Abstract
The early development of many animals relies on the posttranscriptional regulations of maternally stored mRNAs. In particular, the translation of maternal mRNAs is tightly controlled during oocyte maturation and early mitotic cycles in Xenopus. The Embryonic Deadenylation ElemeNt (EDEN) and its associated protein EDEN-BP are known to trigger deadenylation and translational silencing to several mRNAs bearing an EDEN. This Xenopus RNA-binding protein is an ortholog of the human protein CUG-BP1/CELF1. Five mRNAs, encoding cell cycle regulators and a protein involved in the notch pathway, have been identified as being deadenylated by EDEN/EDEN-BP. To identify new EDEN-BP targets, we immunoprecipitated EDEN-BP/mRNA complexes from Xenopus tropicalis egg extracts. We identified 153 mRNAs as new binding targets for EDEN-BP using microarrays. Sequence analyses of the 3′ untranslated regions of the newly identified EDEN-BP targets reveal an enrichment in putative EDEN sequences. EDEN-BP binding to a subset of the targets was confirmed both in vitro and in vivo. Among the newly identified targets, Cdk1, a key player of oocyte maturation and cell cycle progression, is specifically targeted by its 3′ UTR for an EDEN-BP-dependent deadenylation after fertilization.
INTRODUCTION
The posttranscriptional regulation of gene expression relies on specific interactions between mRNAs and trans-acting factors that could be either proteins or microRNA. Known binding sites for regulatory RNA-binding proteins (RNA-BPs) are usually short and degenerated sequences those provide insufficient information to allow a systematic bioinformatics screen to identify the target mRNAs. However, the definition of the RNA targets of a given RNA-BP is crucial for the global understanding of the posttranscriptional processes in which the protein is involved. In amphibians, oocyte growth enables a single cell to contain the information required for the first 12 mitotic cycles and it is even sufficient to allow parthenogenetic development if centrosomes are provided (1). Notably, after an initial accumulation of the so-called maternal mRNAs in the growing oocyte, transcription ceases and the oocyte is blocked in prophase of the first meiotic division (stage VI oocyte). In Xenopus, upon hormonal treatment, meiosis resumes and a matured oocyte blocked in metaphase of the second meiotic division is laid that can be fertilized to allow zygotic mitosis. Transcription only resumes after the 12th cell cycle. This emphasizes the requirement for posttranscriptional regulation in the period from a stage VI oocyte to the 12th cell cycle.
From the hormonal activation of stage VI oocytes to the onset of transcription in the 4000 cell embryo, a cascade of translational regulatory mechanisms takes place that allows meiosis and early zygotic mitosis to occur in a coordinated manner [for a review see (2)]. Posttranscriptional mechanisms involve both global translational inhibition mechanisms as well as mechanisms targeting specific mRNAs for translation or translational silencing at specific time during oogenesis or development. Among the mechanisms involved, the deadenylation targeted by the Embryonic Deadenylation ElemeNt Binding Protein (EDEN-BP) ensures the silencing of c-Mos and Aurora A translation after fertilization by binding to the EDEN motif in their 3′ UTR (3,4). In addition to its role in very early development, EDEN-BP is required for the metamerization of the somites (5) by regulating the accumulation of Xenopus suppressor of hairless mRNA. CUG-BP1/CELF1, the mammalian ortholog of EDEN-BP can regulate both alternative splicing and translation (6). Recently, analysis of CUG-BP1/CELF1−/− mice indicated that CUG-BP1/CELF1 is globally required for the viability of the animals and, more specifically for spermatogenesis (7). However, no known CUG-BP1/CELF1 or EDEN-BP target can explain these observed phenotypes.
So far the various analyses of potential substrates for the EDEN/EDEN-BP-dependent deadenylation pathway have identified five mRNAs as EDEN-BP target: Aurora A/(Eg2), c-Mos, the kinesin-like Eg5, Cdk2/(Eg1) and Xsu(H); (3,5,8,9) the former four having been selected on their deadenylation behavior and the latter by its ability to bind EDEN-BP in UV-induced crosslinking experiments. The proteins encoded by three of these mRNAs (Aurora A, c-Mos and Cdk2) are known cell cycle regulators (10–12). Recently, an in vitro analysis of artificial CUG-BP1/CELF1 RNA ligand was performed and indicated that CUG-BP1/CELF1 binding sites could be represented as the occurrence of five UGU trinucleotides in a 35 nt window. However, such a motif is commonly represented in the genome and is of little predictive value. In the present work we used a combination of RNA–protein complex immunopurification (IP) and hybridization of copurified mRNA onto microarrays, to identify new EDEN-BP targets solely on the base of EDEN-BP binding.
To our knowledge this is the first identification on a large scale of mRNA targets of a RNA-BP involved in the early Xenopus development. Among the 158 EDEN-BP targeted mRNAs, we identified an overrepresented cluster of unfertilized egg (UFE) maternal mRNAs involved in the meiotic process of oocyte maturation. As EDEN-BP activity is turned on at, or just after, fertilization (13), the presence of this cluster suggests that EDEN-BP may be part of a mechanistic switch from ‘meiosis to mitosis’ or from ‘oogenesis to early development’.
MATERIALS AND METHODS
Microarray design
A set of 3000 50mer oligonucleotides was designed from 2898 Xenopus tropicalis gene sequences and spotted in duplicate (14). Oligonucleotides were spotted in 16 blocks of 14 × 14 spots, each containing an Arabidopsis thaliana probe, as well as blank and empty buffer controls. MWG Biotech performed oligonucleotide design, synthesis and spotting. X. tropicalis gene sequences were derived from the assembly of public and in-house ESTs (15). Description of the array has been deposited at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/).
UFE collection and extract
To prepare UFE extracts, eggs were collected from two X. tropicalis females after an injection with 100 U of human chorionic gonadotropin. Extracts were prepared as described for Xenopus laevis (16). Briefly, collected eggs were washed in F1 buffer (N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid 10 mM pH 7.6, NaCl 31.25 mM, KCl 1.75 mM, CaCl2 1 mM and MgCl2 60 mM), dejellied in 2% cysteine pH 7.8 in F1, washed with F1 and sorted. Eggs were then washed with XB (N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid 10 mM pH 7.7, KCl 100 mM, MgCl2 1 mM, CaCl2 0.1 mM and sucrose 50 mM) and placed in a 1.5 ml microtube. Buffer was replaced by XB buffer containing 10 µg/ml chymostatine, 10 µg/ml leupeptine and 100 µg/ml cytochalasine B and eggs were lightly swirled in this buffer. Eggs were packed (500 g, 1 min, 20°C) to remove interstitial buffer and layered by 200 µl of versilube oil (F-50, General Electric). Eggs were packed again (500 g, 1 min, 20°C) then centrifuged at high speed (10 000 g, 10 min, 4°C). The cytoplasmic fraction was collected and protease inhibitor (as above) and RNAsin (100 U/ml) were added. The cytoplasmic extract was centrifuged (10 000 g, 10 min, 4°C) and the supernatant was stored at −80°C. When embryos were used, fertilization was performed according to standard procedures (17).
