Abstract
Antigen receptor genes are assembled by a site-specific DNA rearrangement process called V(D)J recombination. This process proceeds through two distinct phases: a cleavage phase in which the RAG1 and RAG2 proteins introduce DNA double-strand breaks at antigen receptor gene segments, and a joining phase in which the resulting DNA breaks are processed and repaired via the non-homologous end-joining (NHEJ) repair pathway. Genetic and biochemical evidence suggest that the RAG proteins play an active role in guiding the repair of DNA breaks introduced during V(D)J recombination to the NHEJ pathway. However, evidence for specific association between the RAG proteins and any of the factors involved in NHEJ remains elusive. Here we present evidence that two components of the NHEJ pathway, Ku70 and Ku80, interact with full-length RAG1, providing a biochemical link between the two phases of V(D)J recombination.
INTRODUCTION
Immunoglobulins and T-cell receptors are encoded in the vertebrate genome in arrays of variable (V), diversity (D) and joining (J) gene segments which are assembled during lymphocyte development by a process called V(D)J recombination (1). V(D)J recombination proceeds in two phases (2). In the first phase, two different gene segments are brought into close proximity through the assembly of a multiprotein synaptic complex containing two lymphoid cell-specific proteins, called RAG1 and RAG2, which mediate interactions with conserved recombination signal sequences (RSSs) that lie adjacent to each gene segment. Each RSS contains a highly conserved heptamer and nonamer sequence, separated by either 12 or 23 bp of intervening spacer DNA (12-RSS and 23-RSS, respectively); synapsis is generally restricted to RSSs bearing different length spacers (the 12/23 rule). Following synapsis, the RAG proteins introduce a DNA double-strand break precisely between the RSS heptamer and the coding segment via a nick-hairpin mechanism (3,4), yielding four DNA ends: two blunt 5′-phosphorylated signal ends and two coding ends terminating in DNA hairpin structures. In the second phase, the hairpins at the coding ends are nicked by a protein complex containing Artemis and the catalytic subunit of the DNA dependent protein kinase (DNA-PKcs) (5), and nucleotides may be further added or removed before the ends are joined to create coding joints. In contrast to coding joints, which are often imprecise, signal ends are generally fused heptamer-to-heptamer, forming precise signal joints. Genetic evidence suggests that efficient signal and coding joint formation requires an intact non-homologous end-joining (NHEJ) DNA repair pathway, involving the activities of at least five proteins in addition to DNA-PKcs and Artemis, including Ku70, Ku80, XRCC4, DNA Ligase IV (1), and the recently discovered Cernunnos/XLF protein (6,7).
Substantial biochemical evidence indicates that various NHEJ components physically interact with one another. For example, Ku70 and Ku80 form a stable heterodimer, which associates with DNA-PKcs to form DNA-PK (8). In addition, XRCC4 and DNA Ligase IV form a stable complex that also associates with Ku70/Ku80 (9), and Cernunnos/XLF (7,10). Whether any component(s) of the known end-processing and end-joining machinery involved in V(D)J recombination specifically interacts with the RAG proteins remains uncertain. However, several lines of genetic and biochemical evidence provide indirect experimental support for this possibility. First, joining-deficient RAG mutants that support levels of RSS cleavage comparable to wild-type (WT) RAGs have been identified (11,12). Second, evidence from several laboratories suggest the RAG proteins guide repair of the DNA DSBs they introduce to the NHEJ pathway and away from alternative error-prone repair pathways (13–16). Third, the RAG proteins mediate coupled cleavage in vitro with enhanced fidelity to the 12/23 rule when reactions are supplemented with Ku70/Ku80 and DNA-PKcs (17). Fourth, studies demonstrating reconstitution of coding joint formation in vitro show that the continued presence of the RAG proteins in the joining reaction promotes repair of RAG-generated coding ends (18,19). Signal joint formation, by contrast, is inhibited by the presence of the RAG proteins (18–20), perhaps because they impair access to the signal ends by the NHEJ machinery though ongoing association with the RSSs (21). The continued presence of the RAG proteins after cleavage also promotes more efficient hairpin opening mediated by the Artemis/DNA-PKcs complex in vitro (5), raising the possibility that one or both of these factors interact directly or indirectly with the RAG proteins. Finally, there is precedence for Ku association with other recombinases active in vertebrate organisms, such as the Sleeping Beauty transposase (22).
Here, we present biochemical evidence that Ku70/Ku80 associates with full-length RAG1, but not truncated catalytically active ‘core’ RAG1 (residues 384–1040), when RAG1 is co-expressed with ‘core’ RAG2 (residues 1–387) in mammalian cells and recovered using a mild purification procedure. Under these conditions, Ku association with purified core RAG1/full-length RAG2 is also observed. However, Ku70/Ku80 interaction with core RAG1/full-length RAG2, but not full-length RAG1/core RAG2, appears to be DNA-dependent. Ku70/Ku80 is also detected in a novel RAG–RSS complex assembled with full-length RAG1/core RAG2, but not core RAG1/full-length RAG2. Formation of this complex minimally requires the addition of residues 211–383 to core RAG1; RAG1 mutants containing alanine substitutions in the ‘dispensable’ N-terminal domain (NTD) of RAG1 have been identified that impair or promote Ku–RAG–RSS complex formation. V(D)J recombination assays performed in cell culture suggest that Ku association with full-length RAG1 plays a role in facilitating signal joint formation. The implications of Ku70/Ku80 association with pre-cleavage RAG–RSS complexes are discussed.
MATERIALS AND METHODS
DNA constructs
Expression constructs encoding core or full-length RAG1 and RAG2, fused at the amino-terminus to maltose-binding protein (MBP), and human HMGB1 with an amino-terminal hexahistidine tag have been described previously (23) (see Figure 1). Using PCR and subcloning, a total of 12 expression constructs were generated that encode truncated forms of RAG1 in which the amino-terminal third of RAG-1 (residue 1–383) was added back to core RAG-1 (residues 384–1040) in increments of 30 amino acids starting at residue 361. Constructs encoding forms of 181MR1 with alanine substitutions at residues 193–202, 221–230, or 305–314 were generated by inside-out PCR. Further description of the cloning strategies used to generate these truncation and alanine substitution mutant RAG1 expression constructs can be found in Supplementary Data.
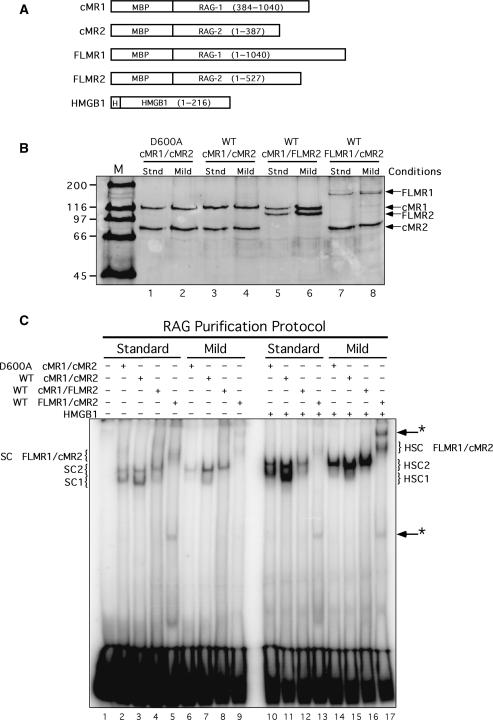
Figure 1.
