Abstract
The emergence of pathogenic strains of enteric bacteria and their adaptation to unique niches are associated with the acquisition of foreign DNA segments termed ‘genetic islands’. We explored these islands for the occurrence of small RNA (sRNA) encoding genes. Previous systematic screens for enteric bacteria sRNAs were mainly carried out using the laboratory strain Escherichia coli K12, leading to the discovery of ∼80 new sRNA genes. These searches were based on conservation within closely related members of enteric bacteria and thus, sRNAs, unique to pathogenic strains were excluded. Here we describe the identification and characterization of 19 novel unique sRNA genes encoded within the ‘genetic islands’ of the virulent strain Salmonella typhimurium. We show that the expression of many of the island-encoded genes is associated with stress conditions and stationary phase. Several of these sRNA genes are induced when Salmonella resides within macrophages. One sRNA, IsrJ, was further examined and found to affect the translocation efficiency of virulence-associated effector proteins into nonphagocytic cells. In addition, we report that unlike the majority of the E. coli sRNAs that are trans regulators, many of the island-encoded sRNAs affect the expression of cis-encoded genes. Our study suggests that the island encoded sRNA genes play an important role within the network that regulates bacterial adaptation to environmental changes and stress conditions and thus controls virulence.
INTRODUCTION
Recent systematic searches for bacterial small RNA (sRNA) genes have led to the discovery of almost 140 bacterial sRNAs, 80 of which were identified in Escherichia coli. The characterization of a subset of these sRNAs indicated that they play important regulatory roles in bacterial physiology affecting iron homeostasis, sugar metabolism, responses to oxidative stress and conditions induced by DNA damage, stationary phase and cell-surface composition (1–6). Many of these sRNAs act as antisense RNAs, by baseparing to their target mRNAs, a step mediated by the Sm-like RNA-binding protein, Hfq. Hfq acts as a chaperone that modulates the translation of many mRNAs and the stability of sRNA–mRNA hybrids in E. coli (7). The other well-characterized class of regulatory sRNAs acts by binding to proteins to modulate their activities (8,9).
Screens for sRNA-encoding genes were conducted in a few other bacteria, some of which were pathogenic. Two different screens have resulted in the discovery of 15 sRNA genes in Listeria monocytogenes (10,11). A dozen sRNA genes were identified in Staphylococcus aureus, including seven in accessory genetic elements (12). Seventeen new transcripts were detected in the opportunistic pathogen Pseudomonas aeruginosa (13). Whether these sRNA genes are involved in virulence is as yet to be discovered.
Many of the searches that were conducted in E. coli were based on conservation with closely related members of the Enterobacteriaceae. Therefore, most of the newly discovered sRNA genes reside within the relatively conserved genetic backbone, which is shared by the pathogenic and the nonpathogenic strains. The adaptation of enteric bacteria to their environment is associated with the acquisition of genes encoding virulence factors, many of which were acquired horizontally via once-mobile genetic elements. The particular repertoire of acquired genes dictates the pathogenic strategy employed. For many enteric bacteria the acquisition of a single DNA segment can enhance the microorganism's virulence. For example, the transfer of Shigella's ‘virulence plasmid’ encoding determinants for invasion and intracellular spreading into a laboratory strain of E. coli, rendered the latter invasive (14). Similarly, the phenotype of attaching and effacing can be reproduced by a laboratory strain, through the introduction of a plasmid carrying the locus of enterocyte effacement (LEE) of enteropathogenic E. coli (15). In fact, recent genome comparisons showed that the chromosomes of enteric bacteria are mosaics, composed of conserved collinear regions interspersed with ‘loops’ or ‘islands’ unique to certain species. The conserved genes may reflect the basic lifestyle of the bacteria, while the unique gene clusters probably contribute to adaptation to environmental niches and to pathogenicity.
Salmonella species are ubiquitous human and animal pathogens that cause a variety of food-borne infections ranging in severity from self-limiting gastroenteritis to life-threatening infections such as typhoid fever (16). During the course of infection, after ingestion in contaminated food, Salmonella passes through the stomach where conditions are very acidic. After passage from the stomach into the distal ileum, the bacteria associates with the epithelial lining, where it adheres to and enters the apical membrane of M cells in the Peyer's patches. Salmonella strains that pass through the basolateral membrane are engulfed by macrophages. The adaptation of Salmonella to the acidic environment of the stomach, and to the intracellular environment of macrophages greatly contributes to its virulence.
Salmonella as well, owes its virulence largely to its pathogenicity islands. Sequence analysis of Salmonella typhi and Salmonella typhimurium genomes revealed the presence of many insertions compared with the E. coli genome, ranging in size from single genes to large islands. Previous genetic studies and current sequence comparisons have demonstrated that Salmonella pathogenicity islands contain many clusters of virulence genes encoding proteins of type III secretion systems (TTSS), a number of fimbriae as well as regulatory proteins of complex networks (15,17–19).
We applied a modified version of our predictive algorithm (20) to the sequences of S. typhimurium genetic islands, in search of unique sRNA-encoding genes. This search has led to the identification of 19 novel island-encoded sRNAs. Several of these genes are expressed when Salmonella resides within macrophages. Characterization of IsrJ sRNA that is expressed under conditions of low oxygen and low magnesium shows that it plays a role in pathogenicity via its effect on the translocation of effector proteins and hence on invasion of Salmonella into nonphagocytic cells. In addition, we characterized the regulation of expression of IsrE, an island-encoded homolog of the iron responsive sRNA, RyhB (21). We found the regulation of these two sRNAs to be considerably different, and their function to be nonredundant, suggesting that each may have individual targets.
MATERIALS AND METHODS
The computational screen and part of the experimental procedures including, construction of plasmids and bacterial strains, northern blots and primer extension assays and the sequences of the oligonucleotides are detailed in Supplementary Data.