EDEN-BP IP
Two polyclonal antibodies detecting EDEN-BP (αE1 or αE2) were purified on the same recombinant protein as that used to immunize rabbits (3). For IP, protein A-purified IgG from an unrelated serum and αE1 or αE2 were covalently bound to protein A/Dynabeads (Dynal). Thirty microliter of antibodies-coupled beads were then incubated (1 h, 20°C) in 50 µl UFE extracts diluted 5-fold in XB buffer to a total volume of 250 µl in presence of RNAsin (0.2 U/µl) (Promega). After binding and extensive washes with PBS 1X (6 × 800 µl), bound protein complexes were eluted by boiling (5 min) in 60 mM Tris pH 6.8, 1% SDS, 10% glycerol and 0.01% bromophenol blue. An aliquot was saved for western blot analysis while RNA extraction was performed on the eluates with Tri-reagent following the manufacturer recommendations (MRC).
RNA labeling
RNAs (250 ng) were reverse transcribed into double-stranded cDNAs by MMLV-RT using an oligodT-T7 promoter primer at 40°C. Labeled cRNAs were transcribed from the cDNAs by T7 RNA polymerase with direct incorporation of Cy5-CTP or Cy3-CTP, using the Low Input Fluorescent Linear Amplification Kit (Agilent). Probes were purified using the RNAeasy kit (Qiagen). Label incorporation and probe yield was controlled on a Nanodrop-1000.
Probing of cDNA microarrays
QMT Epoxy Slides were blocked 15 min at 50°C, in 50 mM ethanolamine and 0.1% SDS in 0.1 M Tris, pH 9.0. Fluorescent probes (700 ng each of tested and reference cRNAs) were mixed with Hybridization Buffer (MWG). The mix was denatured 3 min at 95°C and applied to the microarray. Hybridizations were performed at 42°C overnight and followed by a 15 min wash in 2X SSC with 0.2% SDS at 30°C, then two 15 min washes in 1X SSC at 30°C and two 15 min washes in 0.5X SSC at 20°C. Microarrays were scanned using a GenePix 4000B scanner (Axon Instruments) and GenepixPro was used to acquire spot information. GPR files have been deposited at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/).
Microarray analysis
Two separate IPs were performed with two female frogs using the two different antiEDEN-BP sera (αE1, αE2) and controlled by an IP with rabbit IgG. For each IP experiment, the labeled RNAs were hybridized on two slides, each slide bearing two sets of oligonucleotides. Therefore, for the IP with each antibody, four experimental values were obtained. In the first experiment, the eluted RNA were labeled with CTP-Cy5 while the reference sample composed of UFE total RNA was labeled with CTP-Cy3. In the second experiment, the eluted RNAs were labeled with CTP-Cy3 and the reference sample with CTP-Cy5.
In each experiment, data obtained from microarray scanning were submitted to Significance Analysis of Microarray (SAM) (18) using the TMEV 4 suite (http://www.tm4.org/mev.html) in order to determine the mRNAs specifically enriched in EDEN-BP IP compared to the IgG IP or input samples. In a second step the set of genes specifically enriched in IP 1 (n = 307, δ = 0.588, minimal fold change = 1.81 and false discovery rate =0%) was compared with the set of genes specifically enriched in IP 2 (n = 336, δ = 1.1962, minimal fold change = 1.86 and false discovery rate = 0%) to determine the mRNAs which are enriched in all the IP we performed. All the relative data have been deposited at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/).
Databases and sequences analysis
Taking advantage of the availability of X. tropicalis genomic sequences and partial genome annotations, we recovered 27 916 predicted transcripts (transcript_xt) and predicted proteins from the last X. tropicalis genome release (Version 4.1, August 2005). We could unambiguously match 1954 JGI predicted transcripts to at least one of our 3000 oligos. By comparing the predicted ORF and transcripts we extracted 7474 3′ UTR (UTR_xt); 1236 of the transcripts represented on the microarray contained identified 3′ UTR (UTR_array). The 158 EDEN-BP target mRNA identified in our work are represented by 108 predicted transcripts and 95 3′ UTRs (UTR_target). Two control dataset were derived from the UTR_target dataset. UTR_Target_Rev is the reverse complement of the UTR_target dataset (revseq, EMBOSS suite), this dataset does not retain the nucleotide composition. The UTR_Target_Shuffle dataset is derived from UTR_Target by shuffling (shuffleseq, EMBOSS suite) the sequences retain the nucleotide composition of each sequence. Datasets (Utr_array, UTR_Target, UTR_Target_Rev and UTR_target_Shuffle) are available in Supplementary Data S1.
To define a bonafide EDEN motif, we recovered from (19) the sequences selected for binding in vitro to CUG-BP-1, and identified as family 1. This set of 63 sequences was used to perform a motif search using MEME (http://meme.sdsc.edu/meme/meme.html). A motif of 15 nt (EDEN15) was determined to be present in 39 out of 62 sequences (P < 10−3). This motif was represented as a simple frequency matrix and a sequence logo was generated (20,21). The simple frequency matrix was used with PROFIT (http://emboss.bioinformatics.nl/cgi-bin/emboss) to identify and count transcripts in datasets bearing the defined EDEN15 motif (Supplementary Data S2, EDEN15). For each match, a score was attributed to the identified motif as a percentage of the best possible match. As a control, a motif was generated by randomly permutating nucleotide positions in the EDEN15 motif and ensuring there was no UGU trinucleotide present in the resulting motif; this motif retains the overall nucleotide composition of the EDEN15 motif (Supplementary Data S2, NO_UGU).
RT-PCR and transcription template synthesis
To control the IP of EDEN-BP mRNA targets, the RNA extracted from the IPs was primed with oligo-dT primer and reverse transcribed with Superscript II reverse transcriptase following the manufacturer recommendation (Invitrogen). PCR were performed in standard conditions with the primers presented in Supplementary Data S3.