Core and full-length RAG proteins purified using different conditions exhibit distinct DNA binding properties. (A) Schematic diagrams of RAG1, RAG2 and HMGB1 fusion proteins used in this study. MBP and polyhistidine (H) sequences are also indicated. (B) Purified proteins analyzed by SDS–PAGE. WT or catalytically inactive (D600A) RAG1 and RAG2 fusion proteins shown in (A) were co-expressed in HEK 293 cells in the indicated combinations and purified by amylose affinity chromatography using standard (stnd) or mild buffers (see ‘Materials and Methods’ section). Proteins were fractionated by SDS–PAGE in parallel with protein standards (M) and detected by staining the gel with SYPRO orange. (C) EMSA of RAG protein preparations. Radiolabeled intact 12-RSS substrate was incubated with cMR1/cMR2 (WT or D600A), cMR1/FLMR2 or FLMR1/cMR2 purified using standard or mild conditions in binding reactions lacking or containing HMGB1 as indicated above the gel. Protein–DNA complexes were fractionated by EMSA. The positions of SC1, HSC1, SC2, HSC2 species (described in the text) are indicated at left and right. Novel protein–DNA complexes are denoted by arrows with an asterisk at right.
Protein expression and purification
Full-length HMGB1 was expressed in the Escherichia coli strain BL21(DE3)pLysS and purified by immobilized metal affinity chromatography and ion exchange chromatography as previously described (24). MBP-RAG1 (WT or catalytically inactive [D600A]; either core, full-length, truncated or alanine replacement mutant RAG1) and MBP-RAG2 (either core or full-length RAG2) were co-expressed in 293 or HeLa cells as previously described (23,25), except that RAG proteins were prepared from 20 10 cm dishes of cells. In addition, for the preparation of 151MR1/cMR2 and FLMR1/cMR2, the amount of the RAG1 expression construct used for transfection was increased from 5 µg/plate to 7 µg/plate to improve the yield of these proteins. The transfected cells were harvested 48 h post-transfection and either purified immediately or frozen at −80°C for later purification. Proteins were purified either using the method described previously (23), or using a milder procedure described here. Briefly, each cell pellet was resuspended in 3 ml buffer R [25 mM HEPES [pH 7.4], 150 mM KCl, 10 mM MgCl2, 10% glycerol, 2 mM DTT and protease inhibitors (10 µM leupeptin, 2 µM pepstatin A and 100 µM PMSF)] and subjected to three cycles of sonication for 1.5 min at 22–25% amplitude (Fisher Scientific Sonic Dismembranator Model 500). The lysates were clarified by centrifugation at 46 000 × g (Beckman SW55Ti rotor, 22 000 r.p.m.) at 4°C for 30 min and the supernatants collected from two pellets were passed over amylose resin (New England Biolabs, Ipswich, MA, USA; 1 ml) equilibrated with buffer R. The column was washed with 10 ml buffer R (the final 5 ml lacked protease inhibitors), and the MBP-RAG proteins were eluted with buffer R containing 10 mM maltose (but lacking protease inhibitors).
Oligonucleotide cleavage and binding assays
Radiolabeled 12-RSS and 23-RSS substrates (50 or 62 bp long, respectively) were prepared as described (23). DNA cleavage and binding activities of the RAG preparations were analyzed using in-tube or in-gel cleavage assays, and electrophoretic mobility shift assays (EMSAs) as indicated in the text using published procedures (23), except that binding reactions were incubated on ice instead of 25°C to reduce RSS cleavage levels observed at this temperature for RAG proteins purified using the mild protocol. Selected binding reactions were further supplemented with 5.5 ng purified human Ku70/Ku80 complex (Trevigen, Gaithersburg, MD, USA). To detect Ku70, Ku80 or MBP-RAG fusion proteins using an electrophoretic mobility supershift approach, binding reactions were assembled and incubated on ice for 10 min, and then 1 µg of antibody specific for Ku70 (Clone N3H10; Lab Vision/NeoMarkers, Fremont, CA, USA), Ku80 (Clone 111; BD Biosciences, San Jose, CA, USA), or MBP (Clone 8G1; Cell Signaling Technology, Danvers, MA, USA) was added to the reaction mixture and incubated for an additional 5 min on ice before gel electrophoresis. Purified mouse IgG1 was used as a negative control in these experiments (Cedarlane Laboratories, Burlington, NC, USA).
Detection of transposition and hybrid joint formation
Assays for transposition and hybrid joint formation were performed as described previously (25,26), except that the percentage of polyacrylamide used in gels to assay cleavage products in assays of hybrid joint formation was reduced to 5.5%.
Detection of signal end breaks, and signal and coding joint formation in cells
V(D)J recombination assays were performed as previously described (26) by cotransfecting 293 cells with the inversional V(D)J recombination substrate pJH299 and WT cMR1 or WT or mutant 181MR1 and cMR2 expression constructs. Signal end breaks were detected by ligation-mediated PCR (LM-PCR), and signal and coding joints were amplified and quantified using real-time PCR following published procedures (26).
RESULTS
Full-length RAG1 purified with core RAG2 under mild conditions forms a novel higher-order RAG–RSS complex
Previous studies provided indirect evidence for a physical association between the RAG proteins and one or more factors involved in processing and/or joining V(D)J recombination intermediates. We speculated that RAG protein interactions with these factors could be unstable in our standard purification buffers that contain high concentrations of monovalent salt. We also considered the possibility that unidentified cofactors or non-core portions of the RAG proteins may be required to stabilize RAG association with end-processing or end-joining factors to enable the protein–protein interactions to be detected experimentally. To explore these possibilities, we co-expressed core and full-length RAG1 and RAG2 in 293 cells in various combinations (cMR1/cMR2, cMR1/FLMR2, FLMR1/cMR2, see Figure 1A) and purified them following our standard procedure (23), or a modified protocol that uses a milder buffer containing 10% glycerol, 10 mM MgCl2 and 150 mM KCl. We find that, for a given combination of core and full-length RAG1 and RAG2, protein yields are generally quite similar using either purification method (Figure 1B). No other proteins whose abundance approximates those of the RAG proteins are detected by SDS–PAGE after SYPRO Orange staining.