RNA isolation
Overnight cultures of S. typhimurium SL1344 or SL1344 carrying plasmids were diluted 1/100 in LB medium and grown at 37°C. Samples were taken at 1.5, 2.5 and 8 h after dilution at OD600 values of 0.3, 1 and 4.5, respectively. For oxygen limitation conditions, single colonies were inoculated in 10 ml LB and grown overnight without agitation in 50 ml Falcon tubes to OD600 of 0.9. For anaerobic conditions, overnight cultures (9.5 ml) grown without agitation in 10 ml tubes were diluted 1/50 in 50 ml LB-Amp and grown without agitation in 50 ml Falcon tubes for 4 h to OD600 of 0.3 and 26 h to OD600 of 0.6. All other treatments were conducted at OD600 of 0.3 for 30 min unless indicated otherwise. The treatments included 0.5 M NaCl (osmotic shock), 0.2 mM paraquat or 1 mM hydrogen peroxide (oxidative stress) and 0.2 mM 2,2′-dipyridyl (low iron conditions). For pH stress, cultures were collected by centrifugation, resuspended in LB at pH 4.9 (adjusted with HCl) or LBK medium at pH 8.4 (22) and then grown for additional 30 min. For cold shock treatment, cultures were transferred from 37°C to 15°C. For heat shock treatment, cells grown at 30°C to OD600 of 0.3 were transferred to 42°C for 15 min. To examine the effect of Mg2+, SL1344 cells grown overnight in N minimal medium pH 7.7 supplemented with 0.1% Casamino Acids, 38 mM glycerol and 10 mM MgCl2, were collected by centrifugation, washed three times with N-minimal medium without Mg2+ and then diluted 1/50 into N-minimal medium containing 10 mM or 10 μM MgCl2. Samples were taken at OD600 of 0.3 at 3.5 h and 4.3 h after the cells were diluted in media containing 10 mM MgCl2 and 10 μM MgCl2, respectively. To isolate total RNA, the cultures were pelleted and resuspended in 50 μl 10 mM Tris–HCl (pH 7.5) containing 1 mM EDTA. Lysozyme was added to 0.9 mg/ml and the samples were subjected to three freeze-thaw cycles. Total RNA was purified using Ultraspec RNA according to the manufacturer's protocol (BIOTECX Laboratories, Houston, TX), except that 1 ml of reagent was used for 12–16 OD600 units of the cells. RNA samples used in real-time PCR assays were treated with DNaseI (RQ1 RNase free DNase, Promega) to eliminate DNA contaminations.
Analysis of RNA expression within macrophages by real time PCR
J774 macrophages (0.5 × 106 cells/ml) were seeded in 14-cm-diameter plates containing F-12 medium supplemented with 10% fetal calf serum (FCS). The next day the cells were activated with phorbol 12-myristate 13-acetate (PMA, sigma) (6.5 μg/ml) and infected [multiplicity of infection (MOI), 500:1] with S. typhimurium cultures grown without agitation for 20 h to OD600 of 0.9–1.0 (23,24). At 30 min after infection, the macrophages were washed and incubated for an additional 1 h in medium containing gentamicin at 50 μg/ml or 8 h in medium containing gentamicin at 50 μg/ml for 1 hr and 10 μg/ml gentamicin for an additional 7 h. Thereafter, the cells were washed with phosphate-buffered saline and lysed in 1% Triton X-100 for 10 min. Control RNA was obtained by growing SL1344 for 1.5 h to log phase (OD600 of 0.1) in the cell culture medium. RNA was extracted as described earlier. RNA concentrations were determined using a NanoDrop machine (NanoDrop Technologies). DNA was removed by DNase treatment according to the manufacturer's instructions (RQ1 RNase free DNase, Promega). About 2.5 μg DNA-free total RNA was used for cDNA synthesis using MMLV reverse transcriptase and random primers (Promega), according to the manufacturer's protocol. Quantification of cDNA was performed by real-time PCR using SYBR-green mix (Absolute SYBR GREEN ROX MIX, ABgene) with Rotor gene 3000 A (Corbett) according to manufacturer's instructions. Specific primer pairs were designed according to the Guidelines for Amplicon and Primer Design (http://www.tamar.co.il/site/index.php?ln=en&main_id=1). The level of 16S rRNA (rrsA) was used to normalize the expression data for each target gene. The relative amount of cDNA was calculated using the standard curve method. A standard curve was obtained from PCR on serially diluted genomic DNA as templates and was analyzed using Rotor-gene analysis software 6.0.
Invasion assays
HeLa cells (3–5 × 105 cells/ml) seeded in 24-well microtiter plates containing DMEM medium supplemented with 10% FCS and infected [multiplicity of infection (MOI), 20:1] with S. typhimurium cultures grown without agitation to OD600 of 0.9–1.0. At 30 min after infection, the cells were washed and incubated for an additional 1 h in medium containing gentamicin at 50 μg/ml. Thereafter, the cells were washed with phosphate-buffered saline and lysed in 1% Triton X-100 for 10 min, and aliquots were plated to determine the number of viable intracellular bacteria.
Translocation assay
The assay was carried out as described in (25). Basically, HeLa cells were seeded in 96-well plates at a density of ∼2 × 104 cells/well in DMEM (Sigma), supplemented with 10% FCS and antibiotics (Pen/Strep). Bacterial strains (SL1344, hilA::kan and ΔisrJ::frt) carrying the plasmid pKB3138 were inoculated in LB broth supplemented with tetracycline (5 μg/ml) and grown non-agitated at 37°C, for 21 h. The next day, the HeLa cultures were washed twice with CDMEM, and treated with 120 μl of CDMEM containing 20 μl of 6X CCF2/AM loading solution (CCF2/AM loading kit, Invitrogen; 1 μM CCF2/AM and 2.5 mM probenecid final concentration). The cells were incubated for 60 min, at room temperature, in the dark and then for another 15 min at 37°C. Meanwhile, the bacterial cultures were OD adjusted, centrifuged and their pellet was resuspended in CDMEM to generate a final dilution of 1:4. HeLa cells were infected with 200 μl of the diluted cultures and the plates were immediately placed in a plate reader (SPECTRAFluor Plus, TECAN) that was set at 37°C. Thus, the infection process took place in the plate reader. The cultures were excited at 405 nm and emission at 465 nm and 535 nm was recorded at 150 s intervals. Bacterial growth, OD at 600 nm, was also recorded. Data were collected with Magelan5 Software and processed using the MATLAB computing platform.
The [P] values were calculated as follows:
[P] = [Praw − Pbkg]/[S0 − Sbkg]
Where
Praw = measured product fluorescence,
Pbkg = background fluorescence at 460 nm,
S0 = measured substrate fluorescence at t = 0,
Sbkg = background fluorescence at 530 nm.
S0 normalizes variations (well to well) in number of HeLa cells and/or efficiency of CCF2 loading. Assays were carried out in duplicates or triplicates.