Except for 3′ Eg5 and 3′ Eg5C6 (3), transcription templates for the RNAs to be tested in electrophoretic mobility shift assay (EMSA) and UV-crosslinking experiments were generated by PCR. A forward primer corresponding to a T7 promoter followed by the specific sequences was synthesized (for sequences see Supplementary Data S3). PCR products were purified and confirmed by sequencing from the T7 primer. The transcription templates were subsequently used to synthesize radiolabeled RNA using T7 RNA polymerase (Promega), NTPs and α-[32P]UTP (NEN). Each radiolabeled probe was <200-nt long. For microinjection into embryos, in vitro transcription was realized as previously described (17) in presence of cap analog (Promega).
3′ UTR cloning
pgemGbORFV5 was constructed by inserting the PCR-amplified GbORFV5 (PCR template pGbORF (17), oligonucleotides T7 and ORFV5-R, for oligonucleotide sequences see Supplementary Data S3) into the pgemT easy vector (Promega). pGbORF-AurA was constructed by inserting the GBORFV5 [digested KpnI/SphI (blunt) from pgemGbORFV5] into the vector Gb-Eg2-410-A65 (22) digested KpnI/PstI (blunt). pGbORF-Cdk1 was constructed by inserting the RT-PCR-amplified Cdk1 3′ UTR (for oligonucleotide sequences see Supplementary Data S3) into the NotI (blunt)/BamHI sites of vector pGbORFV5-AurA, therefore, replacing the partial AurA 3′ UTR sequence by the complete Cdk1 3′ UTR. All constructions were controlled by sequencing.
Transcription templates were generated by linearizing the pGbORFV5-AurA or pGbORFV5-Cdk1 with EcoRV to allow synthesis of a RNA bearing a 65 nt poly(A)tail as described previously (17).
Recombinant EDEN-BP synthesis and EMSA
His-tagged EDEN-BP was produced in bacteria and purified on Ni-NTA agarose beads (Qiagen) as described (23). Denaturing electrophoresis and Coomassie blue staining was used to check the purity of recombinant EDEN-BP and to measure its concentration.
EMSA were performed as described (23). Dried gels were submitted to Phosphoimager (STORM 840, Molecular Dynamics) analysis. Quantifications were made using the ImageQuant software (Version 5.4, Molecular Dynamics). The apparent Kd was measured as the concentration of recombinant EDEN-BP required for a ratio of bound to free probe of 1.
Crosslinking experiments
About 100 fmol of radiolabeled RNA were incubated in 10-fold diluted UFE extracts for 30 min at 23°C. Extracts were UV irradiated as described (9). RNAse A was added to the extract (final concentration 2 µg/µl) and incubated 1 h at 37°C. SDS–PAGE loading buffer was added and proteins were separated by SDS–PAGE on a 10% acrylamide gel. Dried gels were submitted to Phosphoimager. When indicated, specific (3′ Eg5) or non-specific (3′ Eg5C6) cold competitor RNA was added to the initial incubation step. 3′ Eg5 contain 40 nt of Eg5 3′ UTR centered on the EDEN motif while 3′ Eg5C6 is identical to 3′ Eg5 except in the substitution of a (UG)3 motif by (C)6 in the EDEN motif [described in (3)].
RESULTS
EDEN-BP IP selectively enriched known EDEN-BP target mRNAs
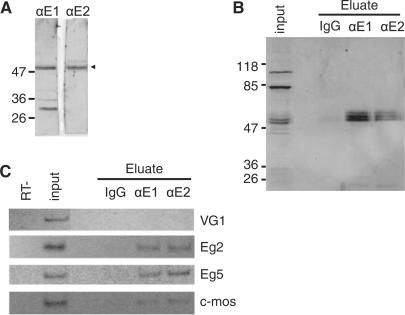
To identify mRNAs bound in vivo to EDEN-BP, we performed IPs from UFE extracts with two different antibodies (αE1 and αE2) raised against recombinant X. laevis EDEN-BP (3). The antibodies were purified on recombinant EDEN-BP protein. Western blot analysis of crude UFE extract (Figure 1A) showed that both serum recognized EDEN-BP as a specific band located at ∼50 kDa (arrowhead) but differed in cross-reacting background signals. A western blot analysis of a typical experiment is presented (Figure 1B) where, EDEN-BP in the immunopurified RNPs was revealed using an antiEDEN-BP antibody raised in guinea pig (3). EDEN-BP was selectively purified with αE1 or αE2 and could be detected as multiple bands representative of the various phosphorylation states of the protein (13). No EDEN-BP could be detected in the control IgG purification. As can be observed in the input lane, the guinea pig antibody recognized, in addition to EDEN-BP, two bands at 80 and 110 kDa that were not present in the immunopurified fractions showing the specificity of the IP procedure. To verify that known EDEN-BP targets were enriched in the EDEN-BP fractions, total RNA was extracted from the eluted IgG, αE1 and αE2 fractions shown in Figure 1B and used in RT-PCR analysis. As shown in Figure 1C, the Vg1 mRNA is detectable in the UFE extract but not detectable in either IgG, αE1 or αE2 eluted samples. In contrast, Aurora A(Eg2), Eg5 and c-Mos mRNAs were all significantly enriched in the αE1 and αE2 samples while not or barely present in the IgG sample. These RNA samples were used to identify new EDEN-BP target mRNAs on microarrays.
Figure 1.
EDEN-BP antibodies and IP specificities. (A) αE1- and αE2-purified polyclonal antibodies were used to detect endogenous EDEN-BP in crude UFE extracts. Protein size (kiloDalton) are indicated on the left side of the panel. The arrowhead indicates EDEN-BP. (B) Western blot analysis of EDEN-BP/RNA complexes immunopurified from UFE extract with control IgG, αE1 or αE2 antibodies as indicated. Input indicates initial sample (1/7.5). Detection is realized with a guinea pig antibody raised against recombinant EDEN-BP. (C) Detection by RT-PCR of the mRNA recovered from the IP eluates presented in B. The tested mRNAs are indicated on the right of the panel. Input is the original UFE extract and RT− corresponds to control reaction with input RNA and no reverse transcriptase.
