressure traits, the genes protecting and contributing to normal fetal renal programming have often been overlooked as potential susceptibility factors.
We showed that the putative upstream NAT8 promoter region is highly diverse compared to the gene's coding region, as well as to the promoters of BP-associated reference genes resequenced for comparison. High diversity may indicate the lack of selective constraint and/or the maintenance of several functioning promoter variants with differential effect on gene expression. For a number of genes shown to contribute to blood pressure regulation, the promoter polymorphisms have been associated with the diagnosis of hypertension. Examples of experimentally confirmed differential effects of alternative alleles on promoter activity are CYP11B2 [42-44], NPR1 (reviewed in [45]), SCNN1A [46] and AGT [47].
In a pilot association study based on a random Estonian population cohort we identified an association between NAT8 promoter SNPs and systolic BP. Consistently, in a separate cohort of essential hypertension patients the same markers showed association with estimated glomerular filtration rate (eGFR). Interestingly, the minor alleles of the same SNPs reveal protective effects towards both, the elevated BP and the risk of kidney failure as the complication to high blood pressure. We are aware that the results of these pilot association studies may be biased due to phenotypic misclassification and/or may be underpowered due to limited sample sizes. Also, multiple testing tends to inflate type 1 error rates. Another limitation may be the lack of sensitivity of the eGFR calculations based on serum creatinine instead of the actual GFR values obtained from measurements using an exogenous filtration marker. Thus, replication of the association of NAT8 with SBP and eGFR in other study populations, as well as functional experiments addressing the effect of the alternative promoter variants are required to ultimately determine the role of this gene in blood pressure regulation and kidney function. While evaluating the data from the association studies, one also has to keep in mind that both BP regulation and kidney metabolic function are complex traits determined by the combination of environmental, individual's life style and heterogenous genetic factors and thus the contribution of each gene variant is expected to be small. Recently, Yamada et al [48] performed a large association study with 150 SNPs in 128 functionally relevant BP regulating genes for 2818 hypertensive Japanese and 2035 matched controls. Using multivariable logistic regression analysis, significant associations (0.002 < p < 0.05) with hypertension were detected only with 10% (n = 15) of the targeted SNPs. Interestingly, seven of these were promoter variants and in 10 SNPs out of the significant 15 the carrier status of the minor allele provided a protective effect against blood pressure elevation.
High homology between NAT8 and its pseudogene NAT8B is due to a recent origin of NAT8B through the duplication of an ancestral gene after the ape-Old World monkey split. In chimpanzee both gene copies are still active, whereas in human NAT8B appears to be inactive due to human-specific nonsense mutations [49]. There are multiple examples of gene conversion events between genes and their pseudogenes causing the dysfunction of the genes [50]. In case of NAT8, the alternative promoter sequence may have arisen through gene conversion from NAT8B leading to differentially expressed gene variants.
Although the exact function of NAT8 is still to be determined, it has recently been shown to participate in detoxification processes of TCE (trichloroethylene) metabolite DCVC (S-(1,2-dichlorovinyl)-L-cysteine) in the kidney. DCVC detoxification in kidneys has two alternative metabolic pathways: (1) bioactivation through B-lyase activity leading to the accumulation of a nephrotoxic reactive sulfhydryl-intermediate (DCVSH – dichlorovinyl mercaptan) and (2) N-acetylation of DCVC resulting in a harmless acetylated product (NacDCVC – N-acetyl-S-(1,2-dichlorovinyl)-L-cysteine) excreted in urine [51]. When human renal proximal tubule cells were exposed to low concentrations of DCVC, NAT8 expression was reduced more than twice [15]. We hypothesize that the transcriptional activation of the common NAT8 promoter variant is repressible by toxic substances (e.g. TCE metabolites) and reduction of N-acetylation activity may lead to favouring of the nephrotoxic pathway. Due to differential cocktail of activating transcription factors for the NAT8 promoter variant with alternative SNP alleles, the minor NAT8 variant may be less sensitive to transcriptional suppression and the carriers may be better protected for the nephrotoxicity. The two disease processes, renal response to nephrotoxins and the pathogenesis of hypertension may be connected through overlapping physiological pathways. A representative example is the enzyme haem oxigenase 1 (HO-1) responsible for the catabolism of free heam, being induced in some models of hypertension as well as up-regulated in a variety of kidney injury models, including the effect of nephrotoxins (reviewed in [52]).
