Abstract
We have used the interaction between the erythroid-specific enhancer in hypersensitivity site 2 of the human β-globin locus control region and the globin gene promoters as a paradigm to examine the mechanisms governing promoter/enhancer interactions in this locus. We have demonstrated that enhancer-dependent activation of the globin promoters is dependent on the presence of both a TATA box in the proximal promoter and the binding site for the erythroid-specific heteromeric transcription factor NF-E2 in the enhancer. Mutational analysis of the transcriptionally active component of NF-E2, p45NF-E2, localizes the critical region for this function to a proline-rich transcriptional activation domain in the NH2-terminal 80 amino acids of the protein. In contrast to the wild-type protein, expression of p45 NF-E2 lacking this activation domain in an NF-E2 null cell line fails to support enhancer-dependent transcription in transient assays. More significantly, the mutated protein also fails to reactivate expression of the endogenous β- or α-globin loci in this cell line. Protein-protein interaction studies reveal that this domain of p45 NF-E2 binds specifically to a component of the transcription initiation complex, TATA binding protein associated factor TAFII130. These findings suggest one potential mechanism for direct recruitment of distal regulatory regions of the globin loci to the individual promoters.
Tissue and developmental specificity of eukaryotic gene expression is influenced by regulatory sequences in core promoter regions and distal enhancer elements (1). The activity of these sequences is dependent on the binding of ubiquitous and tissue-specific transcription factors (1, 2). In the context of proximal promoter sequences, numerous protein-protein interactions have been demonstrated between the activation domains of promoter-bound factors and components of the transcription initiation complex (2–4). These interactions are essential for high levels of gene expression. The general transcription factor complex TFIID, which binds to the TATA box, and an associated component TFIIB, have been identified as the major targets of these upstream transactivators (4, 5). TFIID consists of a core TATA-binding protein (TBP), which is sufficient for promoter recognition and basal transcription, and TBP-associated factors (TAFs) that are required for activated transcription. TAFs bind to TBP in an ordered fashion and provide a physical and functional link between upstream activators and the RNA polymerase holocomplex (6).
In contrast to the well characterized role of factors bound to the core promoter, the mechanism of action of a similar array of factors binding to distal enhancers remains unclear. Many eukaryotic enhancers reside kilobases away from the genes whose expression they influence, and several models have been proposed to explain their long-range action. The scanning model suggests that proteins binding to distal elements track along the DNA until they reach the promoter where they interact with the basal machinery (7). In contrast, the looping model predicts that enhancer-bound transactivators are juxtaposed to proteins bound to the proximal promoter, with looping out or bending of the intervening DNA (8, 9). A third model suggests that binding of these transcription factors to the enhancer alters the tertiary structure of the downstream promoter (chromatin opening), allowing greater access to promoter binding transcriptional activators (9).
We have used the human β-globin cluster as a model to explore the mechanisms of promoter/enhancer interactions in the context of a multigene locus (10, 11). Tissue and developmentally specific expression of the genes of this locus (ɛ, Gγ, Aγ, δ, and β) is dependent on sequences in the core promoters (11). However, high-level expression requires the presence of the powerful regulatory elements of the locus control region (LCR), located 6–20 kb upstream of the ɛ-globin gene (12, 13). The LCR consists of four erythroid-specific DNaseI hypersensitive sites (HS1–4) (14, 15), which contain a highly conserved array of binding sites for hematopoietically restricted and ubiquitous transcription factors (10, 11). Recent evidence indicates that the HSs of the LCR function as a multiprotein holocomplex, interacting with a single gene promoter at any given time point to achieve high-level globin gene expression (16). The concept that no single site is critical for LCR activity and that considerable redundancy exists is further supported by the modest phenotypes observed with deletion of single sites (17, 18).
Using HS2, we have studied enhancer-dependent transcription of the globin genes. A tandem AP-1/NF-E2 binding motif forms the core of the HS2 enhancer and is essential for its function (19, 20). Similar sites are found in all HSs of the LCR in humans and other species, as well as the HS-40 enhancer of the α-globin cluster (21, 22). In contrast, no NF-E2 binding sites have been identified in the globin promoters. The NF-E2 motif binds a heteromeric complex consisting of an hematopoietic-specific 45-kDa subunit (p45 NF-E2) (23, 24), a member of the cap’n-collar family of transcription factors, and a ubiquitously expressed 18-kDa subunit (p18 NF-E2) (25, 26), a member of the NRL/maf family of DNA-binding proteins. Both proteins contain a basic region-leucine zipper motif. The NH2-terminal half of p45 NF-E2 also contains a proline- and serine-rich domain, previously reported to act as a transcriptional activator (27). The DNA binding specificity of the complex is conveyed by the p18 subunit, which has no transactivation potential (25).
Transcriptional activation of the β-globin gene by activator sequences of the LCR is observed in the presence of a minimal β-globin promoter possessing only an intact TATA box (28). In the context of HS2, activation is dependent on the presence of the tandem NF-E2 binding sites (19, 20). These findings suggest that an interaction between NF-E2 and the transcription initiation complex may provide a functional link between the distal LCR and the individual globin gene promoters. We report that the trans-activation domain of p45 NF-E2 is critical for its ability to support enhancer-dependent transcription of the globin genes in transient assays and in the context of the endogenous β- and α-globin loci. We demonstrate that this domain interacts with a component of the human TFIID complex, hTAFII130 (29). These findings suggest a possible mechanism for long-range transcriptional activation of the globin genes.