Microarray identification of mRNAs bound to EDEN-BP
RNAs were extracted from the eluted fractions and from the input egg extracts. In the two experiments reported here, the UFE extracts were obtained from different X. tropicalis females. In one experimental set (αE1, αE2 and IgG, input from female 1 presented in Figure 1) RNAs were labeled with CTP-Cy5 while a reference sample composed of total RNA from a stock of UFEs was labeled with CTP-Cy3 (see Materiels and Methods section). A dye swap was performed for the second experiment (second female). For each sample, hybridization was performed in duplicate. Data were analyzed by SAM algorithm (18) to detect mRNAs selectively enriched in each IP experiments compared with the control IgG experiment. SAM for experiment 1 identified 307 mRNAs specifically overrepresented in the antiEDEN-BP eluates. SAM for experiment 2 identified 336 mRNAs specifically overrepresented in the antiEDEN-BP eluates. Comparing the two dye swap experiments, 162 oligo hits corresponding to 158 different mRNAs were found to be selectively associated with the endogenous EDEN-BP in all four IP experiments (Supplementary Data S4 for the SAM scores and S5 for the annotations of the oligonucleotides). Our positive controls, the five known EDEN-BP targets mRNAs [Cdk2(Eg1), Eg5, Aurora A, Xsu(H) and c-Mos] are among the 158 identified target. Furthermore, it was recently shown that the highly conserved 3′ UTR of LMO4 [a protein involved in modulation of BMP signaling (24)] was bound by CUG-BP1. LMO4 is uncovered in our screen as one of the new target. Therefore, our analysis identified 153 new EDEN-BP targets. It is noteworthy that the same analysis realized by comparing αE1 and αE2 to total input RNA gave similar results.
The EDEN motif is overrepresented in the coimmunopurified mRNAs
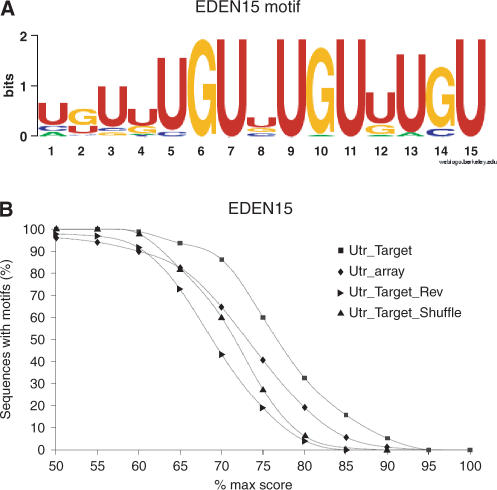
Because it is expected that the identified target mRNAs contained EDEN motifs, we used all annotated transcripts and 3′ UTR from the JGI V4.1 assembly to search for the presence of an EDEN motif. Recently, SELEX experiments to identify in vitro RNA targets for CUG-BP1/CELF1 have been performed (19). Using the MEME algorithm (http://meme.sdsc.edu/meme/meme.html) we identified a motif of 15 nt (EDEN15) as being statistically enriched in this SELEX dataset. This EDEN15 motif was present in 32 out of 63 sequences (P < 0.001). This motif was represented as a simple frequency matrix (Supplementary Data S2, EDEN15) and a sequence logo (Figure 2A). This motif is present in the original targets Aurora A, c-Mos, Eg5, Xsu(H) and Cdk2 with a score of 70, 86, 76, 82 and 83%, respectively, of its best match.
Figure 2.
Overrepresentation of the EDEN15 motif in the 3′UTR of mRNAs complexed to EDEN-BP. (A) MEME analysis of 62 sequences binding EDEN-BP in vitro identified 39 times a 15 nt motif (EDEN15) in 37 sequences (out of 62). These 39 motifs were aligned and used to generate the EDEN15 motif represented here as a sequence logo. (B) Occurences of mRNAs bearing the EDEN15 motif at a given score in the 3′ UTR from identified target mRNAs and control datasets. UTR_array corresponds to all the 3′ UTR available for mRNAs represented on the microarray. UTR_target corresponds to all the 3′ UTR available for the identified EDEN-BP target mRNAs. UTR_target_Rev corresponds to the reverse complement of the UTR_target dataset. UTR_Target_Shuffle corresponds to the UTR_Target dataset after shuffling of the sequences.
Because the three RNA recognition motif (RRM) for the three EDEN-BP orthologs (mm, hs and xt) are highly conserved (almost 100%) and because the human ortholog can functionally and specifically replace the Xenopus protein in egg extract (25) we used the EDEN15 motif definition to count the number of sequence harboring putative EDEN motif.
When comparing the percentage of 3′ UTR sequences harboring an EDEN15 motif above a given score (Figure 2B), the EDEN15 motif was clearly overrepresented in the 3′ UTR sequences of the identified target RNA (UTR_Target, n = 95) when compared to the 3′ UTR of all the RNA represented on the microarray (Utr_array, n = 1236). To ensure the relevance of the EDEN15 motif, we searched for EDEN15 motif in the UTR_target dataset after shuffling it (UTR_Target_shuffle) or using the reverse complement strands (UTR_Target_Rev). We found that no overrepresentation of EDEN15 motif was detected in these datasets when compared with Utr_array. These analyses showed that the EDEN15 motif is clearly overrepresented in the 3′ UTR of the target mRNAs identified with ∼90% of the 3′ UTR harboring an EDEN15 motif with a score above 70%.
As an additional control, we generated a random 15 nt motif with the same base composition as the EDEN15 motif but devoid of any UGU in its consensus. When this motif is searched for in the databases (Supplementary Data S2, NO_UGU motif), only a very limited enrichment was observed between the UTR target and the control dataset. Therefore, the EDEN15 motif is clearly representative of putative binding sites for EDEN-BP and the UGU trinucleotide, a key pattern of this element as can be observed on the sequence logo (Figure 2A).
The intersection between experiments 1 and 2 consists of 158 different mRNAs. These mRNAs were, therefore, isolated with both antiserum and in both experiments and are enriched in the EDEN15 motif. To determine whether the RNA that was purified in only one experiment contain putative EDEN, we searched in the 3′ UTR of these transcripts for the EDEN15 motif. No specific EDEN15 motif enrichment could be observed in these two 3′ UTR populations when compared with the control dataset (Supplementary Data S6). This indicates that the population of RNA purified during one experiment certainly consists of a population directly bound to the EDEN-BP protein and another population probably representative of background signals. This justifies the need to corroborate the results using several serum and repetitions of the IP experiments. Next, we postulated that mRNA specifically immunoprecipitated with the two different serum and presenting EDEN15 motif should bind EDEN-BP in in vitro and in vivo assays.