Conclusion
The full resequencing and pilot association study of a novel positional candidate gene for blood pressure and renal function, human N-acetyltransferase 8, suggested the contribution of highly variable NAT8 promoter polymorphisms in determination of systolic blood pressure and estimated glomerular filtration rate. Based on in silico analysis, we raise the hypothesis that the alternative SNP alleles within NAT8 upstream region may have differential effect on gene expression.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
PJ designed the study, performed resequencing, and contributed to the analysis and interpretation of the data as well as to writing of the manuscript. EO, MV, GV and MR recruited hypertensive individuals for the study, collected the epidemiological data, and revised the manuscript since its early versions. EO, KK, GV and PK assisted in collection of long-term blood donors, preparation of the DNA samples and laboratory experiments. ML contributed in outlining the study design and directing research, participated in the data analysis and drafted the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
PCR and resequencing primers for the promoter regions (+100 bp up to -500 bp relative to mRNA transcription initiation site) of 29 genes associated with blood pressure determination.
Nucleotide sequence alignment between human NAT8 gene and its duplicated pseudogene NAT8B. The alignment includes the entire gene along with 541 bp of 5' upstream and 700 bp of 3' downstream regions. Untranslated mRNA and protein-coding regions are marked with thin and bold lines, respectively. Nucleotides typed in bold indicate SNPs detected by resequencing and their position relative to ATG. Divergent positions in NAT8 and NAT8B, which co-localize with human NAT8 SNPs in NCBI dbSNP, are boxed.
Acknowledgments
Acknowledgements
We thank Drs Aleksei Teor and Karel Tomberg; Anne Speek and Aino Hallik from North Estonia Medical Centre, Tallinn, Estonia; Drs Sirly Tikas, Riho Paesüld and Tatjana Plahhova from North Estonian Blood Center; the personnel and especially Dr Ülle Kuusik from the Blood Center of Tartu University Clinics; Kersti Kivi, Külli Kangur, Dr Tiina Ristimäe from the Department of Cardiology and Dr Külli Kõlvald from the Department of Internal Medicine, University of Tartu Clinics; Tiina Rebane, Kärt Tomberg and Margus Putku for assistance in subject recruitment in the framework of HYPEST study. Viljo Soo is acknowledged for technical assistance in DNA sequencing, and Tõnu Margus for bioinformatics support. M.L. is a Wellcome Trust International Senior Research Fellows (grants no. 070191/Z/03/Z) in Biomedical Science in Central Europe and HHMI International Scholar (grant #55005617). Additionally, the study has been supported by Estonian Ministry of Education and Science core grant no. 0182721s06 and Estonian Science Foundation grant no. 5796.
Contributor Information
Peeter Juhanson, Email: peeter.juhanson@ut.ee.
Katrin Kepp, Email: katike@ut.ee.
Elin Org, Email: elin@ut.ee.
Gudrun Veldre, Email: gveldre@ut.ee.
Piret Kelgo, Email: piret.kelgo@hot.ee.
Mai Rosenberg, Email: mai.rosenberg@kliinikum.ee.
Margus Viigimaa, Email: margus.viigimaa@regionaalhaigla.ee.
Maris Laan, Email: maris@ebc.ee.