MATERIALS AND METHODS
Plasmid Construction.
Plasmids containing the wild-type or TATA box mutated human Aγ-globin promoter (−356 → +35) upstream of the firefly luciferase gene and the 1.9-kb fragment of HS2 of the human β-globin locus have been described previously (30). Yeast two-hybrid plasmids containing the GAL4 DNA-binding domain (DB) fused to polymerase complex components were made by subcloning the coding sequences of human TBP, TFIIB, and TAFs 250, 70 and 55, 30 and Drosophila TAFII110 into PGBT9 (CLONTECH). A similar approach was used for the partial clone of hTAFII130. Yeast NF-E2 expression vectors (AD-NF-E2, and NH2 and COOH terminal mutants were made by subcloning fragments of the human p45NF-E2 cDNA in-frame with the GAL4 activation domain in the pGAD424 vector (CLONTECH). pGEXNF-E2, and NH2 and COOH-terminal mutants of this construct were made in a similar fashion. PCIGAL, a mammalian expression vector containing the GAL4 DNA-binding domain, was made by subcloning the blunted HindIII–SalI fragment of pGBT9 into the SalI-blunted NheI fragment of PCINeo. COOH-terminal truncation mutants of p45 NF-E2 were inserted 3′ to the GAL4 DNA-binding domain, by subcloning the same fragments used to generate the yeast mutants outlined above, into the blunted NotI–EcoRI fragment of PCIGAL. An oligonucleotide encoding a consensus Kozak motif and the sequence of a hemagglutinin (HA) tag was inserted into the mammalian expression vector PCINeo, to generate PCINeoHA. NF-E2 cDNA fragments were subcloned in-frame with the HA tag. pSPTAF110 was made for in vitro transcription/translation studies by subcloning the complete dTAFII110 cDNA into the pSP72 vector (Promega).
DNA Transfection and Transient Assays.
DNA was transfected into K562 human erythroleukemia and CB3 mouse erythroleukemia cells by electroporation as described previously (31), except that CB3 cells were electroporated at a slightly lower cell density of 108/ml. Five micrograms of a β galactosidase-expressing plasmid was included in all transfections in K562 cells and 10 μg in CB3 transfections as an internal control. In transactivation experiments using the chloramphenicol acetyltransferase reporter gene under the control of tandem GAL4 binding sites, 20 μg of pG5EC reporter plasmid was cotransfected with equimolar amounts of the PCIGalNF-E2 mutants equivalent to 1 μg of the wild-type construct in the presence of carrier plasmid (PCINeo) to a total of 50 μg. In CB3 cotransfection experiments, 20 μg of the expression vector was electroporated with 20 μg of the reporter plasmid. CB3 cells were preinduced with 1.8% dimethyl sulfoxide 72 hr before electroporation and K562 cells with 20 μM hemin at the time of electroporation. After 48 hr, cells were harvested, lysed, and assayed as previously described (31). Results shown represent at least six different transfections with two independent plasmid preparations. Statistics were calculated using Student’s t test; values of P < 0.05 were considered significant.
Establishment of Stable CB3 Cell Lines Expressing p45 NF-E2.
CB3 cells (32), a gift of Yacov Ben-David (Sunnybrook Health Science Center, Toronto, Canada), were stably transfected with eukaryotic vectors expressing NF-E2 and selected in 1 mg/ml G418. Clones were isolated and expanded, and expression of the transgene was analyzed by Western blotting and electrophoretic mobility shift assay using previously described procedures (27).
Yeast Two-Hybrid Assays.
Yeast strains HF7c and SFY526 were obtained from CLONTECH and transformations performed using the lithium acetate method (33). All constructs initially were transformed singly into SFY526 and transformants assayed for β-galactosidase to exclude intrinsic activation. Pairs of activation domain and DNA binding domain hybrid constructs then were used to cotransform the host strains, with selection on media lacking both leucine and tryptophan. Double transformants then were assayed for β-galactosidase activity using a filter assay or, in the case of HF7c, were replated on media lacking histidine, leucine, and tryptophan with varying concentrations of 3-aminotriazole.
Expression of Glutathione S-Transferase (GST) Fusion Proteins and Affinity Chromatography.
GST fusion proteins were expressed in BL21 cells. Fusion proteins were purified (34) and quantified using the Bio-Rad method after purification on glutathione-Sepharose beads (Pharmacia). SDS/PAGE gel electrophoresis with Coomassie staining was used to confirm the integrity of the full-length fusion protein. For in vitro protein-protein interaction assays, 1 μg of GST or GST-fusion protein was incubated for 1 hr at 4°C with 10 μl glutathione-Sepharose beads, which had been preblocked with 0.5% milk. After extensive washing, the beads were resuspended in 200 μl binding buffer (10 mM Tris⋅HCl, pH 7.9/500 mM KCl/0.1 mM EDTA/150 μg/ml BSA/0.1% Nonidet P-40/10% glycerol) and incubated for 1 hr at room temperature with 35S methionine-labeled dTAFII110 or hTAFII130. After extensive washing, retained proteins were eluted by boiling in SDS loading buffer and analyzed on a 10% SDS/PAGE gel.
RNase Protection Assays.
Isolation of total RNA and RNase protection analysis were performed as previously reported (27, 34).