We hypothesized that association between the RAG proteins and any interacting factor stabilized by the mild purification conditions might be visualized using an electrophoretic mobility shift assay (EMSA), appearing as a RAG–RSS complex whose mobility is slower than its counterpart purified using the standard procedure. To test this possibility, each of the various RAG1 and RAG2 protein preparations shown in Figure 1B were incubated with a radiolabeled intact 12-RSS substrate in binding reactions containing Ca2+, and RAG–RSS complex formation was analyzed using an EMSA (Figure 1C). As expected from previous results (27), two distinct protein–DNA complexes, called SC1 and SC2, are observed by EMSA when cMR1/cMR2 purified using the standard procedure is incubated with an isolated RSS substrate. The more abundant SC1 complex was previously shown to contain a RAG1 dimer and monomeric RAG2, whereas the less abundant and slower migrating SC2 complex differed from SC1 by the incorporation of a second RAG2 molecule (27). Using the same purification conditions, cMR1/FLMR2 assembled two protein–DNA complexes similar to SC1 and SC2 formed with cMR1/cMR2, but of lower abundance, and FLMR1/cMR2 exhibited poor DNA binding, yielding a smear of complexes migrating more slowly than those formed by cMR1/FLMR2. These results are consistent with previous data (25). Interestingly, when purified using the mild procedure, cMR1/cMR2 predominantly forms the SC2 complex, possibly reflecting more stable RAG2 association with RAG1 and/or the RSS. We find that cMR1/FLMR2 also exhibits the same trend as cMR1/cMR2. However, when similarly purified FLMR1/cMR2 is analyzed by EMSA, we find the SC species are slightly better resolved and we notice the appearance of a novel protein–DNA complex migrating more slowly than the SC species (Figure 1C, lane 9, see upper arrow). Another protein–DNA complex migrating much faster than SC1 and SC2 is also observed by EMSA in binding reactions containing FLMR1/cMR2 purified using either procedure (Figure 1C, lane 9, see lower arrow). When binding reactions are supplemented with purified HMGB1, RAG–RSS complex formation is stimulated (except for FLMR1/cMR2 purified using standard conditions), and the resulting protein–DNA complexes are supershifted slightly relative to their counterparts lacking HMGB1 (designated HSC, HSC1 or HSC2; see Figure 1C, compare lanes 2–9 to 10–17).
Ku70 and Ku80 associate with RAG–RSS complexes containing full-length, but not core RAG1
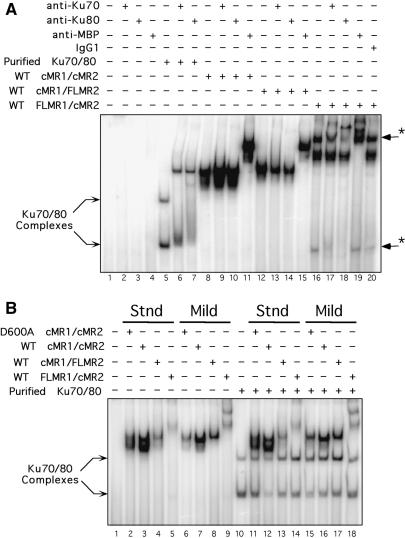
We speculated that the novel higher-order protein–DNA complex formed with FLMR1/cMR2 purified under mild conditions might contain factors associated with the joining phase of V(D)J recombination. To test this possibility, we examined whether this complex could be supershifted with antibodies against proteins known to be required for processing and joining V(D)J recombination intermediates. In preliminary experiments, we found that monoclonal antibodies specific for human Ku70 or Ku80 are capable of supershifting the novel complex (data not shown). To follow-up this observation, we compared the ability of anti-Ku antibodies to supershift RAG–RSS complexes formed when the three different RAG protein preparations purified under mild conditions are incubated with a radiolabeled 12-RSS in the presence of HMGB1. As a control, protein–DNA complexes assembled with purified Ku70/Ku80 were similarly analyzed. In control reactions, we find that anti-Ku70 and anti-Ku80 antibodies do not bind a radiolabeled 12-RSS substrate alone, nor does a monoclonal anti-MBP antibody (Figure 2A, lanes 2–4). When the 12-RSS substrate is incubated with purified Ku70/Ku80, two protein–DNA complexes are formed that are detectable by EMSA. Interestingly, the lower-order Ku–RSS complex co-migrates with the fast migrating protein–DNA complex observed by EMSA in binding reactions containing FLMR1/cMR2 (Figure 2A, compare lanes 5 and 16). Subsequent addition of anti-Ku70 or anti-Ku80 antibodies to the Ku-binding reaction visibly supershifts both the Ku–RSS complexes. The slower-migrating complex is more efficiently supershifted, presumably due to multivalent antibody binding to larger multimeric Ku complexes. Addition of anti-Ku70 or anti-Ku80 antibodies to binding reactions containing HMGB1 and either cMR1/cMR2 or cMR1/FLMR2 does not cause an observable supershift of HSC1 or HSC2 formed with these protein preparations (Figure 2A, compare lane 8 with lanes 9 and 10, and lane 12 with lanes 13 and 14, respectively). In contrast, the novel higher-order protein–DNA complex formed with FLMR1/cMR2 and HMGB1, but not the HSC species in the same lane, are supershifted by anti-Ku70 and anti-Ku80 antibodies, indicating that this higher-order RAG–RSS complex contains Ku70 and Ku80 (Figure 2A, compare lane 16 to lanes 17 and 18). The fast migrating protein–DNA complex in the same lane is also supershifted with anti-Ku antibodies, consistent with its co-migration with a Ku–RSS complex assembled with purified Ku70/Ku80. As a positive control, monoclonal anti-MBP antibodies supershift all HSC complexes formed with cMR1/cMR2, cMR1/FLMR2 and FLMR1/cMR2 protein preparations, as well as the novel higher-order RAG–RSS complex formed with FLMR1/cMR2, but not the fast migrating Ku–RSS complex detected in the same lane (Figure 2A, lanes 11, 15 and 19). However, purified mouse IgG1 antibody used as a negative control does not supershift any of the protein–DNA complexes (Figure 2A, lane 20). Taken together, these data suggest that association of Ku70/Ku80 to the RAG complex is stabilized by the presence of the RAG1 NTD and is sensitive to the conditions used to purify the RAG proteins. The HSC/Ku complex is also observed in mobility shift assays performed using FLMR1/cMR2 proteins prepared from HeLa cells (Supplementary Figure S1), suggesting that Ku association with full-length RAG1 is not a phenomenon unique to 293 cells.
Figure 2.
The RAG1 NTD is necessary, but not sufficient, for stabilizing association of Ku70/Ku80 to a RAG–RSS complex. (A) Anti-Ku70 and anti-Ku80 antibodies supershift FLMR1/cMR2 RAG–RSS complexes. Radiolabeled intact 12-RSS substrate was incubated with purified Ku70/Ku80, or with HMGB1 and the various RAG preparations purified using the mild protocol, and subjected to supershift analysis by EMSA using purified monoclonal antibodies to Ku70, Ku80 or MBP, as indicated above the gel. Protein-DNA complexes supershifted by anti-Ku antibodies are indicated by arrows with an asterisk at right. (B) Supplementing RAG-RSS binding reactions with purified Ku70/Ku80 fails to supershift RAG–RSS complexes. Radiolabeled intact 12-RSS substrate was incubated with the various RAG preparations shown in Figure 3B in binding reactions containing HMGB1 in the absence or presence of purified Ku70/Ku80 as indicated above the gel, and then RAG–RSS complex formation was analyzed by EMSA.
If Ku70/Ku80 association with the RAG proteins is limited by the availability of Ku in the binding reaction, one might expect that supplementing RAG binding reactions with purified Ku70/Ku80 could drive the formation of higher-order RAG–RSS complexes containing Ku. To test this possibility, we incubated the various RAG protein preparations purified under either standard or mild conditions with HMGB1 and a radiolabeled 12-RSS in the presence of purified Ku70/Ku80, and examined protein–DNA complex formation by EMSA. We find that addition of purified Ku70/Ku80 to binding reactions containing cMR1/cMR2 fails to supershift the HSC1 and/or HSC2 complexes formed with this combination of RAG proteins, regardless of how they are purified (Figure 2B). Comparable experiments using cMR1/FLMR2 and FLMR1/cMR2 purified under either standard or mild conditions yield similar results. Taken together, these results suggest that the presence of the RAG1 NTD is necessary but not sufficient to stabilize association of Ku70/Ku80 with a RAG–RSS complex formed with FLMR1/cMR2.