References
- Luft FC. Twins in cardiovascular genetic research. Hypertension. 2001;37:350–356. doi: 10.1161/01.hyp.37.2.350. [DOI] [PubMed] [Google Scholar]
- Snieder H, Harshfield GA, Treiber FA. Heritability of blood pressure and hemodynamics in African- and European-American youth. Hypertension. 2003;41:1196–1201. doi: 10.1161/01.HYP.0000072269.19820.0D. [DOI] [PubMed] [Google Scholar]
- Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/S0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- Klag MJ, Whelton PK, Randall BL, Neaton JD, Brancati FL, Ford CE, Shulman NB, Stamler J. Blood pressure and end-stage renal disease in men. The New England journal of medicine. 1996;334:13–18. doi: 10.1056/NEJM199601043340103. [DOI] [PubMed] [Google Scholar]
- Johnson RJ, Segal MS, Srinivas T, Ejaz A, Mu W, Roncal C, Sanchez-Lozada LG, Gersch M, Rodriguez-Iturbe B, Kang DH, Acosta JH. Essential hypertension, progressive renal disease, and uric acid: a pathogenetic link? J Am Soc Nephrol. 2005;16:1909–1919. doi: 10.1681/ASN.2005010063. [DOI] [PubMed] [Google Scholar]
- Rosario RF, Wesson DE. Primary hypertension and nephropathy. Current opinion in nephrology and hypertension. 2006;15:130–134. doi: 10.1097/01.mnh.0000214771.88737.ee. [DOI] [PubMed] [Google Scholar]
- Brenner BM, Garcia DL, Anderson S. Glomeruli and blood pressure. Less of one, more the other? Am J Hypertens. 1988;1:335–347. doi: 10.1093/ajh/1.4.335. [DOI] [PubMed] [Google Scholar]
- Moritz KM, Dodic M, Wintour EM. Kidney development and the fetal programming of adult disease. Bioessays. 2003;25:212–220. doi: 10.1002/bies.10240. [DOI] [PubMed] [Google Scholar]
- Jensen BL. Reduced nephron number, renal development and 'programming' of adult hypertension. Journal of hypertension. 2004;22:2065–2066. doi: 10.1097/00004872-200411000-00006. [DOI] [PubMed] [Google Scholar]
- Zandi-Nejad K, Luyckx VA, Brenner BM. Adult hypertension and kidney disease: the role of fetal programming. Hypertension. 2006;47:502–508. doi: 10.1161/01.HYP.0000198544.09909.1a. [DOI] [PubMed] [Google Scholar]
- Samani NJ. Genome scans for hypertension and blood pressure regulation. Am J Hypertens. 2003;16:167–171. doi: 10.1016/S0895-7061(02)03244-2. [DOI] [PubMed] [Google Scholar]
- Casari G, Barlassina C, Cusi D, Zagato L, Muirhead R, Righetti M, Nembri P, Amar K, Gatti M, Macciardi F, Binelli G, Bianchi G. Association of the alpha-adducin locus with essential hypertension. Hypertension. 1995;25:320–326. doi: 10.1161/01.hyp.25.3.320. [DOI] [PubMed] [Google Scholar]
- Iwamoto T, Kita S, Zhang J, Blaustein MP, Arai Y, Yoshida S, Wakimoto K, Komuro I, Katsuragi T. Salt-sensitive hypertension is triggered by Ca2+ entry via Na+/Ca2+ exchanger type-1 in vascular smooth muscle. Nature medicine. 2004;10:1193–1199. doi: 10.1038/nm1118. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Fujiwara T, Nakamura Y, Takahashi E. Isolation and mapping of a novel human kidney- and liver-specific gene homologous to the bacterial acetyltransferases. J Hum Genet. 1998;43:255–258. doi: 10.1007/s100380050084. [DOI] [PubMed] [Google Scholar]
- Lock EA, Barth JL, Argraves SW, Schnellmann RG. Changes in gene expression in human renal proximal tubule cells exposed to low concentrations of S-(1,2-dichlorovinyl)-l-cysteine, a metabolite of trichloroethylene. Toxicology and applied pharmacology. 2006;216:319–330. doi: 10.1016/j.taap.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Mueller JC, Lohmussaar E, Magi R, Remm M, Bettecken T, Lichtner P, Biskup S, Illig T, Pfeufer A, Luedemann J, Schreiber S, Pramstaller P, Pichler I, Romeo G, Gaddi A, Testa A, Wichmann HE, Metspalu A, Meitinger T. Linkage disequilibrium patterns and tagSNP transferability among European populations. American journal of human genetics. 2005;76:387–398. doi: 10.1086/427925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Web-based global alignment tool CLUSTALW http://www.ebi.ac.uk/Tools/clustalw2/index.html