Binding of recombinant EDEN-BP to the 3′ UTR of identified target mRNAs
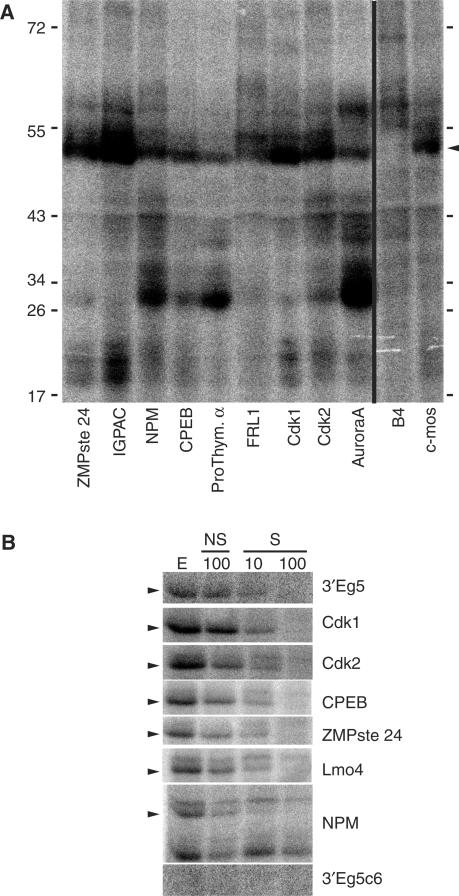
To determine whether the identified target mRNAs could effectively bind EDEN-BP, we performed EMSA analyses with recombinant EDEN-BP. We picked eight new EDEN-BP targets for which the 3′ UTR sequences could be determined that were representatives of the range of enrichment observed in our experiments and with various predicted adenylation profiles (Figure 3A). We also analyzed the previously identified target Eg1/Cdk2. Using the EDEN15 motif we localized the potential EDEN-BP binding site in these mRNAs 3′ UTRs; the match score for the identified motif ranged from 67 to 90% of the maximum match score. Selected portions of candidates 3′ UTRs centered on the EDEN15 motifs were amplified by RT-PCR with the incorporation of a T7 promoter. Radiolabeled RNAs (<200-nt long) from these templates were incubated with recombinant EDEN-BP and complex formation was analyzed by native polyacrylamide gel electrophoresis. As shown on Figure 3B (left panel) two complexes (denoted C1 and C2) were sequentially formed when increasing concentrations of EDEN-BP were added to the incubation mix. The two complexes are compatible with the multimerization of EDEN-BP on its target that has already been reported (23). When the same experiment is conducted with 3′ Eg5C6 (Figure 3B, right panel), a mutated EDEN where a 6 nt substitution in the UGU motifs makes it unable to bind EDEN-BP in UV-crosslinking assay (3), complex C1 is formed but only at higher concentration of recombinant protein and complex C2 is only detectable at the maximum concentration of recombinant protein used (1.25 µM). For all the EDEN-BP target tested, the affinity of EDEN-BP for the RNAs was estimated from the concentration of protein that bound 50% of the RNA probe. Because more than one complex could form on the RNAs this value represents a good indication of the binding affinity for EDEN-BP and is termed here apparent Kd. Average values from three different experiments and for the 10 RNA tested are presented on Figure 3C. As already reported, the mutated version of the Eg5-binding site (3′ Eg5C6) has an apparent Kd value of 700 nM, corresponding to a very low affinity binding sites. Contrariwise, for all of the new target mRNAs, the portions of the 3′ UTR that contained a match for the EDEN15 motif were shifted by recombinant EDEN-BP with apparent Kd below 100 nM indicative of their ability to efficiently bind the recombinant EDEN-BP. These values are similar to the reported apparent Kd of CUG-BP1 for Eg5 that is 34 nM (19).
Figure 3.
EMSA for eight new EDEN-BP target mRNAs. (A) Based on the study by Graindorge et al. (34), inferred adenylation changes for the RNA tested in bandshift. (−) Indicates a decrease and (+) indicates an increase in the poly(A)+ signal of at least 10% after fertilization for the indicated RNA. 0 Indicates an absence of adenylation change. (B) Two representative EMSA with recombinant EDEN-BP and the indicated RNAs are shown; 3′ Eg5C6 is a negative control and ZMPste 24 is a new EDEN-BP target RNA. C1 and C2 indicates the two different complexes formed by multimerization of EDEN-BP, F indicates the free probe, the rEDEN-BP concentrations are indicated on top of each lane. (C) Apparent Kd (nanomolar) and standard deviation was calculated from three different EMSA experiments for each of the indicated RNAs.
In vivo binding to EDEN-BP
To determine whether the identified mRNAs were able to bind in vivo to EDEN-BP we used UV-induced crosslinking to evaluate the association of portion of the 3′ UTR sequences with the endogenous EDEN-BP in UFE extract. As can be observed on Figure 4A, all the tested sequences from our screen present a radioactive signal at a size compatible with EDEN-BP (Figure 4A, arrowhead). This radiolabeled protein migrates at the same position as the one for Aurora A(Eg2) and c-Mos RNA, two known EDEN targets. B4, a RNA that is not bound to EDEN-BP (9) does not present this signal.
Figure 4.
Binding of endogenous EDEN-BP protein to newly identified target RNAs. The indicated RNAs were tested for their ability to bind endogenous EDEN-BP protein by UV crosslinking. (A) The radiolabeled proteins were separated by SDS–PAGE and revealed using a Phosphorimager. (B) UV-crosslinking experiment in presence of specific (S) or non-specific (NS) unlabeled competitor. E corresponds to extract only, NS 100 corresponds to a binding realized in presence of 100-fold excess of non-specific competitor (3′ Eg5C6), S10 and S100 correspond to binding realized in the presence, respectively, of 10- and 100-fold excess of EDEN-BP-specific competitor (3′ Eg5). The arrowhead indicates the EDEN-BP position.
To control the identity and specificity of the observed binding, unlabeled EDEN-BP-specific (3′ Eg5) or non-specific (3′ Eg5C6) RNAs were used as competitors on a subset of sequences. The Eg5 sequence (3′ Eg5) bound a unique protein (Figure 4B, arrowhead) that could be efficiently competed by a 10-fold excess of cold-specific competitor (S10) but not by a 100-fold excess of non-specific-cold competitor (NS100) (Figure 4B). No EDEN-BP signal was observed for the radiolabeled 3′ Eg5C6 RNA. For all the new target RNA tested, the EDEN-BP signal (arrowhead) was efficiently competed by a 10-fold excess of EDEN-BP-specific competitor and totally competed by a 100-fold excess of EDEN-BP-specific cold competitor, while very poorly by a 100-fold excess of the non-specific competitor. These experiments confirmed, therefore, that the identified mRNAs contain a motif able to specifically bind EDEN-BP. Interestingly, when cold-specific competitor is added one can observe the enhancement of a radioactive signal migrating slightly above EDENBP for some RNA (Lmo4, Cdk2, CPEB and ZMPste 24). This suggests that competitive binding between EDEN-BP and another RNA-BP may occur on these RNA.
These results show that the tested RNAs bind both recombinant and endogenous EDEN-BP. Therefore, the combination of EDEN-BP IP and localization of the EDEN-binding site with the defined EDEN15 motif has enabled us to identify new binding targets for EDEN-BP.