RESULTS
Computational screens for sRNAs encoded within the genetic islands of S. typhimurium
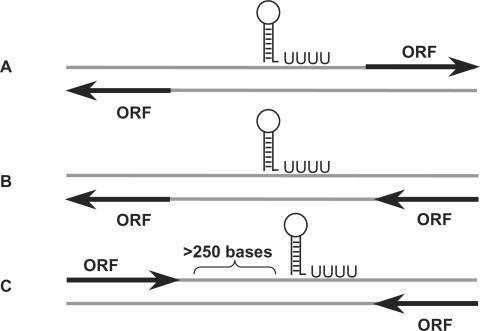
A total of 392 island sequences were determined in S. typhimurium genome (see Materials and Methods section). Subsequent analysis using the annotations of S. typhimurium open reading frames (ORFs) (18) showed that the islands contain 862 ‘empty’ intergenic regions that are longer than 50 base pairs. These sequences were extracted and searched for putative sRNA genes. Since in our previous screen in E. coli (20) we realized that the rho-independent transcription terminator predictions of the sRNA genes were very accurate, we based the current predictions on identification of such terminators in the intergenic regions. We searched the islands both for sRNA genes that are conserved in other bacteria as well as for unique sRNA genes that show little or no conservation in other bacteria. In the first screen we extracted island sequences that were conserved in any of the 102 bacterial genomes and sequences of mobility elements that were available at the time of the analysis, for which rho-independent terminators were predicted. In the second screen, we relied solely on the presence of rho-independent terminators. Three types of rho-independent transcription terminators termed ‘orphan’ terminators were considered (Figure 1). These terminators are less likely to regulate the termination of previously known genes. The first type involved terminators located between pairs of genes that are divergent relative to each other (Figure 1). Such terminators that are located upstream to the 5′ end of one of the two flanking genes could belong to independent sRNAs or sRNAs that act as leaders or riboswitches. The second type regards terminators located in intergenic regions and on the opposite strand of a pair of genes oriented at the same direction. As for the third type, based on the observation that transcription terminators of ORF are usually found within a distance of 250 bases downstream of their genes (26) we searched for terminators located far-downstream, outside the range of 250 bases (Figure 1). In total, 28 putative genes were predicted (Supplementary Data, Table S1), nine by the first screen and 19 by the second. We denoted these candidate genes pisrA1-pisrA28 (predicted island-encoded sRNA).
Figure 1.
Schematic representation of three types of rho-independent transcription terminators of putative sRNAs positioned relative to their flanking genes. These three types of transcription terminators are less likely to function as terminators of previously annotated genes. (A) A transcription terminator located in an intergenic region between two divergently oriented genes. (B) A transcription terminator located in an intergenic region on the opposite strand from its two flanking ORFs. (C) A transcription terminator located in an intergenic region, more than 250 bases downstream from the 3′ end of the nearest ORF.
Experimental identification of sRNAs
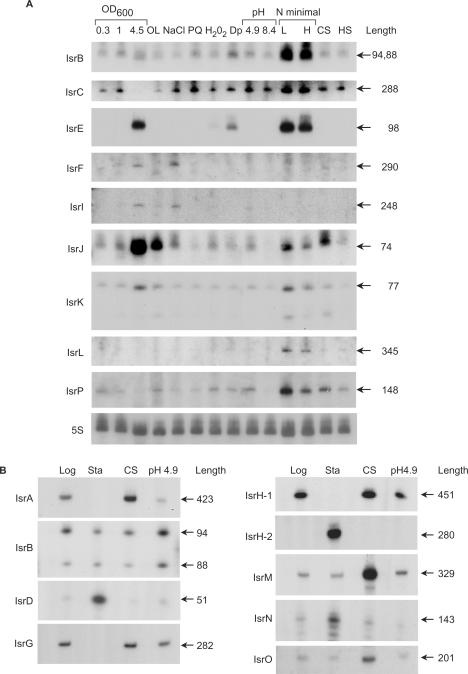
The identification of the corresponding RNA species was carried out in S. typhimurium. We examined expression in cells grown to exponential or stationary phase and in cells subjected to stress conditions of which a few are reminiscent of the environments Salmonella encounters upon invasion and/or within macrophages. These included low oxygen, starvation, extreme acid conditions, oxidative stress, iron limitation, magnesium limitation and osmotic shock. Of the 28 candidate sRNA-genes tested, 19 novel sRNAs were detected (68%), 5 of which were predicted by the first screen, whereas the rest 14 genes were predicted based on transcription terminators. Subsequent sequence conservation analysis with many new relevant fully sequenced genomes demonstrated that of the 19 sRNAs confirmed, all but three were conserved (Table 1). The genes expressing the sRNAs were denoted isr (A, B, etc.) for island-encoded sRNA (Table 1). Figure 2 shows the characterization of these sRNAs with respect to size, abundance and expression pattern. Some of the RNAs were less abundant and thus, we cloned the intergenic regions of these genes on multi copy plasmids for their mapping (Figure 2B). Nevertheless, the transcripts of the majority of these sRNAs were detected from the chromosome by real time PCR (see below). The expression of many of the island-encoded genes was associated with both stationary phase and stress conditions. IsrC RNA for example, was detected under most stress conditions, including oxidative stress, osmotic shock, lack of iron, acid conditions and heat shock as well as, in logarithmic and early stationary phase. Interestingly, the transcript levels of this RNA were further elevated when the cells were grown in minimal media. Unlike IsrC, IsrL RNA was detected almost only upon growth in minimal media. The expression pattern of IsrH-1 was reminiscent of IsrC; it was highly abundant in logarithmic phase and under various stress conditions (Figure 2 and data not shown). RNA mapping of isrH-1 revealed that it encompassed a second sRNA gene denoted isrH-2. A promoter located within the 450-nucleotide long isrH-1 gene directed the synthesis of IsrH-2, a 280 nt long sRNA expressed in stationary phase. A few other sRNAs including IsrF, IsrI, IsrD and IsrN were detected in stationary phase. It is interesting that both IsrF and IsrI were detected also in exponentially growing cells exposed to high osmolarity, suggesting that these genes might be regulated by the stationary phase sigma subunit of RNA polymerase, σs encoded by rpoS (Figure 2A).
Table 1.