RESULTS
HS2 Enhancer-Dependent Transcriptional Activation of the γ-Globin Promoter Requires a Functional TATA Box and Intact NF-E2 Binding Sites.
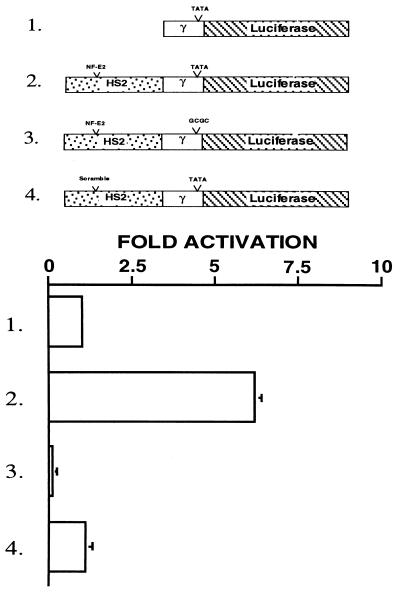
To define the minimal elements necessary for enhancer-dependent transcriptional activation of the γ-globin promoter, we linked a 1.9-kb fragment of HS2, containing intact tandem AP1/NF-E2 binding sites (HS2γTATALUC, Fig. 1, construct 2) or an identical fragment in which the NF-E2 sites had been mutated (HS2SγTATALUC, Fig. 1, construct 4) to a minimal γ-promoter/luciferase gene hybrid. These constructs, and the minimal γ-promoter/luciferase hybrid lacking upstream regulatory sequences (γTATALUC, Fig. 1, construct 1) and a construct in which the TATA box was mutated (HS2γTATASLUC, Fig. 1, construct 3) were transiently transfected into the human erythroid cell line, K562. As seen in Fig. 1, the presence of a functional TATA box was sufficient to allow enhancer-dependent transcription (Fig. 1, constructs 1 and 2). This transcriptional activation was lost with mutation of the TATA box or the NF-E2 binding sites in HS2 (Fig. 1, constructs 3 and 4). Mutation of other protein binding sites within HS2 failed to ablate enhancer activity (P.J.A., J.M.C., and S.M.J., unpublished work), suggesting that the minimal requirements for enhanced transcription are a TATA box in the promoter and intact NF-E2 binding sites in the enhancer.
Figure 1.
A functional TATA box and the core NF-E2 sites are required for enhancer-dependent transcriptional activation. (Upper) The constructs used to examine the interaction between the HS2 enhancer and minimal γ-globin promoter. The open box represents the −356+35 minimal γ- globin promoter. The hatched box represents the luciferase reporter gene. The stippled box represents the 1,900-bp KpnI–BglII fragment of HS2 of the β-globin LCR. The NF-E2 motifs are approximately 1 kb upstream of the TATA box. Constructs 3 and 4 contain mutated TATA box and NF-E2 binding motifs, respectively. (Lower) The fold activation by the enhancer relative to expression of construct 1.
p45 NF-E2 Interacts Specifically with Drosophila TAFII110 and its Human Homologue TAFII130.
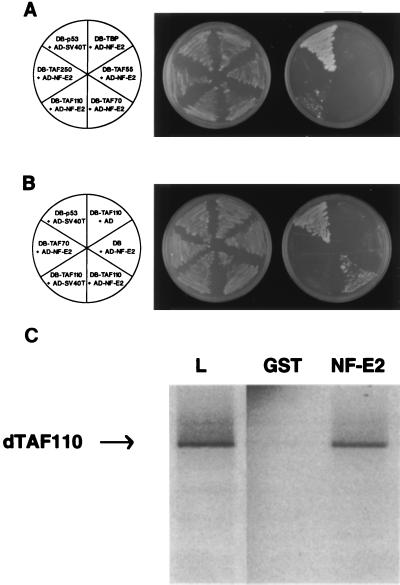
A direct interaction between NF-E2 and a component of the TATA box binding TFIID complex would provide one mechanism explaining the above results (3, 35). To investigate this, we focused on the hematopoietic-specific p45 component of NF-E2, as the transcriptional activity of the complex appears to reside solely in this subunit (25, 26). Using the yeast two-hybrid assay system, we examined protein-protein interactions between p45 NF-E2 and TBP, TFIIB, and TAFIIs 250, 110, 70, 55, and 30. As shown in Fig. 2A, a specific interaction was observed between NF-E2 and the Drosophila TAF, dTAFII110. No interactions were observed with the other components of TFIID, despite immunoblot analysis of yeast extracts confirming high-level expression of all transfected constructs (data not shown). Control experiments, removing either TAFII110 or NF-E2 from their respective yeast plasmids, ablated reporter gene activity (Fig. 2B). We subsequently obtained a partial clone of TAFII130, the human homologue of TAFII110, and confirmed that this interaction also occurred between the two human proteins (data not shown).
Figure 2.