Given the ability of Ku to interact with other NHEJ factors discussed earlier, we considered the possibility that such factors might co-purify with the RAG proteins through Ku. To explore this possibility, we performed a series of immunoblotting experiments to detect whether NHEJ factors in addition to Ku were co-purified with FLMR1/cMR2 or the other RAG protein preparations. As expected, immunoblotting confirmed the presence of Ku70 and Ku80 in the FLMR1/cMR2 preparation (Supplementary Figure S2A). Interestingly, these experiments reveal that Ku70 and Ku80 is slightly more abundant in cMR1/FLMR2 preparations, yet RAG–RSS complexes assembled with this RAG preparation fail to be supershifted by anti-Ku antibodies (Figure 2A). However, other components of the end-processing and NHEJ repair machinery, including human Artemis, DNA-PKcs, XRCC4 or DNA Ligase IV, were not detectable in the RAG preparations by immunoblotting (data not shown). We also failed to detect hSRP1 (also known as nucleoprotein interactor-1 and karyopherin alpha 1), which was previously identified in a yeast two-hybrid screen as a RAG1 interacting protein and shown to associate with RAG1 expressed in HEK293 cells in the absence of RAG2 (28). We also attempted co-immunoprecipitation experiments to validate the association of Ku with endogenous RAG1 in lymphoid cells. However, immunoprecipitating native full-length RAG1 under non-denaturing conditions, from either murine thymocytes or the recombinase-inducible 103/bcl2 cell line (29), proved to be experimentally difficult due to its insolubility, consistent with earlier reports (30).
Because previous studies suggest that the RAG proteins associate with nuclear structures (30), and may bind nucleic acids non-specifically (31), we were concerned that Ku70 and Ku80 may associate with the RAG proteins through interactions with nucleic acids bound non-specifically with the full-length RAG proteins. If so, removal of non-specifically bound nucleic acids should reduce or eliminate Ku association with the full-length RAG proteins. To test this hypothesis, we pretreated clarified supernatants containing cMR1/FLMR2 or FLMR1/cMR2 with DNase I or RNase A and then incubated the samples with ethidium bromide before RAG purification to degrade nucleic acids and inhibit DNA-dependent protein–protein interactions (32). The purified RAG preparations were then analyzed for Ku70/Ku80 association using immunoblotting experiments and RSS binding assays. We find that the pre-treatment regimen slightly reduces the recovery of FLMR1/cMR2, but not cMR1/FLMR2, and significantly reduces the abundance of Ku70/Ku80 in preparations of cMR1/FLMR2, but not FLMR1/cMR2 (Supplementary Figure S1B and C). The RSS binding activity of cMR1/FLMR2 is slightly improved by DNaseI pre-treatment, but FLMR1/cMR2 binding activity is not reproducibly enhanced by either pre-treatment regimen. Notably, however, formation of the higher-order protein–DNA complex containing FLMR1/cMR2 and Ku remains detectable by EMSA (Supplementary Figure S1D). One possible caveat to these experiments is the possibility that DNAse I may have difficulty cleaving DNA if the RAG and Ku proteins were positioned next to one another. Therefore, as an alternative approach to examine whether Ku association with FLMR1/cMR2 is DNA dependent, we challenged RAG–RSS complexes assembled with cMR1/FLMR2 or FLMR1/cMR2 with increasing concentrations of competitor DNA (Supplementary Figure S1E). We find that the abundance of the HSC/Ku complex formed with FLMR1/cMR2 is not selectively diminished relative to the HSC complex in the same sample as a function of competitor DNA concentration. Taken together, these results suggest that Ku association with cMR1/FLMR2 is non-specific, but association with FLMR1/cMR2 is DNA-independent.
Functional activities of the RAG proteins are not significantly altered by the presence of Ku in vitro
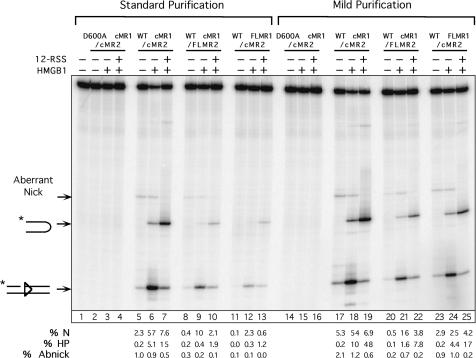
We next wished to determine if and how the mild purification conditions and/or association with Ku affected the DNA strand cleavage and strand transfer activities of the various RAG protein preparations. We first performed an in vitro cleavage assay to compare nicking and hairpin formation catalyzed by the different cMR1/cMR2, cMR1/FLMR2 and FLMR1/cMR2 preparations on an oligonucleotide 23-RSS substrate in the absence or presence of HMGB1 and cold 12-RSS partner DNA [conditions used to promote synapsis and 12/23-regulated cleavage; (33,34)]. We find that the RAG proteins purified using the mild procedure are slightly more active in an in vitro cleavage assay than their counterparts purified using the standard protocol, with the FLMR1/cMR2 preparation showing the greatest increase in cleavage activity (Figure 3). These data suggest that the enhanced activity observed with RAG proteins prepared under mild conditions is most likely attributed to improved RAG stability or RAG1/RAG2 association imparted by the buffers and/or procedures used for purification, rather than any potential association with Ku70/Ku80. This conclusion is further supported by results of in-gel cleavage assays which demonstrate that the RAG–RSS complexes assembled with cMR1/cMR2 and FLMR1/cMR2 purified using the mild protocol are intrinsically more active than their counterparts purified using the standard method (Supplementary Figure S3). Curiously, however, this outcome is not observed with cMR1/FLMR2. Notably, the cleavage activity of HSC and HSC/Ku complexes assembled with FLMR1/cMR2 purified using the mild protocol are not reproducibly different in the in-gel cleavage assay (Supplementary Figure S3), suggesting that Ku association with FLMR1/cMR2 does not enhance its specific activity on oligonucleotide substrates.
Figure 3.
RAG proteins purified using the mild procedure exhibit enhanced activity in RSS cleavage assays. Radiolabeled intact 23-RSS substrate was incubated for 1 h at 37°C with cMR1/cMR2 (WT or D600A RAG1), cMR1/FLMR2 or FLMR1/cMR2 purified using standard or mild conditions in cleavage reactions containing Mg2+ in the absence or presence of HMGB1 or cold 12-RSS partner as indicated above the gel. Reaction products were fractionated by denaturing gel electrophoresis and analyzed using a phosphorimager running the ImageQuant software. The positions of expected products are indicated at right. The asterisk denotes the location of 32P on the top strand. The percentage of correctly nicked (%N), aberrantly nicked (%Abnick) and hairpin (%HP) products in each lane is quantified below the gel.