Summary of the newly identified small RNAs
| sRNA gene | Screen typea | Minutes | Adjacent genes | Strandb | 5′ endc | 3′ endd | Lengthe | Conservationf | Comments |
|---|---|---|---|---|---|---|---|---|---|
| isrA (isrP1) | Conservation | 7.0 | STM0294.1n/STM0295 | → → → | 339338 | 339760 | 423 | A | |
| isrB-1 (isrP6) | Terminator (B) | 22.7 | sbcA/STM1010 | ← → ← | 1104173, 79 | 1104266 | 88, 94 | B | Identical to isrB-2 |
| isrC (isrP7) | Conservation | 27.4 | envF/msgA | ← → ← | 1329145 | 1329432 | 288 | A, B, C, D | |
| isrD (isrP9) | Terminator (B) | 27.7 | STM1261/STM1263 | → ← → | 1345788 | 1345738 | 51 | A, B, C, D | |
| isrE(isrP10) | Terminator (B) | 27.8 | STM1273/STM1274 | → ← → | 1352972 | 1352875 | 98 | A, B, C, D | RyhB ortholog |
| isrF(isrP11) | Terminator (C) | 33.6 | STM1552/STM1554 | → ← ← | 1630160 | 1629871 | 290 | ||
| isrG (isrP14) | Terminator (A) | 48.3 | STM2243/STM2244 | ← → → | 2344732 | 2345013 | 282 | C, D | |
| isrH-1 (isrP15) | Terminator (B) | 49.3 | glpC/STM2287 | → ← → | 2394753 | 2394303 | 451 | A, B, C, D | |
| isrH-2 (isrP15) | Terminator (B) | 49.3 | glpC/STM2287 | → ← → | 2394582 | 2394303 | 280 | A, B, C, D | |
| isrI(isrP16) | Terminator (C) | 56.8 | STM2614/STM2616 | → ← ← | 2761576 | 2761329 | 248 | 62 aa ORFg | |
| isrJ(isrP17) | Conservation | 56.9 | STM2614/STM2616 | → ← ← | 2762030 | 2761957 | 74 | B | |
| isrK(isr18) | Conservation | 56.9 | STM2616/STM2617 | ← ← ← | 2762867 | 2762791 | 77 | E, F, G, H | |
| isrB-2 (isrP19) | Terminator (B) | 57 | STM2631/sbcA | → ← → | 2770959, 65 | 2770872 | 88, 94 | B | Identical to isrB-1 |
| isrL (isrP20) | Terminator (B) | 58.4 | smpB/STM2690 | → ← → | 2839399 | 2839055 | 345 | A, B, C, D | |
| isrM(isrP22) | Terminator (A) | 59.8 | STM2762/STM2763 | ← → → | 2905050 | 2905378 | 329 | A | |
| isrN(isrP23) | Conservation | 59.8 | STM2764/STM2765 | ← → ← | 2906925 | 2907067 | 143 | I, J, K | |
| isrO (isrP25) | Terminator (A) | 65.8 | STM3038/STM3039 | ← → → | 3198380 | 3198580 | 201 | A, B, C, D | |
| isrP (isrP27) | Terminator (B) | 88.7 | STM4097/STM4098 | ← → ← | 4306719 | 4306866 | 148 | A, B, C, D | |
| isrQ (isrP28) | Terminator (A) | 98.0 | STM4508/STM4509 | ← → → | 4762991, 97 | 4763158 | 162, 168 |
asRNAs were predicted based on conservation or based on the presence of rho-independent transcription terminators. Three types of terminators were considered (see Figure 1).
bThe middle arrow represents the sRNA gene, while the flanking arrows indicate the orientation of the adjacent genes, respectively. Genes present on the strand given in the S. typhimurium genome database are indicated by (→), and genes present on the complementary strand are indicated by (←).
cDetermined by primer extension. The additional number represents a second, close start site.
dGiven is the position of the last uridine at the end of the terminator.
eGiven is the length in nucleotides.
fConservation of sRNAs genes in other genomes: (A) S. enterica paratyphi ATCC, (B) S. enterica choleraesius, (C) S. typhi, (D) S. typhi Ty2, (E) E. coli O157:H7, (F) S. dysenteriae. (G) S. flexneri, (H) S. sonnei, (I) E. coli 536, (J) E. coli CFT073, (K) E. coli UTI89.
gRNA transcript predicted to encode 62 aa ORF (No other transcripts were predicted to encode an ORF).
Figure 2.
Detection of novel island-encoded sRNAs. (A) Detection of small RNAs by northern analysis. Total RNA was isolated from S. typhimurium cells grown in rich media (LB; 1.5, 2.5 and 8 h after dilution at OD600 values of 0.3, 1 and 4.5, respectively) or in N minimal medium supplemented with Casamino Acids, glycerol and 10 μM MgCl2 (L; low magnesium) or 10 mM MgCl2 (H; high magnesium) and from cells subjected for 30 min to osmotic shock (NaCl; 0.5 M NaCl), oxidative stress (PQ; 0.2 mM paraquat or H2O2; 1 mM hydrogen peroxide), low iron conditions (Dp; 0.2 mM 2,2′-dipyridyl), pH stress (pH 4.9 or pH 8.4), cold shock (CS; 15°C) and heat shock (HS; 42°C). For oxygen limitation conditions (OL), single colonies were inoculated in 10 ml LB and grown overnight without agitation in 50 ml Falcon tubes to OD600 of 0.9. The RNA lengths (nucleotides) were estimated based on 5′ end mapping by primer extension and the position of the last uridine at the end of the rho-independent transcription terminator. These estimations were confirmed further by the northern blots. There are two IsrB genes each carrying two transcription-start sites (see mapping in B). 5S RNA was used as a control for loading. (B) Detection of small RNAs by primer extension. Total RNA was isolated from S. typhimurium cells harboring pGEM3 carrying the intergenic region of the predicted small RNA. Cells were grown to logarithmic phase (Log), stationary phase (Sta) and subjected to cold shock (CS; 15°C) and acid stress (pH 4.9). IsrD and IsrG were detected using 5 μg RNA and 10 μg RNA, respectively. The RNA lengths (nucleotides) were estimated based on 5′ end mapping by primer extension and the position of the last uridine at the end of the rho-independent transcription terminator.
As we previously observed in E. coli, the levels of some logarithmic-phase sRNAs were affected by cold shock treatment (20). IsrA and IsrG levels, for example, were elevated under cold conditions, while IsrM, IsrO and IsrQ RNAs were almost exclusively present during cold shock (Figure 2B and data not shown).
Two identical isrB genes, surrounded by the same flanking genes were predicted to be located at two different loci on the chromosome (Table S1 see isrP6 and isrP19). The genes were denoted isrB-1 and isrB-2, respectively. The transcript levels of these identical sRNAs were highly induced under starvation conditions in minimal media (Figure 2A). Cloning of the intergenic region encoding one of the two sRNAs to map the 5′-end of its transcript revealed the presence of two species generated either by cleavage or transcription initiation (Figure 2B).