Human NF-E2 interacts specifically with dTAFII110. (A) The yeast two-hybrid assay demonstrates that p45NF-E2 interacts with dTAFII110. The Saccharomyces cerevisiae reporter strain HF7C was transformed with the indicated plasmids. AD-NF-E2 contains the entire coding sequence of human NF-E2 inserted in-frame with the activation domain of GAL4 (amino acids 768–881). DB-TBP, DB-TAFII55, DB-TAFII70, and DB-TAFII250 contain the coding sequence of the human proteins and DB-TAFII110 the coding sequence of the Drosophila protein inserted in-frame to the GAL4 DNA binding domain (amino acids 1–147). A specific interaction between DB-p53 and AD-simian virus 40 T antigen has been reported previously. Leu- and Trp- transformants were streaked onto synthetic medium plates lacking tryptophan, leucine, and histidine to assess potential interactions (Right) and synthetic medium plates lacking tryptophan and leucine to confirm plating efficiency (Middle). The plates were incubated at 30°C for 3 days. (B) The yeast two-hybrid assay demonstrates that the NF-E2/dTAFII110 interaction is specific. DB-TAF110 was plated in combination with the simian virus 40 T antigen or GAL4-AD. AD-NF-E2 was plated in combination with TAFII110, TAFII70, or the GAL4-DB. Transfections were grown in the absence of leucine and tryptophan to assess transformation efficiency (Middle) or in the absence of leucine, tryptophan and histidine to assess protein-protein interactions (Right). Cotransfection of AD-simian virus 40 T-antigen and DB-p53 served as the positive control. (C) GST chromatography confirms that dTAFII110 binds specifically to p45NF-E2. One microgram of GST or GST-NF-E2, bound to glutathione-Sepharose was incubated with in vitro-translated dTAFII110 as described in Materials and Methods. Bound proteins were detected by SDS/PAGE followed by autoradiography. An example of labeled dTAFII110 that was loaded is shown in lane L.
To validate this interaction in an independent system, GST affinity chromatography was performed. GST alone or GST fused in-frame with full-length p45 NF-E2 (GST NF-E2) were coupled to glutathione-Sepharose beads and incubated under stringent conditions with 35S methionine-labeled recombinant dTAFII110. Specific retention of dTAFII110 was observed with the GST NF-E2 beads, but not on control GST beads (Fig. 2C). In contrast, incubation of GST NF-E2 with the other components of the TFIID complex failed to show a specific interaction, in accord with our studies in the yeast two-hybrid assay (data not shown).
The Transactivation Domain of NF-E2 Colocalizes with its TAF Interacting Domain.
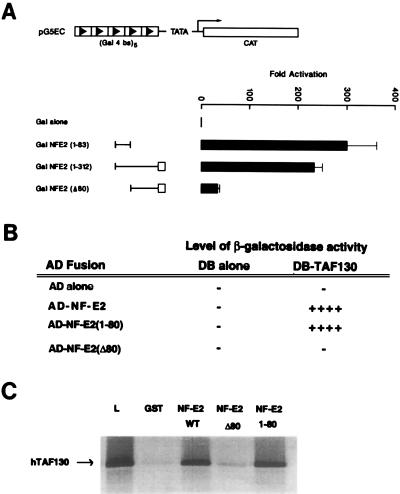
Numerous transcription factors have been shown to interact with their cognate TAFs through their transactivation domains (1). Previous studies had suggested that this domain in NF-E2 was located in the first 206 amino acids (27). This proline-rich sequence shares no structural homology with other proteins. To further localize the transactivation domain, we used mutants of NF-E2 fused to the GAL4 DNA binding domain in a eukaryotic expression vector. These constructs were cotransfected into K562 cells with a reporter plasmid containing five tandem GAL4 binding sites upstream of the chloramphenicol acetyltransferase gene. As seen in Fig. 3A, full transcriptional activation was observed in the presence of the NH2 terminal 80 amino acids of NF-E2 [GALNF-E2 (1–80)] alone. Deletion of this domain (GALNF-E2 Δ80) reduced reporter gene expression to the levels of vector control. To assess whether this minimal activation domain was sufficient for the interaction with TAFII130 we examined NF-E2(1–83) in the yeast two-hybrid assay. As seen in Fig. 3B, a GALNF-E2 (1–80) fusion protein activated the β-galactosidase reporter gene when cotransfected with TAFII130. In contrast, GALNF-E2Δ80 was incapable of interacting. To confirm these observations, we incubated 35S methionine-labeled hTAFII130 with GST, GST NF-E2, and selected mutants (Fig. 3C). Beads loaded with GST NF-E2 and GST NF-E2 (1–80) specifically retained hTAFII130. In contrast, beads linked to GST or GST NF-E2Δ80 failed to bind significant amounts of hTAFII130.
Figure 3.
Colocalization of the activation domain of NF-E2 with the TAFII130 binding domain. (A) Localization of activation domain of NF-E2. K562 cells were cotransfected by electroporation with expression vectors for GAL4 fusion proteins containing the indicated regions of NF-E2 and a reporter construct containing five GAL4 DNA binding sites regulating the expression of the chloramphenicol acetyltransferase gene (pG5EC). The first 312 amino acids of NF-E2 previously have been shown to contain a proline-rich activation domain lacking structural homology to other proteins denoted by a line, and a basic region similar to other cap’n-collar transcriptional activators denoted by an open box. The expression vector used in this study was pCINeo, containing the cytomegalovirus promoter. The fold activation represents the chloramphenicol acetyltransferase activity relative to the expression of the expression vector containing the GAL4 DNA binding domain alone. (B) hTAFII130 interacts with the NF-E2 activation domain in the yeast two-hybrid assay. The S. cerevisiae reporter strain SFY526, which contains a β-galactosidase reporter construct under the control of a GAL4 binding motif multimer, was transformed with the indicated plasmids. The AD-NF-E2 constructs contain the stated regions of the coding sequence of human NF-E2 inserted in-frame to the activation domain of GAL4 (amino acids 768–881). DB-TAFII130 contains the partial coding sequence of the human protein inserted in-frame to the GAL4 DNA binding domain (amino acids 1–147). (C) The amino terminal 80 amino acids of NF-E2 interacts specifically with hTAFII130 in the GST chromatography system. Fusion proteins containing GST fused to amino acids 1–374, 1–80, or 80–374 of NF-E2 or GST alone were expressed in Escherichia coli and bound to glutathione-Sepharose beads. The beads then were incubated with 35S-labeled in vitro-translated hTAFII130. Other experimental details are as stated in Fig. 2C.