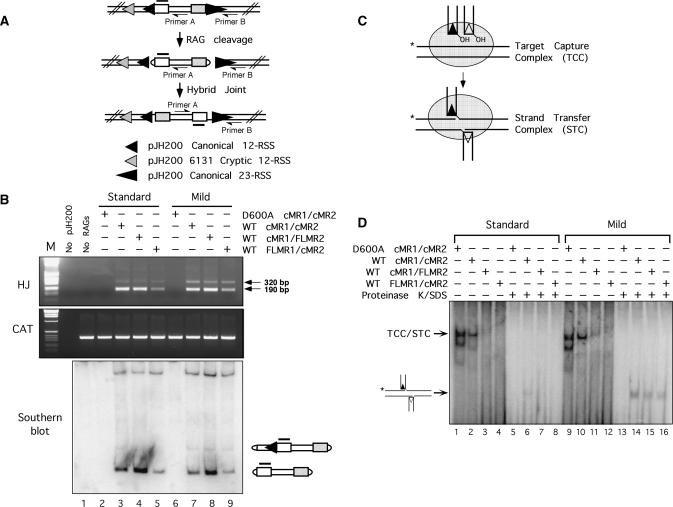
Next, we tested the activity of the various RAG protein preparations in two different alternative strand transfer reactions: hybrid joint formation (35) and transposition (36,37). RAG-mediated hybrid joint formation in vitro was assayed using the plasmid V(D)J recombination substrate pJH200. Hybrid joints were detected using a PCR-based approach (diagrammed in Figure 4A), which we have previously shown can reveal not only hybrid joints involving the canonical 12- and 23-RSSs, but also those involving the canonical 23-RSS and a cryptic 12-RSS in the pJH200 backbone (25). In general, we find that the RAG proteins purified under mild conditions support comparable levels of canonical hybrid joint formation, but slightly higher levels of ‘cryptic’ hybrid joint formation, to their counterparts purified under standard conditions in vitro (Figure 4B). This outcome can be explained by the observation that RAG proteins purified using either method exhibit a similar efficiency of canonical RSS cleavage, but the RAG proteins purified using the mild procedure display a selective but modest increase in cryptic RSS cleavage, as assessed by Southern hybridization (Figure 4B). To compare the ability of the different RAG protein preparations to support transposition in vitro, a pre-assembled signal end complex (SEC) containing the RAG proteins bound to a cleaved 12- and 23-RSS was incubated with a 32P-labeled DNA target in reaction buffer containing Ca2+, and protein–DNA complexes were fractionated on a non-denaturing polyacrylamide gel (diagrammed in Figure 4C). An SEC that has captured a DNA target is termed a target capture complex (TCC); if the RAG proteins transpose the signal ends into the target DNA, the complex is termed a strand transfer complex (STC). The TCC and STC co-migrate on a native polyacrylamide gel, but by treating the samples proteinase-K and SDS to remove the RAG proteins, the transposition product can be visualized on a native gel as a band that migrates faster than the TCC/STC. We find that RAG proteins purified using either procedure exhibit a comparable ability to form the TCC (Figure 4D, lanes 1–4 and 9–12). However, RAG proteins purified under mild conditions appear to more readily integrate the signal ends into target DNA, as higher levels of the transposition product are detected after treating the TCC/STC with proteinase-K and SDS (Figure 4D, lanes 5–8 and 13–16).
Figure 4.
RAG proteins purified using the mild protocol support slightly higher levels of noncanonical hybrid joint formation and transposition. (A) Diagram of hybrid joint assay using the pJH200 substrate. The position of PCR primers A and B (half arrowhead), and the Southern hybridization probe (shaded overline) are shown. The relative position and orientation of the canonical 12- and 23-RSS are indicated by filled small and large triangles; the ‘6131’ cryptic 12-RSS (54) is indicated by a shaded triangle. (B) Hybrid joint assay. The plasmid V(D)J recombination substrate pJH200 was linearized by AatII digestion and incubated with the various RAG protein preparations described in Figure 1 under conditions that permit coupled cleavage. PCR was performed on a portion of the reaction products using primers designed to detect hybrid joints (HJ, top panel) or chloramphenicol acetyltransferase (CAT, middle panel). PCR products were separated on an agarose gel with molecular sizing markers (M); the ∼190 and 320 bp amplicons reflect hybrid joints involving the 23-RSS and either the canonical or cryptic 12-RSS, respectively. The portion of the cleavage reactions not used for PCR were fractionated on a 5.5% non-denaturing polyacrylamide gel and analyzed by Southern hybridization using the probe shown in (A) (lower panel). The composition of the major cleavage products is shown at right. (C) Diagram of TCC/STC formation. (D) Donor and target DNA were incubated with the various RAG protein preparations described in Figure 3 in reaction buffer containing Ca2+ in the presence of HMGB1 as indicated above the gel. Untreated samples (lanes 1–4 and 9–12) and samples further incubated with proteinase K and SDS (lanes 5–8 and 13–16) were fractionated on a native 4% polyacrylamide gel. The positions of the mixed TCC/STC species and the transposition products released by proteinase K/SDS treatment are shown at left.
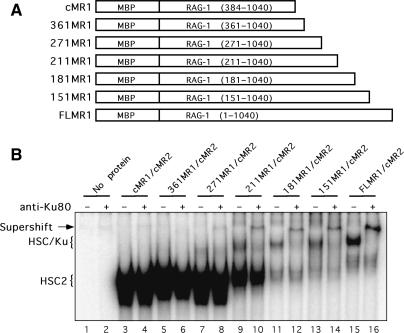
Ku70/Ku80 association with RAG1 is stabilized by inclusion of residues 211–383 to core RAG1, and requires an intact Zn-RING domain
To identify the region in the non-core portion of RAG1 required for Ku70/Ku80 association, we generated a series of RAG1 truncation mutants in which the non-core portion of RAG1 was added back to core RAG1 (residues 384–1040) in increments of 30 amino acids starting at residue 361. The designations and compositions of five representative RAG1 mutants analyzed in this study are shown in Figure 5A. Four RAG1 truncation mutants (301MR1, 241MR1, 91MR1 and 61MR1) were expressed poorly and not analyzed further, and one mutant, 331MR1, has not been tested since it exhibits an expression profile similar to 361MR1 (Supplementary Figure S4A). Variable expression of RAG1 NTD truncation mutants has also been reported by others (38). The selected RAG1 NTD truncation mutants were co-expressed with cMR2 (the amount of RAG1 NTD truncation mutant expression construct used for transfection varied based on its expression profile), and purified by amylose affinity chromatography; yields of recovered proteins were similar (Supplementary Figure S4B). We then analyzed the RAG preparations by immunoblotting (normalizing for RAG1) for the presence of Ku70 and Ku80 (Supplementary Figure S4B). We find that Ku70 and Ku80 is more abundant in preparations of 181MR1/cMR2 and 151MR1/cMR2 than the other three RAG1 NTD truncation mutant preparations, and is found at comparable levels to those observed in the FLMR1/cMR2 preparation. Correspondingly, when the RSS binding activity of the purified mutant RAG1 preparations in the presence of HMGB1 are compared by EMSA, we find that 361MR1/cMR2, 271MR1/cMR2 and 211MR1/cMR2 form mainly the HSC2 complex observed with cMR1/cMR2 (Figure 5B). The higher-order HSC/Ku complex is faintly seen in the sample containing 271MR1/cMR2 or 211MR1/cMR2, but is more abundant in samples containing the larger forms of RAG1 (181MR1, 151MR1 and FLMR1). Supershift analysis using an anti-Ku80 antibody verifies that the HSC/Ku complex observed in these samples indeed contains Ku (Figure 5B). We also notice that as the level of the HSC/Ku complex increases, the abundance of HSC2 observed in the same sample decreases.
Figure 5.
