The cytochrome P450 enzyme CYP203A1 from Rhodopseudomonas palustris binds a wide range of highly substituted aromatic compounds and may play an important role in the astonishing metabolic diversity of this organism. Crystals of CYP203A1 that diffract to 2.0 Å resolution have been obtained.
Keywords: cytochrome P450 203A1, Rhodopseudomonas palustris
Abstract
Cytochrome P450 enzymes constitute a large family of haemoproteins that catalyze the monooxygenation of a great variety of endogenous and exogenous organic compounds. Cytochrome P450 203A1 (CYP203A1, RPA1009) from the metabolically versatile organism Rhodopseudomonas palustris binds a broad range of substrates, in particular substituted aromatic compounds. Crystals of CYP203A1 suitable for X-ray crystallography have been obtained and diffraction data were collected in-house to 2.0 Å resolution from a single crystal. The crystals belong to space group P222, with unit-cell parameters a = 40.1, b = 95.1, c = 99.0 Å, α = β = γ = 90°. There is one protein molecule per asymmetric unit.
1. Introduction
Cytochrome P450 enzymes are haem-dependent monooxygenases that catalyze a reaction in which one atom of molecular oxygen is inserted into a carbon–hydrogen bond of a great variety of endogenous and exogenous organic compounds (Ortiz de Montellano, 2005 ▶). They constitute a complex superfamily of proteins found throughout the biosphere from bacteria to humans and participate in the biosynthesis of physiologically important compounds such as steroids and hormones, in fatty-acid oxidation and in detoxification of exogenous compounds from the environment (Sligar, 1999 ▶; Denisov et al., 2005 ▶; Guengerich, 2001a ▶). P450s additionally catalyze a number of other reactions, including epoxidation, peroxidation, dealkylation, oxidative cleavage of esters, aryl dehalogenation, hydrolysis of phosphatidylcholine etc. (Guengerich, 1991 ▶, 2001b ▶; Zhao et al., 2005 ▶; Ortiz de Montellano, 2005 ▶).
The metabolically diverse organism Rhodopseudomonas palustris has been studied for its biodegradation ability and is a model for studying the biochemistry and metabolic pathways of aromatic ring degradation (Egland et al., 1997 ▶). It has four distinct ring-cleavage pathways for the aerobic degradation of aromatic compounds (Larimer et al., 2004 ▶). There are seven P450 genes in R. palustris (http://genome.ornl.gov/microbial/rpal/). These enzymes belong to different P450 families and bind a different range of substrates. Of these, cytochrome P450 203A1 (CYP203A1) has a wide range of substrates, in particular substituted aromatic compounds such as 4-hydroxybenzoic acid, 3,4-dichlorophenol, 1,2,4-trichlorobenzene, 2,3,5-trichlorobenzoic acid and pentachlorobenzene (Bell et al., 2006 ▶). Being hazardous and recalcitrant environment contaminants, polychlorinated aromatic compounds have been classified as priority pollutants by the environment agencies of the United States and Europe (Aust et al., 1994 ▶; Chen et al., 2002 ▶). As the degree of chlorination increases, the compounds become more inert and the heavily chlorinated homologues are partially degraded at slow rates (Chen et al., 2002 ▶). Interestingly, the haem spin-state shift upon binding of aromatic compounds to CYP203A1 increases with the degree of substitution, suggesting tighter binding of more highly substituted aromatics (Bell et al., 2006 ▶), and thus CYP203A1 has potential biotechnological applications such as the degradation of polychlorinated aromatic compounds in the environment.
A recent report showed that a CYP203 enzyme from an environmental metagenome library oxidized 4-hydroxybenzoate to protocatechuate (3,4-dihydroxybenzoate), the first P450 enzyme to show such activity. The CYP203 catalytic activity was reconstituted in vitro with the electron-transfer proteins putidaredoxin reductase and putidaredoxin from Pseudomonas putida and in vivo with the electron-transfer protein from P450Rhf (Nodate et al., 2006 ▶; Uchiyama et al., 2005 ▶). R. palustris CYP203A1 shares 71% sequence identity with this metagenomic CYP203 enzyme and it also binds 4-hydroxybenzoic acid, but its catalytic activity with the putidaredoxin reductase/putidaredoxin electron-transfer chain has been shown to be slow and the formation of products has yet to be confirmed (Bell et al., 2006 ▶). We have crystallized the substrate-free form of CYP203A1, the first P450 protein from the CYP203 family to be crystallized. The three-dimensional structure will provide valuable information on substrate binding and electron transfer for this new P450 enzyme.