The TAFII130 Interacting Domain of NF-E2 Is Required for Enhancer-Dependent Transcription of the Globin Genes.
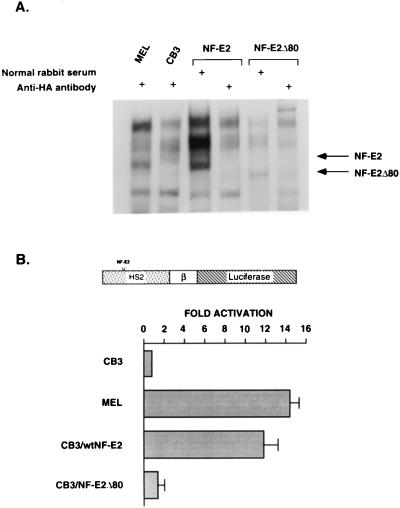
To ascertain whether the TAFII130 binding domain of NF-E2 was necessary for the enhancer activity of HS2, we constructed eukaryotic expression vectors containing wild-type NF-E2, or NF-E2 lacking the TAFII130 interaction domain (NF-E2Δ80). These were stably transfected into CB3 cells, a murine erythroleukemia (MEL) cell line in which both p45 NF-E2 alleles are nonfunctional (32). As a consequence, this line is NF-E2 null and fails to express significant levels of endogenous α- or β-globin chains (27, 32). Expression of wild-type and mutant forms of NF-E2 was verified in individual clones using electrophoretic gel mobility shift assay (Fig. 4A). The parental CB3 line, two derived lines for each NF-E2 expression vector, and a control MEL cell line (which constitutively expresses α- and β-globin) were then transiently transfected with a plasmid containing a β-promoter/luciferase gene hybrid linked to HS2. Enhancer-dependent activation of the β-globin promoter was observed in the MEL line and in the CB3 lines expressing wild-type NF-E2 (Fig. 4B). In contrast, the CB3 lines expressing NF-E2Δ80 failed to support HS2 enhancer activity, suggesting that enhancer function is dependent on the integrity of the TAFII130 binding domain of NF-E2.
Figure 4.
The TAFII130 binding domain of NF-E2 is essential for enhancer-dependent activation of the β-globin gene. (A) Wild-type NF-E2 and NF-E2Δ80 expression in selected CB3 clones as demonstrated in a gel mobility shift assay. CB3 cells were stably transfected with pCINeoHA vector alone or pCINeoHA vectors in which the human NF-E2 cDNA or NF-E2Δ80 was fused in-frame with the HA tag. Nuclear extract was prepared from individual clones and analyzed by gel mobility shift assay, dimethyl sulfoxide-induced MEL extract serving as a control. 32P-labeled DNA fragments containing the tandem NF-E2 binding sites in HS2 were incubated with individual extracts and electrophoresed on a 4% polyacrylamide gel. Lanes 1 and 2 show MEL cell and CB3 control extracts, the latter lacking the characteristic NF-E2 complex. Lane 3 shows a representative CB3 clone transfected with pCINeoHANF-E2 demonstrating a complex that comigrates with the NF-E2 complex observed in MEL cells. Lane 4 verified the presence of HA-tagged NF-E2 with specific ablation of the NF-E2 complex by the addition of anti-HA antibody (anti-HA). Lanes 5 and 6 show similar results for a clone expressing pCINeoHANF-E2Δ80. (B) High-level expression of a transiently transfected β-globin promoter containing construct is dependent on wild-type NF-E2 expression in CB3 cells. CB3 clones expressing wild-type NF-E2 or NF-E2Δ80 or CB3 cells transfected with the pCINeo vector (control) were transiently transfected with a plasmid containing HS2 linked to a β-promoter/luciferase gene hybrid. Whole cell extracts were assayed for luciferase activity. The results shown are representative of the three transfection experiments. MEL cells were transfected as a positive control. The fold activation observed was calculated relative to the expression of the HS2βLuciferase construct in CB3 cells.
The TAFII130 Interacting Domain of NF-E2 Is Required for Endogenous β- and α-Globin Gene Expression.