Residues 211–384 of RAG1 stabilize Ku70/Ku80 association with a RAG–RSS complex. (A) Diagram of RAG1 NTD truncation mutants used in these experiments, labeled as described in Figure 3. (B) EMSA of RAG1 NTD truncation mutant protein preparations. WT cMR1, FLMR1 or the RAG1 NTD mutants shown in (A) were co-expressed with, or mutant forms of 181MR1 were co-expressed with cMR2 in 293 cells and purified using the mild protocol. Radiolabeled intact 12-RSS substrate was incubated with HMGB1 and these various RAG preparations and protein–DNA complexes were subjected to supershift analysis by EMSA using a monoclonal anti-Ku80 antibody as indicated above the gel.
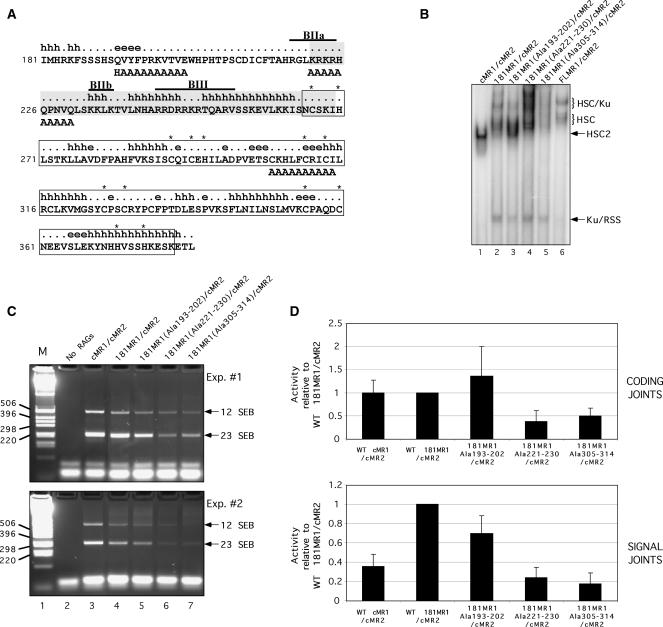
To further characterize the requirements for Ku association, we performed additional alanine scanning mutagenesis within the non-core portion of RAG1. We chose to introduce alanine substitutions in the 181MR1 protein because its yield and DNA binding properties resemble FLMR1. For these experiments, we targeted three different regions within the portion of RAG1 spanning residues 181–383 for disruption by substituting 10 consecutive residues with alanine (Figure 6A). In the RAG1 mutant 181MR1(Ala305-314), the Zn-RING finger motif that promotes RAG1 homodimerization (39) is disrupted by replacement of key cysteine residues involved in Zn2+ binding with alanine. The second RAG1 mutant, 181MR1(Ala 221–230), carries alanine substitutions at the beginning of a putative ‘hot loop’ that is considered intrinsically disordered (40). Hot loops, a subset of random coils, are predicted to possess a high degree of mobility, which may facilitate association with multiple interaction partners. The alanine substitutions also neutralize part of a small basic motif, called BIIa, that overlaps the predicted hot loop sequence and has been shown previously to augment V(D)J recombination (38). In the third RAG1 mutant, 181MR1(Ala193–202), a putative beta strand and its transition to random coil located between residues 181–226 is altered by alanine replacement. This mutant contains an additional Q192H mutation inadvertently introduced by PCR, but reversion of this mutation resulted in poor protein expression (data not shown).
Figure 6.
Identification of RAG1 NTD alanine replacement mutants that exhibit altered association with Ku70/Ku80. (A) Diagram of part of the RAG1 NTD. The amino acid sequence of murine RAG1 residues 181–383 are shown with secondary structure state predicted using the APSSP2 server (h, alpha helix; [.], coil; e, beta strand) (55). A structurally disordered region by the ‘hot-loops’ definition as determined using DisEMBL version 1.4 is shaded (40). The Zn-RING dimerization motif characterized by crystallography is boxed (39). Cysteine and histidine residues coordinating zinc ions are indicated by asterisks. The locations of small basic motifs are indicated by overlines and identified as BIIa, BIIb and BIII as described previously (38). Alanine substitutions in the three 181MR1 mutants are positioned beneath the residues targeted for replacement. (B) EMSA of 181MR1 mutant RAG protein preparations. WT cMR1, FLMR1 or 181MR1, or mutant forms of 181MR1 were co-expressed with cMR2 and purified from 293 cells using the mild procedure. Radiolabeled intact 12-RSS substrate was incubated with HMGB1 and the various RAG preparations and RAG–RSS complex formation was analyzed by EMSA. (C–D) V(D)J cleavage and recombination activity of 181MR1 alanine replacement mutants in cell culture. HEK293 cells were cotransfected with WT or mutant cMR1 or 181MR1 and cMR2 expression constructs together with the plasmid V(D)J recombination substrate pJH299 in the combinations indicated. (C) SEBs at the 12-RSS and 23-RSS were detected by LM-PCR and indicated by arrows at right. (D) Formation of coding joints (upper panel) and signal joints (lower panel) was analyzed using real-time PCR. Data was analyzed using the comparative threshold approach using amplification of a fragment of the chloramphenicol acetyltransferase gene as a calibrator and PCR reactions using template DNA recovered from panel E, lane 2 (‘No RAGs’) for normalization. The data is presented as the mean fold difference in the 2−ΔΔCt value between a given combination of RAG proteins and WT 181MR1/cMR2 (hence, the value obtained for WT 181MR1/cMR2 is always ‘1’). The error bars represent the standard deviation of the mean fold difference obtained from four independent experiments.
We find that all three 181MR1 mutants are recovered with yields similar to 181MR1 when they are co-expressed with cMR2 (Supplementary Figure S5A). Interestingly, when the RSS binding activities of the 181MR1 mutant protein preparations are compared by EMSA, we find that 181MR1(Ala193-202)/cMR2 exhibits a selective reduction in the ability to form the higher-order HSC/Ku complex, with substantially enhanced formation of HSC2 compared to 181MR1/cMR2 (Figure 6B). In contrast, we observe increased formation of several higher-order RAG–RSS complexes with 181MR1(Ala 221–230)/cMR2, which contains RAG1 mutations in the ‘hot loop’ that neutralize several basic residues (Figure 6B). Despite the distinct profiles of protein–DNA complexes observed by EMSA, these two mutant 181MR1/cMR2 preparations exhibit 23-RSS cleavage activity comparable to each other and to WT 181MR1/cMR2 in vitro (Supplementary Figure 5B). However, 181MR1(Ala305-314)/cMR2, in which the Zn-RING finger domain is disrupted, exhibits much poorer RAG–RSS complex formation compared to WT 181MR1/cMR2 (Figure 6B), and lower RSS cleavage activity as well (Supplementary Figure S5B). Notably, all 181MR1 mutants still retain some ability to associate with Ku70/Ku80, as binding of free Ku to the RSS is apparent in all binding reactions containing WT or mutant 181MR1 assayed by EMSA (Figure 6B). To probe the functional consequences of altering Ku association with RAG1, we examined the ability of the 181MR1 mutants to support cleavage and rearrangement of the inversional plasmid V(D)J recombination substrate pJH299 in cell culture. In plasmid DNA isolated from 293 cells expressing cMR2 and either WT cMR1, 181MR1 or 181MR1(Ala193–202), we find that the abundance of signal end breaks (SEBs) at the 12-RSS and 23-RSS detected by LM-PCR is quite similar, but these levels are reproducibly lower in cells expressing 181MR1 (Ala 221–230) or 181MR1(Ala305–314) (Figure 6C). Interestingly, analysis of signal and coding joint formation by real-time PCR indicates that coding joint formation in cells expressing cMR1, 181MR1 or 181MR1(Ala193–202) is similar, but cMR1 and 181MR1(Ala193–202) support less signal joint formation than WT 181MR1, with the former exhibiting a greater decrease than the latter (Figure 6D). Both 181MR1(Ala221–230) and 181MR1(Ala305–314) mediate less cleavage and less rearrangement than WT cMR1 or 181MR1. The latter results are consistent with previously published reports on the recombination activity of RAG1 BIIa and Zn-RING domain mutants (38,41–43).