2. Materials and methods
2.1. Protein expression and purification
The plasmid pET26CYP203A1 originally used to express the CYP203A1 protein (Bell et al., 2006 ▶) was subcloned into the vector pET28a(+) (Novagen Inc.) with NdeI and XhoI restriction sites. The new recombinant plasmid pET28a(+)CYP203A1, which introduced a N-terminal His tag for protein purification by metal-affinity chromatography, was transformed into Escherichia coli strain BL21 (DE3).
Cells were grown in LB medium with 25 µg ml−1 kanamycin and were induced with 0.5 mM IPTG at OD600 ≃ 0.7. After growth for another 8 h at 298 K, the red cells were harvested, resuspended in buffer A (40 mM potassium phosphate pH 7.4, 10 mM β-mercaptoethanol) and lysed by sonication. The lysate was centrifuged at 27 000g at 277 K for 30 min and the cell debris was discarded. The supernatant was loaded onto an Ni–NTA–agarose Superflow column equilibrated with buffer A. Elution of the His-tagged protein was achieved with a linear gradient of 10–200 mM imidazole in buffer A. Fractions containing CYP203A1, as revealed by SDS–PAGE, were pooled, concentrated and desalted by ultrafiltration and then loaded onto a Resource Q column (Amersham-Pharmacia Biotech), washed with five column volumes of buffer B (20 mM Tris–HCl pH 8.0, 10 mM β-mercaptoethanol) and eluted with a linear gradient of 0–500 mM NaCl in buffer B. All red fractions with A 418/A 280 > 1.25 were collected and concentrated. The pooled fractions were then further purified by size-exclusion chromatography on a Superdex 200 (Amersham-Pharmacia Biotech) column, eluting with buffer A. Fractions containing CYP203A1 were pooled, buffer-exchanged into 50 mM HEPES pH 7.4, 200 mM KCl by multiple cycles of ultrafiltration and then concentrated to 34 mg ml−1 for crystallization.
2.2. Dynamic light scattering (DLS)
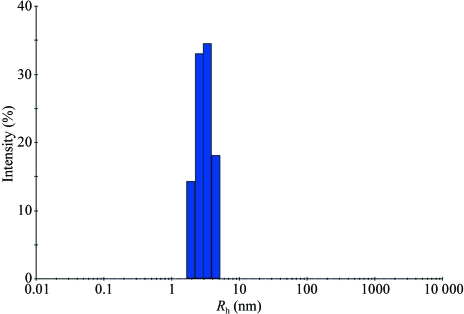
DLS measurements were performed at 289 K on a Wyatt Technologies DynaPro Titan dynamic light-scattering instrument operating at a wavelength of 829 nm. DLS data were recorded at CYP203A1 concentrations of 3–5 mg ml−1 in 50 mM HEPES pH 7.4, 200 mM KCl. Data were collected and analyzed using DYNAMICS software for the DynaPro Titan instrument (Wyatt Technology Corporation). As shown in Fig. 1 ▶, the average hydrodynamic radius (R h) of CYP203A1 was 3.1 nm, corresponding to a molecular weight of 49 kDa. As the calculated molecular weight is 46 174.3 kDa (including the His-tag sequence MGSSHHHHHHSSGLVPRGSH), we conclude that CYP203A1 is a monomer, which agrees with the results from size-exclusion chromatography (data not shown).
Figure 1.
Dynamic light-scattering measurement of CYP203A1 (3.5 mg ml−1) in 50 mM HEPES pH 7.4, 200 mM KCl. The calculated hydrodynamic radius of the protein was 3.1 nm.