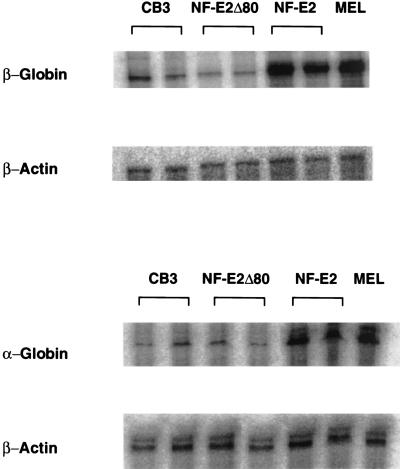
Although our studies have used HS2, the presence of NF-E2 binding sites in all HS of the β-globin locus, and in HS-40 of the α-globin locus (10, 11, 21, 22) suggested that the amino terminal 80 amino acids of NF-E2 may be critical for activation of all globin genes by upstream regulatory sequences in the endogenous loci. To address this, we examined the expression of the endogenous globin genes in the CB3 lines expressing wild-type NF-E2 or NF-E2Δ80. As previously reported, activation of the β-globin gene to levels equivalent to those seen in MEL cells was observed in the presence of wild-type NF-E2 (Fig. 5, Table 1, and refs. 27 and 32). In comparison, NF-E2Δ80 failed to reactivate the endogenous β-globin gene with comparable RNA levels observed between these clones and the parental CB3 line (Fig. 5, compare CB3 and NF-E2Δ80; Table 1). Interestingly, activation of the endogenous α-globin locus (Fig. 5 and Table 1) also was observed only in clones expressing wild-type NF-E2. These results indicate that in CB3 cells β-globin LCR- or α-locus HS-40-enhanced expression of their respective globin genes is dependent on the activation domain of NF-E2.
Figure 5.
High-level endogenous globin gene expression is observed only with wild-type p45NF-E2 expression in erythroid cells. RNase protection assays for α-globin, β-major mRNAs from CB3 cells, CB3 cells stably transfected with various p45 NF-E2 expressing constructs, and MEL cells. Actin levels were measured in parallel in all samples.
Table 1.
Globin gene expression in transfected CB3 cell lines
| Fold activation
|
||
|---|---|---|
| β-globin | α-globin | |
| Empty vector | 1 | 1 |
| NF-E2 wild type | 5.3 ± 0.4* | 4.1 ± 0.2* |
| NF-E2Δ80 | 0.9 ± 0.3 | 0.85 ± 0.2 |
Individual CB3 clones expressing wild-type or mutant NF-E2 cDNAs were assayed for endogenous globin gene expression using specific α- and β-globin specific probes. Variability in the amount of RNA loaded per assay was corrected internally by analyzing the expression of β-actin (see Fig. 5). Comparison of activation of the globin genes were corrected with respect to untransfected CB3 cells. The results shown are the average of at least three independent experiments.
Significantly different from CB3 cells alone (P < 0.01).
DISCUSSION
We have demonstrated that enhancer-dependent transcription by HS2 is observed in the context of a minimal promoter containing only a TATA box. In addition, we have shown that the integrity of the tandem NF-E2 binding sites is essential for this transcriptional activation. Both of these findings are in accord with observations made in different experimental systems (19, 20, 28) and can be linked by a model postulating a direct interaction between NF-E2 and the TFIID complex. We have demonstrated such an interaction between the activation domain of p45 NF-E2 and human TAFII130. Deletion of this domain in p45 NF-E2 has dramatic functional sequelae with loss of enhancer-dependent transcription in transient transfection assays. More significantly, it also results in an inability of NF-E2 to reactivate either the endogenous α- or β-globin loci in NF-E2 null cells.
These findings support a model of DNA looping, with juxtaposition of enhancer and promoter sequences through protein-protein interactions. Previous studies have postulated the existence of a DNA loop anchored by a protein bridge between two known factors as the mechanism mediating enhancer/promoter interactions in vivo (7, 36). The demonstration that the simian virus 40 enhancer could stimulate transcription in vitro when noncovalently linked to a β-globin promoter by the protein streptavidin strengthened this model (8). This stimulation in trans mimics transvection, a natural phenomenon in which one chromosome affects gene expression in a paired homologue. Subsequent studies of the transcription factors Sp1 and p53 demonstrated oligomerization of these factors with coincident loop formation of the intervening DNA (37). This multimerization resulted in synergistic activation of a linked reporter gene. Recently, Cullen et al. (38) demonstrated that juxtaposition of the distal enhancer and proximal promoter regions of the rat prolactin gene occurred in response to estrogen. By acting through its receptor bound to the distal enhancer, estrogen stimulated the interaction between distal and proximal regulatory regions with resultant transcriptional activation. This interaction was postulated to occur through “tethering” mediated by unknown protein-protein bridges.
Studies in the β-globin locus also have suggested that DNA looping is inherent to LCR/promoter interactions. A single gene promoter interacts with the LCR directly at any given time point. During the fetal/adult switch, the LCR appears to flip-flop between the γ- and β-promoters under the influence of the trans-acting environment. Our studies identify a protein-protein interaction that may provide a functional link between the LCR and individual promoters. This is evident in the context of a transient enhancer assay (Fig. 4B) and also in the setting of the native globin loci (Fig. 5). Our findings do not account for the developmental specificity of gene expression, which appears to be mediated through binding sites for developmental proteins in the individual globin promoters (31, 39, 40). Support for this conclusion is observed in studies of transgenic mice lacking the LCR, which retain a normal developmental profile of globin gene expression, albeit at low levels (reviewed in ref. 11). Our recent studies, demonstrating that transcriptional activation of globin promoters by the factors binding to sequences adjacent to the NF-E2 core of HS2 is dependent on the binding of the developmental proteins to promoter sequences, provides a link between the transcriptional activity of the LCR and the temporal regulation of the globin genes (P.J.A., J.M.C., and S.M.J., unpublished work).The importance of NF-E2 as a cornerstone of LCR/promoter interactions is suggested by numerous studies in cell lines and transgenic mice (11, 19, 20, 41). The distribution of binding motifs for this protein in all human HSs, the HSs in many other species, and the α-locus HS-40 further underscores this conclusion (21, 22, 42, 43). The topology of NF-E2 motifs with respect to other transcriptional activators binding to the HSs is also highly conserved (41), suggesting that this array of proteins may function as a multi-unit complex or enhanceosome. This arrangement is analogous to the stereospecific array of transcriptional activators found in the interferon β and IgH enhancers (44, 45). In the β-globin cluster, looping of the DNA stabilized by a direct interaction between NF-E2 and TAFII130 would result in alignment of other enhancer proteins, eg., GATA-1, with the transcription initiation complex. This would facilitate an interaction between these activators and their cognate TAFs, focusing the transcriptional potential of the LCR on the polymerase complex. This hypothesis is supported by the recent demonstration of synergistic interactions between multiple enhancer-bound proteins and their respective TAFs in Drosophila (4).