The expression patterns of IsrK and IsrJ are similar. These genes are expressed in late stationary phase cells as well as in exponential phase cells grown under conditions of low oxygen or low magnesium. Also intriguing is the expression pattern of IsrP that is almost exclusively expressed under low magnesium and extreme acid conditions of pH 2.5 (Figure 2A and data not shown). These expression patterns suggest that the sRNAs might be involved in the regulation of S. typhimurium virulence affecting invasion and/or survival within macrophages.
The sRNA RyhB (also known as SraI) that is expressed under conditions of iron limitation was initially discovered in E. coli (20,27). Salmonella typhimurium carries two RyhB orthologs; a ryhB gene that is surrounded by the same flanking genes as in E. coli, and isrE that is island-encoded. Similar to ryhB, isrE too is induced under conditions of iron limitation, during late stationary phase, and under starvation conditions in N minimal media (Figure 2). Nevertheless, we find IsrE expression regulation to be different from that of RyhB (see below).
Differential regulation of isrE and ryhB
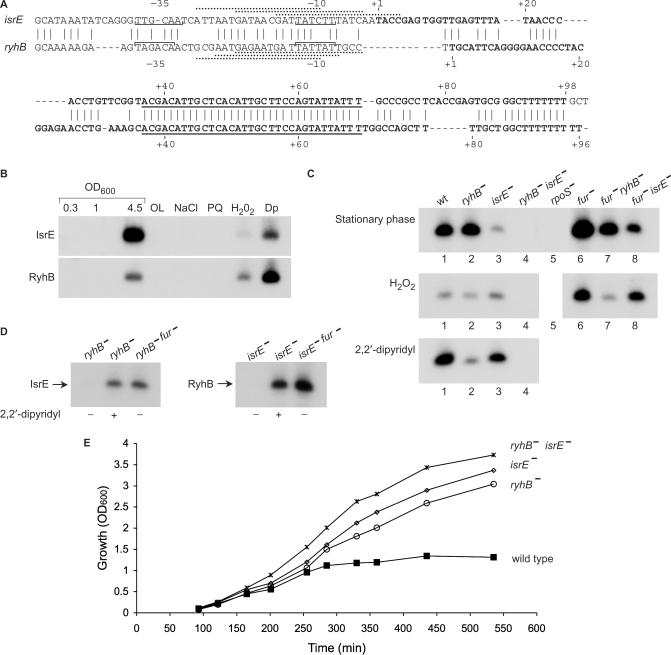
As was observed for ryhB homologs in other bacteria (21,28,29), the sequences of the S. typhimurium ryhB gene and its paralog isrE contain a highly conserved core region spanning nucleotides 37–69 and consensus binding sites for Fur (ferric uptake regulator) that overlap their promoter regions (Figure 3A). Comparing the pattern of expression of these sRNAs in wild type cells by northern blots probed with RyhB and IsrE specific primers, revealed that both genes were induced under conditions of iron limitation, in the presence of an iron chelator, upon exposure to hydrogen peroxide and during stationary phase (Figures 2 and 3B). These blots also showed that, whereas at stationary phase IsrE level was much higher than that of RyhB, in exponential phase cells, under conditions of iron limitation or oxidative stress, RyhB levels exceeded those of IsrE, indicating that the expression control of these genes is different (see also real time PCR Figure S1B Supplementary Data).
Figure 3.
Differential expression of isrE and ryhB. (A) Sequence of isrE and ryhB genes. The start sites observed by primer extension are indicated (+1). Brackets indicate putative −10 and −35 regions of the promoters mapped. The core sequences (nucleotides 37–69 relative to the start sites) are underlined. Three possible Fur binding sites that match Fur consensus (gataatgataatcattatc) at 15, 16 and 13 of 19 bases are indicated by dashed lines above the sequence of IsrE. Fur sites at ryhB, matching 14 of 19 bases of the consensus are indicated under the sequence of ryhB. (B) Detection of IsrE and RyhB in wild type cells. Northern blots as in Figure 2 of RyhB and IsrE using RyhB and IsrE specific primers (1271 and 797, respectively). The conditions used were; rich media (LB; at OD600 values of 0.3, 1 and 4.5, respectively), oxygen limitation (OL), osmotic shock (NaCl; 0.5 M NaCl), oxidative stress (PQ; 0.2 mM paraquat or H2O2; 1 mM hydrogen peroxide) and low iron conditions (Dp; 0.2 mM 2,2′-dipyridyl). (C) Late stationary phase cultures of wild type and mutants of isrE, ryhB, fur and rpoS were analyzed for isrE and ryhB levels by primer extension (top panel). Exponentially growing cultures were exposed to H2O2 (1 mM for 30 min. middle panel) or to 2,2′-dipyridyl (0.2 mM for 30 min. bottom panel). The primer extension reactions (0.5 μg RNA) were carried out using a primer (470) that is complementary to the core sequence of isrE and ryhB. (D) Fur regulation of isrE and ryhB as detected by primer extension. Exponentially growing cells were treated as in C. (E) Growth curves. Overnight cultures of wild type S. typhimurium and isrE and/or ryhB mutants were diluted 1/100 in LB medium and incubated shaking at 37°C. At 30 min after dilution the cultures were treated with 0.2 mM 2,2′-dipyridyl. Optical density was measured at the indicated time intervals.
To accurately compare between the levels of the two genes, we examined IsrE and RyhB levels in mutants of ryhB and isrE by primer extension using a primer that is complementary to the conserved core sequence of both. These data further demonstrated that the increase in the levels of RyhB in response to iron limitation or hydrogen peroxide was indeed greater than the increase observed for IsrE (Figure 3C, middle and bottom panels, lanes 2 and 3). In contrast, at late stationary phase, IsrE levels were significantly higher than the levels of RyhB (Figure 3C, top panel, lanes 2 and 3), and yet, the expression at stationary phase of both genes was dependent on σs, the stationary phase sigma factor of RNA polymerase (Figure 3C, top panel, lane 5 and data not shown).
Although to a different extent, both isrE and ryhB are regulated by iron metabolism and harbor consensus sites for Fur binding. Thus, the increase in isrE and ryhB levels at stationary phase and under oxidative stress conditions could be indirect, possibly due to a decrease in the levels of free iron that in turn leads to a decrease in the activity of Fur as a transcriptional repressor. We examined isrE and ryhB transcription at stationary phase and upon exposure to hydrogen peroxide in the absence of Fur and found that while RyhB levels were further elevated in a fur mutant (Figure 3C compare lanes 3 and 8 top and middle panels), no increase in IsrE RNA could be detected in fur-deficient cells at both hydrogen peroxide treated and stationary phase cells (Figure 3C compare lanes 2 and 7 top and middle panels), further confirming that the expression control of these genes differs. Most intriguing is the observation that in fur deficient cells, without any treatment, RyhB levels are higher than those of IsrE (Figure 3D compare lanes 1 and3), indicating that the regulation of the isrE promoter by iron metabolism is minor relative to its activation during stationary phase.