Cdk1 is deadenylated by an EDEN-BP-dependent mechanism
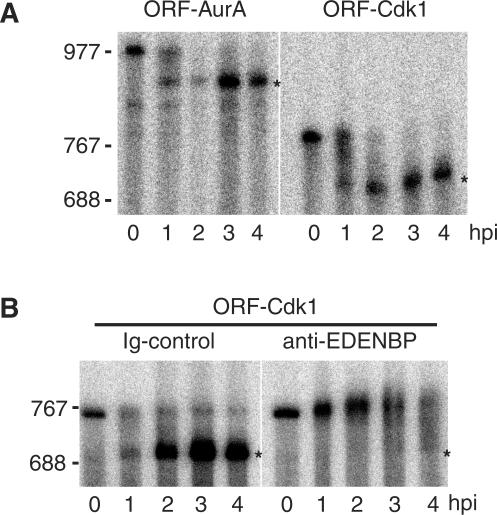
Among the RNA identified as target for EDENBP, Cdk1 was reported earlier to be translationally silent after fertilization (26,27); therefore, we cloned the entire 3′ UTR of X. tropicalis Cdk1 (ENSXETG00000003123) and analyzed its adenylation behavior after injection into X. laevis embryos. As can be observed in Figure 5A, 2 h after injection, both the reporter with the Cdk1 3′ UTR (ORF-Cdk1) and the reporter with the Aurora A 3′ UTR region (ORF-AurA) have accumulated as deadenylated mRNAs. It is clear that Cdk1 3′ UTR targets a reporter RNA for rapid deadenylation with kinetics similar to those observed for Aurora A, a known target of EDEN-BP-dependent deadenylation. It was previously showed that coinjection of αE2 antibodies specifically inhibited EDEN-BP-dependent deadenylation on a reporter RNA containing an EDEN sequence (5,28). Therefore, to verify that the 3′ UTR of Cdk1 was targeting the mRNA for EDEN-BP-dependent deadenylation, we coinjected the ORF-Cdk1 reporter RNA with either purified antiEDEN-BP antibodies (αE2) or control IgG. While ORF-Cdk1 is deadenylated by 2 h postinjection in presence of control IgG, the immunoneutralization of EDEN-BP led to a complete inhibition of deadenylation with no accumulation of poly(A)− reporter RNA by 4 h postinjection. These experiments clearly demonstrate that Cdk1 mRNA, that is among the newly identified binding target for EDEN-BP, is specifically deadenylated by an EDEN-BP-dependent mechanism.
Figure 5.
Cdk1 3′ UTR target a reporter mRNA for EDEN-BP-dependent deadenylation in Xenopus embryos. (A) The indicated capped poly(A)+ and radiolabeled mRNAs (ORF-Cdk1, ORF-AurA) were injected into two-cell embryos and samples were harvested at the indicated time after injection (hpi). Total RNA were extracted and separated by denaturing polyacrylamide gel electrophoresis. Radiolabeled RNAs were revealed by Phosphorimaging. For ORF-Cdk1, one can observe a small smiling of the gel. (B) ORF-Cdk1 mRNA was coinjected with either αE2 (antiEDEN-BP) antibody or control IgG as indicated. Samples were harvested at the indicated time after injection. Radiolabeled RNAs were analyzed as described for A). RNA size markers are positioned on the left side of the panel. The asterisk (*) denote the position of the poly(A)− mRNAs.
DISCUSSION
In the present work, we have identified 158 maternal mRNAs that are new binding targets for EDEN-BP (5% of the tested RNAs on the microarray); five mRNAs are the previously identified EDEN-BP targets c-Mos, Cdk2(Eg1), Aurora A(Eg2), Eg5/KLP and XSU(H), and 153 are newly identified target. Among the newly identified target, xLMO4, a maternally expressed member of the LIM-domain-only protein family was independently shown to bind CUG-BP1/EDEN-BP in mice (24). Interestingly, when we analyzed the X. tropicalis LMO4 3′ UTR that is highly conserved with the human LMO4 3′ UTR, the EDEN15 motif could be detected with a best match score of 90%. This suggests that the binding of CUG-BP1/EDENBP to LMO4 3′ UTR is conserved from Xenopus to human. Furthermore, we showed that a small region of xtLMO4 was bound to CUG-BP1 both in vitro (Figure 3) and in vivo (Figure 4B). This region corresponds to the region independently identified as binding CUG-BP1/EDEN-BP in mice (24).
Among the mRNAs copurified with EDEN-BP, those encoding proteins involved in cell cycle control or oocyte maturation are common when compared with the functional categories of the gene products represented on the array; in particular, Aurora A(Eg2), Aurora B, CPEB, c-Mos, Cdk1, Cdk2(Eg1), casein Kinase 2 beta, Bub 3, Wee1, MELK(Eg3), Eg5/KLP and NEK2B are among the mRNAs recovered in our screen. Interestingly, an analysis of the expression profiles of these mRNAs, based on EST representation, indicated that the majority of these mRNAs (Aurora A(Eg2), Aurora B, CPEB, c-Mos, Cdk2(Eg1), Bub 3, Wee1A, MELK(Eg3), Eg5/KLP and NEK2B) are mainly restricted to the germline and expression levels are usually low throughout development. This could indicate that binding of EDEN-BP to these mRNAs contribute to their low expression levels after fertilization. For example, CPEB protein is degraded during oocyte maturation with only 25% of the protein present in stage VI oocytes remaining after maturation (29). Destruction of CPEB is required for meiotic progression and expression of a stable form of CPEB in embryos blocks cell divisions (30). Another intriguing observation comes from the simultaneous presence of EDEN motifs in both Cdk1 and Cdk2. The amount of the proteins encoded by these mRNAs increases during oocyte maturation and then remain constant after fertilization and until the MBT while their mRNAs are not translated as observed by S35 labeling (26,27). During the cleavage stage, Cdk2 is apparently in limiting amount compared with cyclin E levels until the MBT (27,31). Hence, the presence of the EDEN motif in the Cdk2 3′ UTR would reduce the translation of this mRNA and, therefore, assist or even cause the maintenance of the balance between Cdk2 and cyclin E. Here, we demonstrated that Cdk1 3′ UTR is a new binding target for EDEN-BP and that EDEN-BP activity is required for the rapid deadenylation driven by the Cdk1 3′ UTR. It is therefore likely that EDEN-BP acts as a translational repressor on both Cdk1 and Cdk2. As Ueno and Sagata (32) presented evidence that, in the case of c-Mos, the ability of EDEN to trigger translational silencing and deadenylation was potentiated by AUUUA tetranucleotides, it must be noted that the Cdk1 3′ UTR is devoid of any AUUUA but contain some AU element located 3′ of the EDEN. These observations reinforce the hypothesis that the presence of an EDEN motif may be part of a mechanism to ensure a limited expression of some important cell cycle regulators in the cleavage stage embryos. EDEN-BP protein is maternally expressed but is activated only after fertilization (13). This particularity may ensure that mRNAs containing EDEN motifs are translationally repressed either to inhibit the overaccumulation of stable protein encoded by maternal mRNAs and required for early development, or to ensure reduced expression of proteins that are unstable and mainly required during oocyte maturation (such as c-Mos or CPEB).