The p45NF-E2 protein previously has been shown to be essential for high-level globin gene expression in the MEL-derived CB3 cell line (32). Similarly, expression of a dominant negative p18 mutant significantly reduced the expression of α- and β-genes in MEL cells (27). Globin expression in these cells is fully restored upon the introduction of a tethered p45-p18 heterodimer. In accord with our findings, introduction of the NH2-terminal half of the protein also restored globin expression. These findings are in contrast to the subtle defects in hemoglobin production observed in mice with an NF-E2 null phenotype (46) and suggest that the repertoire of functional NF-E2 binding factors may be more limited in MEL cells than in the developing mouse. Although additional NF-E2-like proteins have been identified (47, 48), their ability to mediate enhancer-dependent transcription is not established. The discrepancy in the action of NF-E2 also may be influenced by the developmental stage with MEL cells arrested at a time in erythroid maturation in which NF-E2 is critical (27, 32).
Examination of the transactivation domain of NF-E2 reveals that it is proline-rich (20%) and serine-rich (10%). Previous studies have demonstrated that this class of domain is associated with factors that act as both “proximal and distal” activators of transcription (49). In contrast, glutamine-rich domains function only as proximal activators, stimulating transcription only from a position close to the TATA box. Interestingly, glutamine-rich factors have been demonstrated to transactivate via an interaction with TAFII130. The binding of this TAF to the proline/serine-rich transactivation domain of NF-E2 suggests that the differential enhancer effect of transactivator domains is not due solely to a specificity of coactivator interaction but may be influenced by other factors, such as the region of interaction of the TAF protein. We currently are defining the domains of TAFII130 necessary for interaction with glutamine- and proline-rich transactivators.
Drosophila and human in vitro transcription systems show an absolute requirement for TAFs (reviewed in ref. 1). Cellular studies in yeast suggested that TAFs may not be required for the activation of many genes (50). However, recent studies in Drosophila embryos demonstrate an absolute requirement for TAFIIs in the developmental function of the activator, bicoid (51). Thus, identification of activator/coactivator interactions are critical to our understanding of the β-globin locus. The studies described here focus on one potential mechanism of LCR/promoter interactions. However, modulation in chromatin structure mediated by the LCR, in part by p45NF-E2 (52, 53), upstream promoter-bound activators, and other components of the transcriptional machinery are necessary for optimal globin gene expression. Integration of these diverse mechanisms will be required to understand the dynamics of globin gene expression.
Acknowledgments
We thank A.W. Nienhuis, S.H. Orkin, R. Tjian, and J.H. Ihle for helpful discussions. We thank R. Tjian, R. Roeder, and P. Ney for the gift of plasmids and N. Tran and A. Becher for technical assistance. This work was supported by the National Institutes of Health, an American Society of Hematology Junior Faculty Scholars award (J.M.C.), the Cooleys Anemia Foundation (L.R.), the American Lebanese Syrian Associated Charities, the Australian National Health and Medical Research Council, and the Wellcome Trust.