The difference in their expression patterns prompted us to examine the relative physiological significance of these two sRNAs in the response of S. typhimurium to iron limitation. We noticed that under conditions of iron depletion, the growth of wild type S. typhimurium was inhibited whereas, the ryhB, isrE double mutant grew well. Interestingly, the growth inhibition of the wild type strain required the expression of the two genes together, and the expression of neither one alone was sufficient to inhibit the growth of S. typhimurium in the absence of iron, indicating that the two genes are not functionally redundant (Figure 3E).
Island-encoded sRNAs affect the expression of genes encoded in cis
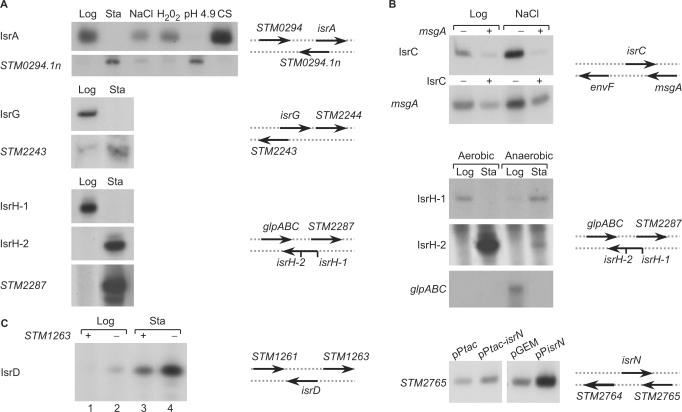
Although the computational screen focused on orphan terminators located in intergenic regions, the experimental mapping of the sRNAs showed that 11 out of the 19 that were confirmed share sequences with at least one of the flanking genes. Five of the genes (i.e. isrA, isrB, isrG, isrH and isrL) overlap their flanking genes at the 5′- end, as cis-antisense, while the other five (i.e. isrB, isrC, isrN, isrH and isrP) overlap the nearby gene at its 3′- end. One sRNA (isrD) shares promoter sequences with the flanking divergent gene.
To learn about the correlation between the expression of the sRNAs and their overlapping genes, we examined their expression under a variety of environmental conditions. Figure 4A shows that the cis-encoded sense-antisense RNAs, IsrA and its overlapping gene STM0294.1n, encoding a protein with no clear functional annotation, exhibit an inversed expression pattern. IsrA levels are high under conditions in which STM0294.1n levels are low and vice versa. Likewise, IsrG and STM2243, encoding a putative phage tail fiber protein, are expressed inversely. IsrG is associated with exponential growth phase, while STM2243 is a stationary phase RNA. STM2287 (sseL), encoding a deubiquitinase and IsrH-1 share overlapping sequences at their 5′-ends and are expressed inversely. STM2287 is expressed at stationary phase, while IsrH-1 is an exponential phase RNA. It is interesting to note that STM2287 shares no sequences with IsrH-2 that is encoded within isrH-1 and is expressed in stationary phase, as is STM2287.
Figure 4.
Cis-encoded targets. Shown are expression data and the schematic representation of the sRNAs and their flanking genes. Total RNA was isolated from S. typhimurium cells grown in rich media to logarithmic phase (Log), stationary phase (Sta) and form exponential cells subjected for 30 min to osmotic shock (NaCl; 0.5 M NaCl) oxidative stress (H2O2; 1 mM hydrogen peroxide), acid stress (pH 4.9) and cold shock (CS; 15°C) as described in the legend of Figure 2. All RNAs were detected by primer extension except for glpABC and STM0294.1n that were detected by northern analysis. Ordinarily, 30 μg of total RNA were used except for IsrG and IsrD that were detected using 10 μg RNA and 2.5 μg RNA, respectively. (A) 5′-end overlap. IsrA was detected using RNA samples isolated from cultures harboring pGEM3 carrying the intergenic region of isrA (pGEM3-isrA). STM0294.1n (chromosomal) was detected by northern analysis. IsrG and STM2243 were detected using RNA isolated from S. typhimurium cells carrying pGEM-isrG. IsrH-1, IsrH-2 and STM2287 were detected using RNA isolated from cells carrying pGEM-isrH. Two different primers were used to examine expression of IsrH-1 and IsrH-2. The primer of IsrH-2 also detects IsrH-1 since it is contained within IsrH-1. The region of the 230 nt long cDNA of IsrH-1 produced by IsrH-2 primer detecting IsrH-1 was left out. This region was included in the following Figure B. The transcription start sites of isrH-1 and isrH-2 are denoted schematically by vertical lines. (B) 3′-end overlap. IsrC and msgA RNAs were detected using RNA isolated from cells carrying pGEM-isrC or pGEM-msgA with (+) and without (−), in cis, msgA or isrC, respectively. IsrH-1 and IsrH-2 were detected using RNA isolated from cells carrying pGEM-isrH. The region of the 230 nt long cDNA of IsrH-1 produced by IsrH-2 primer is included. glpABC (chromosomal) was detected by northern analysis. Cells were grown with aeration (Aerobic) and without aeration (Anaerobic). STM2765 was detected using RNA isolated from cells carrying pPtac-isrN expressing isrN from an inducible promoter or the vector plasmid pPtac (pKK177-3) and from cells carrying pPisrN expressing isrN from its own promoter or the vector plasmid (pGEM). (C) Promoter overlap. IsrD was detected using RNA isolated from wild type carrying pGEM-isrD with (+) and without (−) STM1263 (in cis).
At its 3′-end, isrH-1 overlaps the sequence of glpC (STM2286), encoding sn-Glycerol-3-phosphate dehydrogenase small subunit. glpC expression (as part of the operon glpABC) is induced under anaerobic conditions at exponential phase (Figure 4B). Under these conditions IsrH-1 RNA can hardly be detected at exponential phase and is mainly expressed at stationary phase. IsrC and IsrN overlap their flanking genes msgA (macrophage survival gene) and STM2765 (transposase) at their 3′- ends, respectively. The expression pattern of IsrC sRNA resembles that of msgA mRNA (data not shown). The two RNAs undergo mutual degradation when expressed in cis from a multi copy number plasmid (Figure 4B). In contrast, trans expression of IsrN from a multi copy plasmid resulted in an increase in the steady state levels of the chromosome-encoded STM2765.