DISCUSSION
Here, we present biochemical evidence that Ku70/Ku80 associates with full-length RAG1 when FLMR1/cMR2 is prepared using a mild purification procedure. This interaction is DNA-independent, and Ku70/Ku80 remains associated with FLMR1/cMR2 when the RAG proteins are incubated with a radiolabeled RSS substrate to form a stable Ku–RAG–RSS complex that can be visualized by EMSA and supershifted with anti-Ku antibodies. The formation of this complex is observed by EMSA using FLMR1/cMR2 purified from both 293 cells and HeLa cells, and is facilitated by the presence of residues 211–384 of RAG1, but is impaired by RAG1 mutations at residues 193–202.
The association between Ku and full-length RAG1 is labile, as binding of free Ku to the RSS is observed in binding reactions containing FLMR1/cMR2 prepared using either standard or mild purification procedures. This observation explains why Ku association with RAG1 is not stoichiometric and why protein–DNA complexes assembled with FLMR1/cMR2 are observed that either lack or contain Ku. One might argue that these results favor an interpretation that Ku co-purifies with full-length RAG1 as a non-specific aggregate. However, if this were true, addition of purified Ku to binding reactions containing FLMR1/cMR2 would be expected to promote formation of higher-order protein–DNA aggregates that could be visualized by EMSA, which is not observed. On the contrary, addition of Ku to FLMR1/cMR2 (prepared using either method of purification) fails to promote formation of FLMR1/cMR2 complexes bound to an RSS that contain Ku (Figure 2B). One possible explanation for this outcome is that Ku association with RAG1 is not direct, perhaps requiring a bridging molecule that is limiting in the binding reaction. Alternatively, or in addition, Ku dissociation from the FLMR1/cMR2 protein complex may be accompanied by RAG1 conformational changes and/or protein unfolding that precludes subsequent stable interaction with Ku. In support of this latter possibility, protein secondary structure prediction algorithms, such as DisEMBL, suggest that the RAG1 NTD contains regions that are intrinsically disordered and could lose structural integrity if an interacting protein is disassociated (Figure 6A). Further evidence against the interpretation that Ku association with full-length RAG1 is non-specific is the observation that mutations in RAG1 that disrupt Ku association also impair signal joint formation (discussed further below).
Implications for the processing and repair of V(D)J recombination intermediates
Genetic evidence suggests that the RAG proteins play an active role in guiding the DNA ends generated by RAG-mediated cleavage to the NHEJ pathway for repair (13–16). Biochemical studies have also provided experimental evidence that the RAG proteins mediate coupled cleavage in vitro with greater fidelity to the 12/23 rule when Ku70/Ku80 and DNA-PKcs are present in the cleavage reaction, but these studies did not formally demonstrate that the RAG proteins and NHEJ factors directly interact with one another (17). The data presented here provide a mechanism to explain these previous observations; namely, that there is a physical association between RAG1 and the Ku70/Ku80 complex. We recognize that earlier reports suggest that the core RAG proteins may be sufficient to interact with components of the NHEJ machinery (15,17). However, we argue here that such interactions are too weak to be maintained through the procedures normally used to purify the RAG proteins, but that addition of the NTD to core RAG1 stabilizes its association with Ku70/Ku80. That Ku is implicated here as an interaction partner of the RAG proteins seems plausible for two reasons. From a practical standpoint, the Ku70/Ku80 complex is known to associate directly or indirectly through its interaction partners with all known components of the NHEJ pathway required for V(D)J recombination (5,7,9,10,44,45). By positioning Ku proximal to the RSS before it is cleaved, the RAG proteins can ensure that Ku is poised to capture the nascent DNA double-strand break introduced by the RAG complex and direct to the NHEJ pathway for repair. Second, there is precedence for Ku associating with recombinases active in vertebrate organisms, as Ku has been shown to interact with the Sleeping Beauty transposase (22).
Functional analysis of 181MR1 alanine replacement mutants provides some new insights into how the RAG1 NTD contributes to the generation and repair of DNA breaks. For example, we show here that disruption of the Zn-RING finger (Ala305–314) motif impairs RAG binding and cleavage of RSS substrates in vitro, and cleavage and recombination of plasmid substrates in cell culture. This result suggests that even though the RAG1 NTD is not necessary for the basic enzymatic activity of the RAG complex, NTD mutations can perturb the DNA binding and cleavage activity supported by the ‘core’ portion of RAG1. This may partly explain why RAG1 NTD mutations in the Zn-RING domain cause severe immunodeficiency (46), rather than a milder recombination defect as is observed in core RAG1 ‘knock-in’ mice (47). In contrast, disruption of a putative ‘hot loop’ in RAG1 that includes the BIIa motif (Ala 221–230) impairs RAG-mediated cleavage and recombination of plasmid substrates in cell culture, consistent with results reported by others (38), but does not adversely affect the ability of the RAG proteins to bind or cleave RSS substrates in vitro. We speculate that the differential cleavage activity of this RAG1 mutant in vitro and in cell culture may be attributed to its propensity toward self-aggregation and/or association with other proteins reflected by the formation of multiple higher-order protein–DNA complexes by EMSA. Such protein complexes may remain competent to cleave simple oligonucleotide substrates but may be unable to assemble functional complexes on longer plasmid DNA substrates, a possibility that will be explored in future studies. Finally, RAG1 NTD mutations that selectively impair the formation of higher-order RAG–RSS complexes containing Ku (Ala193–202), do not impair the intrinsic DNA binding or cleavage activity of the RAG complex in vitro or in cell culture, but diminish the efficiency of signal joint formation with little affect on coding joint formation. This apparent bias may reflect differences in how signal and coding ends are processed and joined. Available evidence suggests that coding ends are readily released from a post-cleavage RAG complex, where they are rapidly subjected to hairpin opening, end processing, and joining, whereas signal ends remain bound by the RAG proteins after cleavage, protecting the ends from degradation until the protein–DNA complex is disassembled (21,48–50). We speculate that full-length RAG1, through its association with Ku, promotes signal end joining that is coordinated with the disassembly of the RAG complex and release of the signal ends. Such coordination may be important for limiting potential insertion of signal ends elsewhere in the genome. This scenario also provides a possible explanation for why core RAG1 knock-in mice display an increased frequency of deletions in Dβ–Jβ signal joints (which are normally precise) (51). Under normal circumstances, the facilitated transfer of signal ends from the RAG proteins to Ku through full-length RAG1 may limit the time the signal ends are accessible for modification. In core-RAG1 knock-in mice, by contrast, the released DNA ends may have to await capture by the NHEJ apparatus (or an alternative repair pathway) by a diffusion-mediated process, rendering them more available and susceptible to extensive modification before they are eventually joined. We recognize that Ku deficiency also impairs coding joint formation (52,53). However, since the release of signal and coding ends from the post-cleavage RAG complex is asynchronous, it seems plausible that Ku recruitment to coding ends could occur through a mechanism that is independent of Ku association with full-length RAG1. We are working to identify RAG1 mutants with a more severe deficit in Ku association than we observe with the 181MR1(Ala193–202)/cMR2 preparation described here in order to further test the functional role of Ku association with RAG1 in V(D)J recombination.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