ABBREVIATIONS
- HS
hypersensitivity site
- LCR
locus control region
- TAF
TATA binding protein associated factor
- TBP
TATA-binding protein
- DB
DNA-binding domain
- HA
hemagglutinin
- GST
glutathione S-transferase
- MEL
murine erythroleukemia
References
- 1.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 2.McKnight S L. Genes Dev. 1996;10:367–381. doi: 10.1101/gad.10.4.367. [DOI] [PubMed] [Google Scholar]
- 3.Tanese N, Tjian R. Cold Spring Harb Symp Quant Biol. 1993;58:179–185. doi: 10.1101/sqb.1993.058.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Sauer F, Hansen S K, Tjian R. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 5.Chiang C M, Roeder R G. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 6.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 7.Brent R, Ptashne M. Nature (London) 1984;312:612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- 8.Mueller-Storm H P, Sogo J M, Schaffner W. Cell. 1989;58:767–777. doi: 10.1016/0092-8674(89)90110-4. [DOI] [PubMed] [Google Scholar]
- 9.Felsenfeld G. Nature (London) 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 10.Stamatoyannopoulos G, Nienhuis A W. In: The Molecular Basis of Blood Diseases. Stamatoyannopoulos G, Nienhuis A W, Majerus P J, Varmus H, editors. Philadelphia: Saunders; 1994. pp. 107–156. [Google Scholar]
- 11.Dillon N, Grosveld F. Trends Genet. 1993;9:134–137. doi: 10.1016/0168-9525(93)90208-y. [DOI] [PubMed] [Google Scholar]
- 12.Grosveld F, Blom van Assendelft G, Greaves D R, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 13.Forrester W C, Takegawa S, Papayannopoulou T, Stamatoyannopoulos G, Groudine M. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuan D, Solomon W, Li Q, London I M. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talbot D, Collis P, Antoniou M, Vidal M, Grosveld F, Greaves D R. Nature (London) 1989;338:352–355. doi: 10.1038/338352a0. [DOI] [PubMed] [Google Scholar]
- 16.Wijgerde M, Grosveld F, Fraser P. Nature (London) 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 17.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 18.Hug B A, Wesselschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ney P A, Sorrentino B P, McDonagh K T, Nienhuis A W. Genes Dev. 1990;4:993–1006. doi: 10.1101/gad.4.6.993. [DOI] [PubMed] [Google Scholar]
- 20.Talbot D, Grosveld F. EMBO J. 1991;10:1391–1398. doi: 10.1002/j.1460-2075.1991.tb07659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gumucio D L, Shelton D A, Blanchard-McQuate K, Gray T, Tarle S, Heilstedt-Williamson H, Slightom J L, Collins F, Goodman M. J Biol Chem. 1994;269:15371–15380. [PubMed] [Google Scholar]
- 22.Vyas P, Vickers M A, Simmons D, Ayyub H, Craddock C F, Higgs D R. Cell. 1992;69:781–793. doi: 10.1016/0092-8674(92)90290-s. [DOI] [PubMed] [Google Scholar]
- 23.Andrews N C, Erdjument-Bromage H, Davidson M B, Tempst P, Orkin S H. Nature (London) 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 24.Ney P A, Andrews N C, Jane S M, Safer B, Purucker M E, Weremowicz S, Morton C C, Goff S C, Orkin S H, Nienhuis A W. Mol Cell Biol. 1993;13:5604–5612. doi: 10.1128/mcb.13.9.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews N C, Kotkow K J, Ney P A, Erdjument-Bromage H, Tempst P, Orkin S H. Proc Natl Acad Sci USA. 1993;90:11488–11492. doi: 10.1073/pnas.90.24.11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Nature (London) 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- 27.Kotkow K J, Orkin S H. Mol Cell Biol. 1995;15:4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoniou M, Grosveld F. Genes Dev. 1990;4:1007–1013. doi: 10.1101/gad.4.6.1007. [DOI] [PubMed] [Google Scholar]
- 29.Tanese N T, Saluja D, Vassallo M F, Chen J L, Admon A. Proc Natl Acad Sci USA. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amrolia P J, Cunningham J M, Ney P, Nienhuis A W, Jane S M. J Biol Chem. 1995;270:12892–12898. doi: 10.1074/jbc.270.21.12892. [DOI] [PubMed] [Google Scholar]
- 31.Jane S M, Ney P A, Vanin E F, Gumucio D L, Nienhuis A W. EMBO J. 1992;11:2961–2969. doi: 10.1002/j.1460-2075.1992.tb05366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu S, Rowan S, Bani M R, Ben-David Y. Proc Natl Acad Sci USA. 1994;91:8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fields S. Methods Companion Methods Enzymol. 1993;5:116–124. [Google Scholar]
- 34.Jane S M, Nienhuis A W, Cunningham J M. EMBO J. 1995;14:97–105. doi: 10.1002/j.1460-2075.1995.tb06979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez N. Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 36.Carey M, Lin Y S, Green M R, Ptashne M. Nature (London) 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 37.Stenger J E, Tegtmeyer P, Mayr G A, Reed M, Wang Y, Wang P, Hough P V, Mastrangelo I A. EMBO J. 1994;13:6011–6020. doi: 10.1002/j.1460-2075.1994.tb06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cullen K E, Kladde M P, Seyfred M A. Science. 1993;261:203–206. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- 39.Perkins A C, Sharpe A H, Orkin S H. Nature (London) 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 40.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Nature (London) 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 41.Stamatoyannopoulos J A, Goodwin A, Joyce T, Lowrey C H. EMBO J. 1995;14:106–116. doi: 10.1002/j.1460-2075.1995.tb06980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, Zhou B, Powers P, Enver T, Stamatoyannopoulos G. Proc Natl Acad Sci USA. 1990;87:8207–8211. doi: 10.1073/pnas.87.21.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss E C, Andrews N C, Higgs D R, Orkin S H. Mol Cell Biol. 1992;12:2135–2142. doi: 10.1128/mcb.12.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thanos D, Maniatis T. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 45.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 46.Shivdasani R A, Rosenblatt M F, Zucker-Franklin D, Jackson C W, Hunt P, Saris C J, Orkin S H. Cell. 1995;81:695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- 47.Caterina J J, Donze D, Sun C W, Ciavatta D J, Townes T M. Nucleic Acids Res. 1994;22:2383–2391. doi: 10.1093/nar/22.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moi P, Chan K, Asunis I, Cao A, Kan Y W. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seipel K, Georgiev O, Schaffner W. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker S S, Reece J C, Apone L M, Green M R. Nature (London) 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 51.Sauer F, Wassarman D A, Rubin G M, Tjian R. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 52.Armstrong J A, Emerson B M. Mol Cell Biol. 1996;16:5634–5644. doi: 10.1128/mcb.16.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong Q, McDowell J, Dean A. Mol Cell Biol. 1996;16:6055–6064. doi: 10.1128/mcb.16.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]