IsrD and STM1263 share promoter sequences in a divergent orientation. RNA analysis of this locus indicated that STM1263 inhibits isrD transcription (Figure 4C; lanes 3 and 4). In the presence of the inhibitor STM1263, IsrD expression at stationary phase is weak, whereas in its absence, IsrD transcript levels increase significantly. These data suggest that IsrD expression is regulated by a cis-encoded factor, STM1263.
Together our data suggest that a significant number of the island-encoded sRNAs modulate the expression of their flanking genes and are in turn affected by those same genes.
Island-encoded sRNA gene expression in macrophages
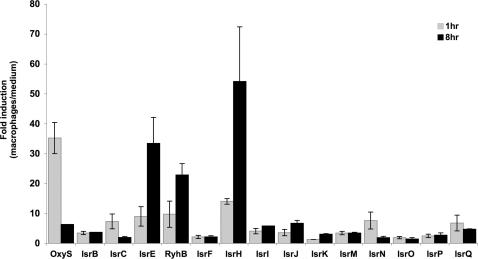
The ability to survive and multiply within macrophages is an intriguing characteristic of Salmonella species (30). Within macrophages, Salmonella cells encounter a variety of stress conditions such as oxidative burst, nutrient limitation, acidification and more. These environmental conditions develop and change as the infection process progresses (31). To learn whether the island-encoded sRNAs participate in the infection process, we examined the expression of these genes early and late in infection, by studying their transcripts at 1 and 8 h post infection. To monitor expression, activated J774 macrophage-like cells were infected with S. typhimurium and incubated for the defined periods of time in medium containing gentamicin to eliminate extracellular bacteria. At the end of the incubation periods, the intracellular bacteria were recovered and total RNA was extracted. Because many of the island-encoded sRNAs are expressed at low levels, we monitored the expression of these genes by real-time PCR, an accurate and sensitive technique for measuring low-quantity RNAs. We first examined the transcription of OxyS sRNA, previously shown to be induced in response to oxidative stress and following entry into macrophages (6,32). The monitoring of OxyS transcription by real-time PCR demonstrated that OxyS levels increased 35-fold within the first hour post infection (Figure 5). OxyS levels declined during infection and at 8 h post infection the ratio of induction decreased to 5-fold (Figure 5). Analysis of OxyS expression in macrophages by primer extension further confirmed that OxyS levels increase within the first hour and decrease as the infection progresses (data not shown). The monitoring of 12 of the island-encoded sRNAs by real-time PCR revealed that the expression patterns of two of them, IsrC and IsrN, resembled the expression pattern observed with OxyS. Both IsrC and IsrN levels increased 7-fold within the first hour of infection, and declined thereafter. A slight increase in expression upon infection was observed with sRNAs, IsrB, IsrM, IsrP and IsrQ. However, unlike the former, the levels of these sRNAs were maintained unchanged throughout the infection. The expression pattern of isrH and the iron-regulated sRNA genes, isrE and ryhB was significantly different inside macrophages; the transcript levels of these genes increased dramatically during infection. In contrast, IsrI, IsrJ and IsrK sRNAs that are expressed in stationary phase and under conditions of low oxygen and low magnesium (IsrJ and IsrK) (Figure 2A) exhibited only a slight increase in expression within macrophages (Figure 5). Interestingly, the difference in expression between IsrE and RyhB is apparent also within macrophages.
Figure 5.
Expression of island-encoded sRNA genes in J774 macrophages. Activated macrophages and infected with S. typhimurium were lysed at 1 and 8 h post infection. Thereafter, the intracellular bacteria were recovered, from which total RNA was isolated and subjected to assays of Real-Time PCR as described. To quantify changes in the expression of sRNAs within macrophages, the levels of the RNAs obtained 1 and 8 h post infection were compared with the levels of these genes as detected in S. typhimurium grown in the cell culture medium for 1.5 h.
IsrJ affects invasion of Salmonella into nonphagocytic cells
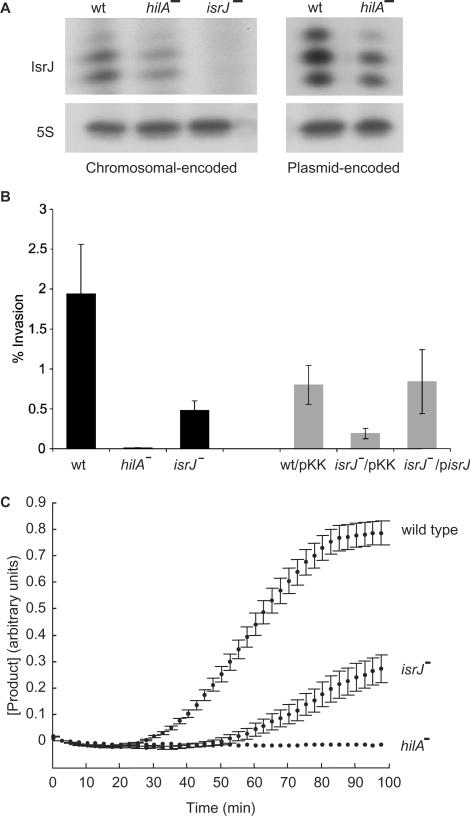
As a pathogenic bacterium, Salmonella is capable of active invasion of the intestinal epithelium. This process requires the functions of a TTSS, encoded within pathogenicity-island 1 (SPI-1) and regulators encoded outside of SPI-1 (33). In addition, a number of environmental conditions are known to regulate invasion, including pH, osmolarity, oxygen tension and more. The transcriptional activator HilA is a central regulator of invasion, which in response to multiple environmental and genetic signals directly regulates the transcription of the SPI-1 genes. The unique expression pattern observed for a few of the newly discovered sRNAs, such as IsrJ, suggest that they might be part of the SPI-1 regulatory circuit that controls infection. Thus, we examined whether the isrJ expression is regulated by HilA. RNA analysis of the chromosomally encoded isrJ gene in wild type and hilA mutant cells shows that IsrJ levels are low in the absence of HilA (Figure 6A). Similarly, RNA analysis of a plasmid expressing IsrJ from its own promoter, exhibited a decrease in IsrJ expression in the absence of HilA (Figure 6A).
Figure 6.