- The Primer3 primer designing software http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
- National Centre for Biotechnology Information (NCBI BLAST) http://www.ncbi.nlm.nih.gov/BLAST
- Hallast P, Nagirnaja L, Margus T, Laan M. Segmental duplications and gene conversion: Human luteinizing hormone/chorionic gonadotropin beta gene cluster. Genome research. 2005;15:1535–1546. doi: 10.1101/gr.4270505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North Estonian Blood Center http://www.verekeskus.ee
- Blood Centre of the Tartu University Clinics http://www.kliinikum.ee/verekeskus
- Diagnostics Division Laboratory of the North Estonia Medical Centre http://www.regionaalhaigla.ee/?op=body&id=50
- Estonian Accreditation Centre http://www.eak.ee/index_eng.php?pageCus=head&head=3
- European co-operation for Accreditation http://www.european-accreditation.org
- EURACHEM guidelines http://www.eurachem.org/
- Levey AS, Coresh J, Balk E, Kausz AT, Levin A, Steffes MW, Hogg RJ, Perrone RD, Lau J, Eknoyan G. National Kidney Foundation practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Annals of internal medicine. 2003;139:137–147. doi: 10.7326/0003-4819-139-2-200307150-00013. [DOI] [PubMed] [Google Scholar]
- Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Annals of internal medicine. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- Kepp K, Juhanson P, Kozich V, Ots M, Viigimaa M, Laan M. Resequencing PNMT in European hypertensive and normotensive individuals: no common susceptibilily variants for hypertension and purifying selection on intron 1. BMC Med Genet. 2007;8:47. doi: 10.1186/1471-2350-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallast P, Rull K, Laan M. The evolution and genomic landscape of CGB1 and CGB2 genes. Molecular and cellular endocrinology. 2007;260–262:2–11. doi: 10.1016/j.mce.2005.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Web-based interface for Mann-Whitney two-sided U-test http://elegans.swmed.edu/~leon/stats/utest.cgi
- National Centre for Biotechnology Information (NCBI dbSNP database) http://www.ncbi.nlm.nih.gov/SNP/
- Crawford DC, Bhangale T, Li N, Hellenthal G, Rieder MJ, Nickerson DA, Stephens M. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nature genetics. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- Genomatix company webpage http://www.genomatix.de/products/MatInspector
- Zhang Z, Teng CT. Interplay between estrogen-related receptor alpha (ERRalpha) and gamma (ERRgamma) on the regulation of ERRalpha gene expression. Molecular and cellular endocrinology. 2007;264:128–141. doi: 10.1016/j.mce.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhtouche F, Mariani J, Tedgui A. The "CholesteROR" protective pathway in the vascular system. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:637–643. doi: 10.1161/01.ATV.0000119355.56036.de. [DOI] [PubMed] [Google Scholar]
- Ito T, Kawahara K, Nakamura T, Yamada S, Nakamura T, Abeyama K, Hashiguchi T, Maruyama I. High-mobility group box 1 protein promotes development of microvascular thrombosis in rats. J Thromb Haemost. 2007;5:109–116. doi: 10.1111/j.1538-7836.2006.02255.x. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ortega M, Esteban V, Egido J. The regulation of the inflammatory response through nuclear factor-kappab pathway by angiotensin IV extends the role of the renin angiotensin system in cardiovascular diseases. Trends in cardiovascular medicine. 2007;17:19–25. doi: 10.1016/j.tcm.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Transcription factor binding site prediction program AliBaba 2.1 http://darwin.nmsu.edu/~molb470/fall2003/Projects/solorz/aliBaba_2_1.htm
- Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends in cell biology. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Lim PO, Macdonald TM, Holloway C, Friel E, Anderson NH, Dow E, Jung RT, Davies E, Fraser R, Connell JM. Variation at the aldosterone synthase (CYP11B2) locus contributes to hypertension in subjects with a raised aldosterone-to-renin ratio. The Journal of clinical endocrinology and metabolism. 2002;87:4398–4402. doi: 10.1210/jc.2001-012070. [DOI] [PubMed] [Google Scholar]
- Nicod J, Bruhin D, Auer L, Vogt B, Frey FJ, Ferrari P. A biallelic gene polymorphism of CYP11B2 predicts increased aldosterone to renin ratio in selected hypertensive patients. The Journal of clinical endocrinology and metabolism. 2003;88:2495–2500. doi: 10.1210/jc.2002-021598. [DOI] [PubMed] [Google Scholar]
- Yu HM, Lin SG, Liu GZ, Zhang YQ, Ma WJ, Deng CY. Associations between CYP11B2 gene polymorphisms and the response to angiotensin-converting enzyme inhibitors. Clinical pharmacology and therapeutics. 2006;79:581–589. doi: 10.1016/j.clpt.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocrine reviews. 2006;27:47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- Iwai N, Baba S, Mannami T, Ogihara T, Ogata J. Association of a sodium channel alpha subunit promoter variant with blood pressure. J Am Soc Nephrol. 2002;13:80–85. doi: 10.1681/ASN.V13180. [DOI] [PubMed] [Google Scholar]
- Jain S, Li Y, Patil S, Kumar A. A single-nucleotide polymorphism in human angiotensinogen gene is associated with essential hypertension and affects glucocorticoid induced promoter activity. Journal of molecular medicine (Berlin, Germany) 2005;83:121–131. doi: 10.1007/s00109-004-0621-5. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Matsuo H, Segawa T, Watanabe S, Kato K, Hibino T, Yokoi K, Ichihara S, Metoki N, Yoshida H, Satoh K, Nozawa Y. Assessment of the genetic component of hypertension. Am J Hypertens. 2006;19:1158–1165. doi: 10.1016/j.amjhyper.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Hahn Y, Lee B. Human-specific nonsense mutations identified by genome sequence comparisons. Human genetics. 2006;119:169–178. doi: 10.1007/s00439-005-0125-6. [DOI] [PubMed] [Google Scholar]
- Bischof JM, Chiang AP, Scheetz TE, Stone EM, Casavant TL, Sheffield VC, Braun TA. Genome-wide identification of pseudogenes capable of disease-causing gene conversion. Human mutation. 2006;27:545–552. doi: 10.1002/humu.20335. [DOI] [PubMed] [Google Scholar]
- Cummings BS, Zangar RC, Novak RF, Lash LH. Cytotoxicity of trichloroethylene and S-(1, 2-dichlorovinyl)-L-cysteine in primary cultures of rat renal proximal tubular and distal tubular cells. Toxicology. 2000;150:83–98. doi: 10.1016/S0300-483X(00)00252-3. [DOI] [PubMed] [Google Scholar]
- Hill-Kapturczak N, Agarwal A. Haem oxygenase-1--a culprit in vascular and renal damage? Nephrol Dial Transplant. 2007;22:1495–1499. doi: 10.1093/ndt/gfm093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR and resequencing primers for the promoter regions (+100 bp up to -500 bp relative to mRNA transcription initiation site) of 29 genes associated with blood pressure determination.
Nucleotide sequence alignment between human NAT8 gene and its duplicated pseudogene NAT8B. The alignment includes the entire gene along with 541 bp of 5' upstream and 700 bp of 3' downstream regions. Untranslated mRNA and protein-coding regions are marked with thin and bold lines, respectively. Nucleotides typed in bold indicate SNPs detected by resequencing and their position relative to ATG. Divergent positions in NAT8 and NAT8B, which co-localize with human NAT8 SNPs in NCBI dbSNP, are boxed.