National Institutes of Health (AI055599 to P.C.S.); laboratory renovation supported by the Research Facilities Improvement Program of the National Institutes of Health National Center for Research Resources (C06 RR17417-01). Funding to pay the Open Access publication charges for the article was provided by the National Institutes of Health and the LB692 Nebraska Tobacco Settlement Biomedical Research Program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 2.Schatz DG, Spanopoulou E. Biochemistry of V(D)J recombination. Curr. Top. Microbiol. Immunol. 2005;290:49–85. doi: 10.1007/3-540-26363-2_4. [DOI] [PubMed] [Google Scholar]
- 3.van Gent DC, McBlane JF, Ramsden DA, Sadofsky MJ, Hesse JE, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 4.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 5.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 6.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 9.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP. Cernunnos interacts with the XRCC4 x DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J. Biol. Chem. 2006;281:13857–13860. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
- 11.Schultz HY, Landree MA, Qiu J, Kale SB, Roth DB. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol. Cell. 2001;7:65–75. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 12.Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinstock DM, Jasin M. Alternative pathways for the repair of RAG-induced DNA breaks. Mol. Cell Biol. 2006;26:131–139. doi: 10.1128/MCB.26.1.131-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 15.Cui X, Meek K. Linking double-stranded DNA breaks to the recombination activating gene complex directs repair to the nonhomologous end-joining pathway. Proc. Natl Acad. Sci. USA. 2007;104:17046–17051. doi: 10.1073/pnas.0610928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 17.Sawchuk DJ, Mansilla-Soto J, Alarcon C, Singha NC, Langen H, Bianchi ME, Lees-Miller SP, Nussenzweig MC, Cortes P. Ku70/Ku80 and DNA-dependent protein kinase catalytic subunit modulate RAG-mediated cleavage: implications for the enforcement of the 12/23 rule. J. Biol. Chem. 2004;279:29821–29831. doi: 10.1074/jbc.M403706200. [DOI] [PubMed] [Google Scholar]
- 18.Leu TM, Eastman QM, Schatz DG. Coding joint formation in a cell-free V(D)J recombination system. Immunity. 1997;7:303–314. doi: 10.1016/s1074-7613(00)80532-4. [DOI] [PubMed] [Google Scholar]
- 19.Ramsden DA, Paull TT, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 20.Cortes P, Weis-Garcia F, Misulovin Z, Nussenzweig A, Lai JS, Li G, Nussenzweig MC, Baltimore D. In vitro V(D)J recombination: signal joint formation. Proc. Natl Acad. Sci. USA. 1996;93:14008–14013. doi: 10.1073/pnas.93.24.14008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 22.Izsvak Z, Stuwe EE, Fiedler D, Katzer A, Jeggo PA, Ivics Z. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell. 2004;13:279–290. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- 23.Bergeron S, Anderson DK, Swanson PC. RAG and HMGB1 proteins: purification and biochemical analysis of recombination signal complexes. Methods Enzymol. 2006;408:511–528. doi: 10.1016/S0076-6879(06)08032-3. [DOI] [PubMed] [Google Scholar]
- 24.Bergeron S, Madathiparambil T, Swanson PC. Both high mobility group (HMG)-boxes and the acidic tail of HMGB1 regulate recombination-activating gene (RAG)-mediated recombination signal synapsis and cleavage in vitro. J. Biol. Chem. 2005;280:31314–31324. doi: 10.1074/jbc.M503063200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanson PC, Volkmer D, Wang L. Full-length RAG-2, and not full-length RAG-1, specifically suppresses RAG-mediated transposition but not hybrid joint formation or disintegration. J. Biol. Chem. 2004;279:4034–4044. doi: 10.1074/jbc.M311100200. [DOI] [PubMed] [Google Scholar]
- 26.Kriatchko AN, Anderson DK, Swanson PC. Identification and characterization of a gain-of-function RAG-1 mutant. Mol. Cell. Biol. 2006;26:4712–4728. doi: 10.1128/MCB.02487-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanson PC. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 2002;22:7790–7801. doi: 10.1128/MCB.22.22.7790-7801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortes P, Ye ZS, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc. Natl Acad. Sci. USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YY, Wang LC, Huang MS, Rosenberg N. An active v-abl protein tyrosine kinase blocks immunoglobulin light-chain gene rearrangement. Genes Dev. 1994;8:688–697. doi: 10.1101/gad.8.6.688. [DOI] [PubMed] [Google Scholar]
- 30.Leu TM, Schatz DG. rag-1 and rag-2 are components of a high-molecular-weight complex, and association of rag-2 with this complex is rag-1 dependent. Mol. Cell Biol. 1995;15:5657–5670. doi: 10.1128/mcb.15.10.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spanopoulou E, Cortes P, Shih C, Huang CM, Silver DP, Svec P, Baltimore D. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 1995;3:715–726. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 32.Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl Acad. Sci. USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 35.Melek M, Gellert M, van Gent DC. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 1998;280:301–303. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- 36.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 37.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 38.McMahan CJ, Difilippantonio MJ, Rao N, Spanopoulou E, Schatz DG. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol. Cell Biol. 1997;17:4544–4552. doi: 10.1128/mcb.17.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA. Crystal structure of the RAG1 dimerization domain reveals multiple zinc- binding motifs including a novel zinc binuclear cluster. Nat. Struct. Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- 40.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silver DP, Spanopoulou E, Mulligan RC, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl Acad. Sci. USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirch SA, Sudarsanam P, Oettinger MA. Regions of RAG1 protein critical for V(D)J recombination. Eur. J. Immunol. 1996;26:886–891. doi: 10.1002/eji.1830260425. [DOI] [PubMed] [Google Scholar]
- 44.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl Acad. Sci. USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 46.Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, Arkwright PD, Baniyash M, Brooks EG, Conley ME, et al. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 47.Dudley DD, Sekiguchi J, Zhu C, Sadofsky MJ, Whitlow S, DeVido J, Monroe RJ, Bassing CH, Alt FW. Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J. Exp. Med. 2003;198:1439–1450. doi: 10.1084/jem.20030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roth DB, Nakajima PB, Menetski JP, Bosma MJ, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 49.Ramsden DA, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 50.Jones JM, Gellert M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl Acad. Sci. USA. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talukder SR, Dudley DD, Alt FW, Takahama Y, Akamatsu Y. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic Acids Res. 2004;32:4539–4549. doi: 10.1093/nar/gkh778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng HL, Sekiguchi JM, Frank K, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 53.Zhu C, Bogue MA, Lim DS, Hasty P, Roth DB. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 54.Lewis SM, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol. Cell Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raghava GPS. 2002. APSSP2: A combination method for protein secondary structure prediction based on neural network and example based learning. CASP5. A-132. [Google Scholar]