This does not mean that all the mRNAs identified are rapidly deadenylated after fertilization. Posttranscriptional processes are the results of a complex interplay between RNA-BP with different affinity and different activity. Notably some of the newly identified binding targets, such as xFRL1, are actually polyadenylated after fertilization and their encoded protein accumulates only after fertilization [(33,34), Figure 3 and data not shown]. Whether EDEN-BP is implicated in this polyadenylation possibly by associating with different partners or other sequences specify the postfertilization polyadenylation of xFRL1 requires further study.
The SELEX information used to build a model for the EDEN motif is coroborated by the enrichment of this motif in the population of mRNA coimmunoprecipitated with antiEDEN-BP antibodies. Furthermore, earlier experiments [(35) and data not shown] indicated that while a synthetic (UGUA)4 RNA was able to efficiently crosslink EDEN-BP in Xenopus extracts, an (UAUG)4 motif was not. As defined by the scoring matrix, the best score for the (UGUA)4 motif is 84% (420/495) while it is only 63% (313/495) for the (UAUG)4. Interestingly, it was recently reported that Maskin and Cyclin B1 3′ UTR bind EDEN-BP in X. laevis oocytes (36). We found that the EDEN15 motif was present in the 3′ UTR of these mRNAs (Cyclin B1 (NM_001088495), Maskin (NM_001088495) with a match score of 74 and 83%, respectively.
The EDEN15 motif with a score of at least 70% is found in the 3′ UTR of >90% the coimmunoprecipitated RNA. When a subset of the newly identified targets was analyzed for in vivo and in vitro binding to EDEN-BP, the localization of the EDEN motif was realized solely on the base of the best EDEN15 motif in the 3′ UTR and all the tested RNA were effectively associated to EDEN-BP in our assays. Alternatively, and as has been shown for CUG-BP1, binding sites may be located not only in the 3′ UTR (28) but also in the introns (37), or the 5′ UTR (38). It is therefore possible that some mRNAs are actually associated to EDEN-BP by intronic sites. However, this is very unlikely in the context of this study as there is no ongoing transcription at the stages studied and therefore no pre-mRNA should be present. It is also possible that EDEN-BP is associated to mRNA by sites located in the 5′ UTR, but this region being poorly annotated, the analysis of the 5′ UTR is restricted to a very small number of sequences.
Some mRNAs may be enriched in the immunoprecipitated pool due to their association in a RNP complex containing EDEN-BP and other RNA-BPs that specifically bind to the RNA so that the contribution of EDEN binding motif is only weak or even absent. Such a situation has already been described for the mRNAs that bind both Pumilio-2 (Pum-2) and deleted in azoospermia-like (DAZL) (39). However, as the tested RNAs bind EDEN-BP both in vitro and in vivo, we think that this will be a minor contribution to the pool of 158 maternal mRNAs selected with both antibodies.
We have identified 158 mRNAs as binding targets for the endogenous EDEN-BP protein. The identified mRNAs contain proportionally more potential EDEN motifs in their 3′ UTR than the currently available X. tropicalis 3′ UTRs analyzed as a whole. EMSA experiments with recombinant EDEN-BP and UV-crosslinking experiments with egg extract indicates that all the tested (12) potential EDEN containing RNAs can bind endogenous EDEN-BP. We have therefore identified on a large-scale targets for a RNA-BP known for its role in controlling specifically mRNA translation. Furthermore, we demonstrated that Cdk1 3′ UTR, one of the newly identified target is rapidly and specifically deadenylated by and EDEN-BP-dependent mechanism. It would be of interest to determine to what extent the binding of EDEN-BP controls the translation of the newly identified targets by comparing large-scale polysomal analyses in native conditions and when EDEN-BP is knocked down.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the Association pour la Recherche contre le Cancer (ARC 4791), Ligue Contre le Cancer (comité d’Ille et Vilaine), the French Ministère de la Recherche (ACI BCMS 369) and the Région Bretagne (PRIR–560–412). The production of Xenopus tropicalis microarray was made possible by the « Action Puces à ADN » of the CNRS. The authors would like to thank member of the transcriptomic facilities of the IFR 140 in Rennes where they did the microarray experiments. Funding to pay the Open Access publication charges for this article was provided by the CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klotz C, Dabauvalle M, Paintrand M, Weber T, Bornens M, Karsenti E. Parthenogenesis in xenopus eggs requires centrosomal integrity. J. Cell. Biol. 1990;110:405–415. doi: 10.1083/jcb.110.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasudevan S, Seli E, Steitz J. Metazoan oocyte and early embryo development program: a progression through translation regulatory cascades. Genes Dev. 2006;20:138–146. doi: 10.1101/gad.1398906. [DOI] [PubMed] [Google Scholar]
- 3.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne H. EDEN and EDEN-BP, a cis element and an associated factor that mediates sequence-specific mRNA deadenylation in xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paris J, Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early xenopus development. Dev. Biol. 1990;140:221–224. doi: 10.1016/0012-1606(90)90070-y. [DOI] [PubMed] [Google Scholar]
- 5.Gautier-Courteille C, Le Clainche C, Barreau C, Audic Y, Graindorge A, Maniey D, Osborne H, Paillard L. EDEN-BP-dependent post-transcriptional regulation of gene expression in xenopus somitic segmentation. Development. 2004;131:6107–6117. doi: 10.1242/dev.01528. [DOI] [PubMed] [Google Scholar]
- 6.Barreau C, Paillard L, Méreau A, Osborne HB. Mammalian celf/bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Kress C, Gautier-Courteille C, Osborne H, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol. Cell. Biol. 2007;27:1146–1157. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvet P, Omilli F, Arlot-Bonnemains Y, Legagneux V, Roghi C, Bassez T, Osborne H. The deadenylation conferred by the 3′ untranslated region of a developmentally controlled mRNA in xenopus embryos is switched to polyadenylation by deletion of a short sequence element. Mol. Cell. Biol. 1994;14:1893–1900. doi: 10.1128/mcb.14.3.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legagneux V, Bouvet P, Omilli F, Chevalier S, Osborne H. Identification of RNA-binding proteins specific to xenopus Eg maternal mRNAs: association with the portion of Eg2 mRNA that promotes deadenylation in embryos. Development. 1992;116:1193–1202. doi: 10.1242/dev.116.4.1193. [DOI] [PubMed] [Google Scholar]