2.3. Crystallization
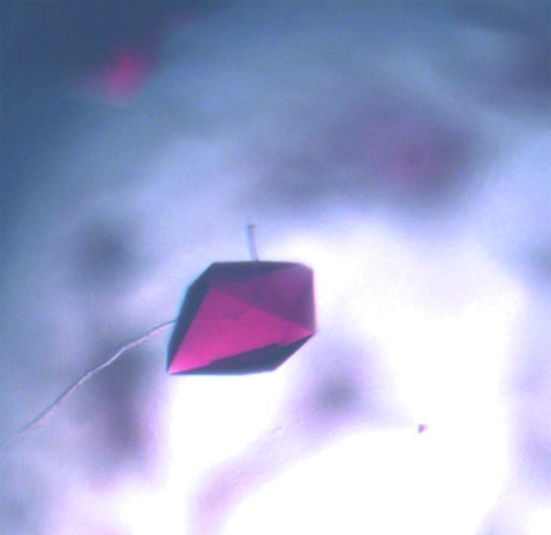
Crystals of CYP203A1 were obtained by the hanging-drop vapour-diffusion method at 291 K. A 1 µl aliquot of protein solution (34 mg ml−1 protein in 50 mM HEPES pH 7.4, 200 mM KCl) was mixed with an equal volume of reservoir solution and equilibrated against 200 µl reservoir solution. Initial screening was performed using Crystal Screens I and II (Hampton Research) at 291 K. Crystals appeared in 2 d from Crystal Screen I condition No. 20 (0.2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 4.6, 25% PEG 4000) and Crystal Screen I condition No. 10 (0.2 M ammonium acetate, 0.1 M sodium acetate trihydrate pH 4.6, 30% PEG 4000). After optimization by variation of precipitant concentration and buffer pH, good diffraction-quality crystals were obtained under the conditions 0.2 M ammonium acetate, 0.1 M sodium acetate trihydrate pH 4.3, 23% PEG 4000 (Fig. 2 ▶).
Figure 2.
Crystals of CYP203A1 grown in 0.2 M ammonium acetate, 0.1 M sodium acetate trihydrate pH 4.3, 23% PEG 4000.
2.4. Data collection and processing
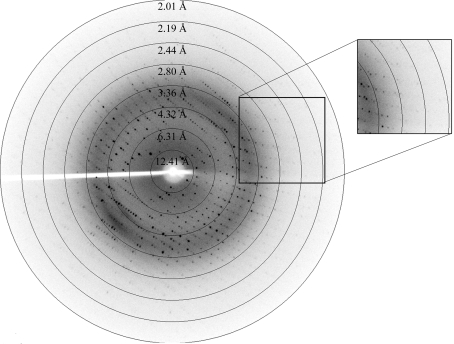
Immediately prior to data collection, crystals were soaked in a cryoprotectant solution consisting of 70 mM sodium cacodylate pH 6.5, 0.98 M sodium acetate, 30%(v/v) glycerol and were flash-frozen at 100 K in a stream of nitrogen gas. Complete diffraction data were collected at 100 K on an R-AXIS IV++ image plate using an in-house Rigaku MicroMax-007 rotating-anode X-ray generator (Fig. 3 ▶). The data set was indexed and scaled with the HKL-2000 program package.
Figure 3.
A typical diffraction pattern of a CYP203A1 crystal collected on an R-AXIS IV++ image-plate detector.
3. Results and discussion
The initial crystals had high mosaicity and were unsuitable for data collection. X-ray diffraction-quality crystals were eventually grown by reducing the concentration of the precipitating agent and lowering the pH value. Diffraction data collected in-house extended to 2.0 Å resolution (Fig. 3 ▶ and Table 1 ▶). The crystals belonged to space group P222, with unit-cell parameters a = 40.1, b = 95.1, c = 99.0 Å, α = β = γ = 90° (see Table 1 ▶). Initial analysis of the crystal solvent content using the Matthews coefficient (Matthews, 1968 ▶) suggested that the asymmetric unit contains one protein molecule, with 42.7% solvent content. Although a set of models such as cytochrome P450terp (PDB code 1cpt), which shares 31% sequence identity to CYP203A1, cytochrome P450epok (PDB code 1q5e), epothilone D-bound cytochrome P450epok (PDB code 1pkf) and P450eryF–ketoconazole (PDB code 1jin) have been tried with MOLREP, CNS and Phaser software, the phases cannot be solved by molecular replacement owing to large diversities in structure. Thus, we only report our work in the purification, crystallization and preliminary crystallographic analysis of cytochrome P450 203A1 here.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P222 |
| Crystal system | Primitive orthorhombic |
| Unit-cell parameters (Å, °) | a = 40.1, b = 95.1, c = 99.0, α = 90, β = 90, γ = 90 |
| Wavelength (Å) | 1.5418 |
| Resolution range (Å) | 50–2.02 (2.09–2.02) |
| Completeness (%) | 99.2 (93.0) |
| Redundancy | 6.8 (4.3) |
| Rmerge† (%) | 5.1 (25.8) |
| Average I/σ(I) | 39.7 (4.6) |
| Total reflections | 172208 |
| Unique reflections | 25435 |
| Molecules per asymmetric unit | 1 |
R
merge = 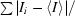
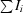 , where Ii is the intensity of the ith observation and 〈I〉 is the mean intensity of reflections.
, where Ii is the intensity of the ith observation and 〈I〉 is the mean intensity of reflections.
Acknowledgments
The work was supported by grants from Project 863 of the Ministry of Science and Technology of China (Grant No. 2005BA711A05-02) and the National Science Foundation of China (grant No. 30221003 to ZR), by grant EP-D048559-1 from the EPSRC and BBSRC, UK and by the Higher Education Funding Council for England (to LLW).
References
- Aust, S. D., Bourquin, A., Loper, J. C., Salanitro, J. P., Suk, W. A. & Tiedje, J. (1994). Environ. Health Perspect.102, Suppl. 1, 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S. G., Hoskins, N., Xu, F., Caprotti, D., Rao, Z. & Wong, L.-L. (2006). Biochem. Biophys. Res. Commun.342, 191–196. [DOI] [PubMed] [Google Scholar]
- Chen, X., Christopher, A., Jones, J. P., Bell, S. G., Guo, Q., Xu, F., Rao, Z. & Wong, L.-L. (2002). J. Biol. Chem.277, 37519–37526. [DOI] [PubMed] [Google Scholar]
- Denisov, I. G., Makris, T. M., Sligar, S. G. & Schlichting, I. (2005). Chem. Rev.105, 2253–2277. [DOI] [PubMed] [Google Scholar]
- Egland, P. G., Pelletier, D. A., Dispensa, M., Gibson, J. & Harwood, C. S. (1997). Proc. Natl Acad. Sci. USA, 94, 6484–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich, F. P. (1991). J. Biol. Chem.266, 10019–10022. [PubMed] [Google Scholar]
- Guengerich, F. P. (2001a). Chem. Res. Toxicol.14, 611–650. [DOI] [PubMed] [Google Scholar]
- Guengerich, F. P. (2001b). Curr. Drug Metab.2, 93–115. [DOI] [PubMed] [Google Scholar]
- Larimer, F. W., Chain, P., Hauser, L., Lamerdin, J., Malfatti, S., Do, L., Land, M. L., Pelletier, D. A., Beatty, J. T., Lang, A. S., Tabita, F. R., Gibson, J. L., Hanson, T. E., Bobst, C., Torres, J. L., Peres, C., Harrison, F. H., Gibson, J. & Harwood, C. S. (2004). Nature Biotechnol.22, 55–61. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Nodate, M., Kubota, M. & Misawa, N. (2006). Appl. Microbiol. Biotechnol.71, 455–462. [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano, P. R. (2005). Editor. Cytochrome P450: Structure, Mechanism and Biochemistry, 3rd ed. New York: Kluwer Academic Press/Plenum.
- Sligar, S. G. (1999). Essays Biochem.34, 71–83. [DOI] [PubMed] [Google Scholar]
- Uchiyama, T., Abe, T., Ikemura, T. & Watanabe, K. (2005). Nature Biotechnol.23, 88–93. [DOI] [PubMed]
- Zhao, B., Guengerich, F. P., Bellamine, A., Lamb, D. C., Izumikawa, M., Lei, L., Podust, L. M., Sundaramoorthy, M., Kalaitzis, J. A., Reddy, L. M., Kelly, S. L., Moore, B. S., Stec, D., Voehler, M., Falck, J. R., Shimada, T. & Waterman, M. R. (2005). J. Biol. Chem.280, 11599–11607. [DOI] [PubMed] [Google Scholar]