- 10.Brittle A, Ohkura H. Centrosome maturation: aurora lights the way to the poles. Curr. Biol. 2005;15:R880–R882. doi: 10.1016/j.cub.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt A, Rauh N, Nigg E, Mayer T. Cytostatic factor: an activity that puts the cell cycle on hold. J. Cell. Sci. 2006;119:1213–1218. doi: 10.1242/jcs.02919. [DOI] [PubMed] [Google Scholar]
- 12.Elledge S, Spottswood M. A new human p34 protein kinase, cdk2, identified by complementation of a cdc28 mutation in saccharomyces cerevisiae, is a homolog of xenopus Eg1. Embo J. 1991;10:2653–2659. doi: 10.1002/j.1460-2075.1991.tb07808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Detivaud L, Pascreau G, Karaiskou A, Osborne H, Kubiak J. Regulation of EDEN-dependent deadenylation of aurora A/Eg2-derived mRNA via phosphorylation and dephosphorylation in xenopus laevis egg extracts. J. Cell. Sci. 2003;116:2697–2705. doi: 10.1242/jcs.00477. [DOI] [PubMed] [Google Scholar]
- 14.Graindorge A, Thuret R, Pollet N, Osborne H, Audic Y. Identification of post-transcriptionally regulated xenopus tropicalis maternal mRNAs by microarray. Nucleic Acids Res. 2006;34:986–995. doi: 10.1093/nar/gkj492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fierro AC, Thuret R, Coen L, Perron M, Demeneix BA, Wegnez M, Gyapay G, Weissenbach J, Wincker P, Mazabraud A, et al. Exploring nervous system transcriptomes during embryogenesis and metamorphosis in xenopus tropicalis using est analysis. BMC Genomics. 2007;8:118. doi: 10.1186/1471-2164-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray A, Solomon M, Kirschner M. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 17.Audic Y, Omilli F, Osborne HB. Postfertilization deadenylation of mRNAs in xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Mol. Cell. Biol. 1997;17:209–218. doi: 10.1128/mcb.17.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tusher V, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne H. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crooks GE, Hon G, Chandonia J, Brenner SE. Weblogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legagneux V, Omilli F, Osborne HB. Substrate-specific regulation of RNA deadenylation in xenopus embryo and activated egg extracts. RNA. 1995;1:1001–1008. [PMC free article] [PubMed] [Google Scholar]
- 23.Cosson B, Gautier-Courteille C, Maniey D, Aït-Ahmed O, Lesimple M, Osborne HB, Paillard L. Oligomerization of EDEN-BP is required for specific mRNA deadenylation and binding. Biol. Cell. 2006;98:653–665. doi: 10.1042/BC20060054. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Xu J, Safarpour F, Stewart AFR. Lmo4 mRNA stability is regulated by extracellular ATP in F11 cells. Biochem. Bioph. Res. Co. 2007;357:56–61. doi: 10.1016/j.bbrc.2007.03.113. [DOI] [PubMed] [Google Scholar]
- 25.Paillard L, Legagneux V, Osborne HB. A functional deadenylation assay identifies human CUG-BP as a deadenylation factor. Biol. Cell. 2003;95:107–113. doi: 10.1016/s0248-4900(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi H, Minshull J, Ford C, Golsteyn R, Poon R, Hunt T. On the synthesis and destruction of A- and B-type cyclins during oogenesis and meiotic maturation in xenopus laevis. J. Cell. Biol. 1991;114:755–765. doi: 10.1083/jcb.114.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howe J, Newport J. A developmental timer regulates degradation of cyclin E1 at the midblastula transition during xenopus embryogenesis. Proc. Natl Acad. Sci. USA. 1996;93:2060–2064. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paillard L, Legagneux V, Maniey D, Osborne HB. C-jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J. Biol. Chem. 2002;277:3232–3235. doi: 10.1074/jbc.M109362200. [DOI] [PubMed] [Google Scholar]
- 29.Groisman I, Huang Y, Mendez R, Cao Q, Theurkauf W, Richter J. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- 30.Mendez R, Barnard D, Richter J. Differential mRNA translation and meiotic progression require cdc2-mediated CPEB destruction. Embo J. 2002;21:1833–1844. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richard-Parpaillon L, Cosgrove R, Devine C, Vernon A, Philpott A. G1/S phase cyclin-dependent kinase overexpression perturbs early development and delays tissue-specific differentiation in xenopus. Development. 2004;131:2577–2586. doi: 10.1242/dev.01121. [DOI] [PubMed] [Google Scholar]
- 32.Ueno S, Sagata N. Requirement for both EDEN and AUUUA motifs in translational arrest of mos mRNA upon fertilization of xenopus eggs. Dev. Biol. 2002;250:156–167. doi: 10.1006/dbio.2002.0787. [DOI] [PubMed] [Google Scholar]
- 33.Dorey K, Hill CS. A novel cripto-related protein reveals an essential role for egf-cfcs in nodal signalling in xenopus embryos. Dev. Biol. 2006;292:303–316. doi: 10.1016/j.ydbio.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Graindorge A, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of post-transcriptionally regulated xenopus tropicalis maternal mRNAs by microarray. Nucleic Acids Res. 2006;34:986–995. doi: 10.1093/nar/gkj492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Audic Y, Omilli F, Osborne H. Embryo deadenylation element-dependent deadenylation is enhanced by a cis element containing auu repeats. Mol. Cell. Biol. 1998;18:6879–6884. doi: 10.1128/mcb.18.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meijer HA, Radford HE, Wilson LS, Lissenden S, de Moor CH. Translational control of maskin mRNA by its 3′ untranslated region. Biol. Cell. 2007;99:239–250. doi: 10.1042/BC20060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philips A, Timchenko L, Cooper T. Disruption of splicing regulated by a cug-binding protein in myotonic dystrophy (see comments) Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 38.Timchenko N, Iakova P, Cai Z, Smith J, Timchenko L. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell. Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore F, Jaruzelska J, Fox M, Urano J, Firpo M, Turek P, Dorfman D, Pera R. Human pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with daz (deleted in azoospermia) and daz-like proteins. Proc. Natl Acad. Sci. USA. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]