IsrJ is part of the SPI-1 TTSS regulon. (A) isrJ regulation by hilA. RNA samples from cultures of S. typhimurium, wild type, hilA (hilA::kan) and isrJ (ΔisrJ::cm) mutants grown under conditions of low oxygen were subjected to primer extension using an isrJ specific primer. The strains in the right panel carry a multi copy number plasmid expressing IsrJ (pisrJ). IsrJ expression from pisrJ was detected using 1 μg RNA. 5S RNA was used as a control for loading. (B) IsrJ affects invasion of Salmonella into nonphagocytic cells. HeLa cells infected with S. typhimurium strains for 1 h in medium containing gentamicin were washed, lysed and aliquots were plated to determine the number of viable intracellular bacteria. Black bars; wild type, hilA (hilA::kan) and isrJ (ΔisrJ::cm). Grey bars; strains carrying a plasmid expressing IsrJ (pisrJ) or the vector plasmid (pKK). The results are means of three independent experiments, each carried out with four cultures of each strain. (C) IsrJ affects translocation into nonphagocytic cells. HeLa cells preloaded with CCF2 were infected with wild type, hilA and isrJ mutant strains expressing the fusion protein SptP-BlaM. The decrease in CCF2 fluorescence and the increase in the fluorescence emitted by the CCF2 hydrolysis product were recorded at real time.
We next examined the effect of this sRNA on the Salmonella invasion rate into nonphagocytic cells. In this assay, wild type S. typhimurium and mutants of hilA and isrJ were used to infect HeLa cultures in vitro. We found the isrJ mutant to exhibit decreased invasion rates, while hilA mutant failed to invade epithelial cells completely (Figure 6B). Additionally, isrJ-deficient mutant expressing IsrJ from a multi copy plasmid exhibited restored invasion rates, similar to wild type (Figure 6B). Taken together, these results demonstrate that IsrJ affects the invasion process.
The SPI-1 TTSS is a virulence determinant that enables the injection of bacterial effector proteins into the cytosol of eukaryotic cells. To further characterize the role of IsrJ in the regulon of SPI-1 TTSS, we examined the effect of IsrJ expression on the translocation of the SptP effector protein of S. typhimurium. We made use of a system that monitors the kinetics of translocation in real time (25,34). Towards this end, the sptP gene was fused to the blaM (β-lactamase) reporter gene, downstream of the tac promoter (pKB3138; Ptac-sptP-blaM). The system measures the entry of SptP-BlaM fusion protein into HeLa cells, preloaded with CCF2. CCF2 consists of a cephalosporin core linking two fluorophores: 7-hydroxycoumarin and fluorescein. Excitation of the 7-hydroxycoumarin at 409 nm results in fluorescence resonance energy transfer (FRET) to the fluorescein moiety that emits at 520 nm (green). On the other hand, cleavage of the intracellular CCF2 by the translocated SptP-BlaM fusion protein results in the disruption of FRET and thus, the excitation of 7-hydroxycoumarin results in emission at 447 nm (blue). To estimate intracellular cleavage by the fusion protein and thus translocation levels, HeLa cells preloaded with CCF2 were infected with wild type S. typhimurium cells or with hilA and isrJ mutant strains expressing SptP-BlaM. Both the decrease in CCF2 fluorescence and the increase in the fluorescence emitted by the CCF2 hydrolysis product were then recorded in real time. We found the kinetics of product accumulation in isrJ mutant cells to be significantly slow, compared with the wild type (Figure 6C). No cleavage of intracellular CCF2 could be detected in hilA-deficient cells. These results demonstrate that IsrJ affects the translocation process of effector proteins into eukaryotic cells, further confirming that IsrJ sRNA is part of the SPI-1 TTSS regulon.
DISCUSSION
Novel sRNA genes encoded within horizontally transferred genetic islands of S. typhimurium were identified in a systematic screen for unique sRNAs. The characterization of the newly discovered island-encoded sRNAs demonstrated that many of the island-encoded genes are expressed under unique stress conditions. Some of the conditions such as high osmolarity, extreme pH, starvation, oxygen limitation, oxidative stress and conditions of iron and magnesium limitation are reminiscent of the environments S. typhimurium encounters upon invasion of intestinal epithelium, or within macrophages. Furthermore, real-time expression analyses demonstrated that numerous sRNA genes of S. typhimurium are induced within the first hour of infection of J774 macrophage-like cells. The level of expression of a few of the sRNAs such as IsrB, IsrM, IsrQ and IsrI remains unchanged throughout infection, while the levels of others including IsrC and IsrN, decline with time, as the infection progresses, very much like the oxidative stress induced OxyS sRNA. In contrast, IsrE, RyhB and IsrH levels increase dramatically as the infection progresses. These changes in the intracellular expression of the sRNAs reflect the changes in the environments inside macrophages and suggest that sRNA genes play an important role in the life cycle and pathogenicity of Salmonella.
The computational search focused on orphan terminators located in intergenic regions, and yet, our experimental mapping of the confirmed sRNAs showed that many of them share sequences with at least one of their flanking genes. Expression analyses of the 5′-end overlapping genes revealed an inverse expression pattern of the sRNAs versus their flanking mRNAs. Accordingly, under conditions leading to elevated sRNA levels, the levels of the flanking mRNA were low and vise versa. It is possible that the transcription of the sRNA gene inhibits the synthesis of the complementary transcript by steric hindrance (35). Alternatively, the binding of the sRNA to the complementary mRNA, either at the ribosome-binding site to inhibit translation or at the coding region could result in the exposure of a ribonuclease sensitive site, leading to its degradation. A similar inverse behavior was observed in E. coli for SraC sRNA (also known as RyeA) and its antisense sRNA gene ryeB (20,27,36). For these genes, it was suggested that duplex formation between these two RNAs could result in hybrid-dependent RNase III processing. Analyses of the two pairs of RNAs sharing 3′-end sequences showed that while the RNAs of isrC/msgA undergo mutual degradation, overexpression of IsrN sRNA results in an increase in STM2765 mRNA levels, reminiscent of the control of gadX by GadY sRNA (37,38). The glpABC operon that overlaps IsrH-1 and IsrH-2 at the 3′-end, encodes a three-subunit enzyme that functions mostly under anaerobic conditions. It is interesting that under aerobic conditions IsrH-1 is expressed mainly at exponential phase, while under anaerobic conditions, transcript levels of this gene increase at stationary phase, leading to an inverse expression pattern between glpABC and isrH-1. The transcription of IsrD is regulated by STM1263, a cis-encoded factor located upstream and divergent of this sRNA. Their opposite promoters overlap in a way that is reminiscent of two other gene pairs that exhibit sRNA regulation by transcription factors, including oxyS control by OxyR in response to oxidative stress, and GcvA regulating gcvB in response to glycine levels (6,39). It is also possible that the divergently oriented promoters affect the activity of one another by transcription-induced negative DNA supercoiling as described for the E. coli ilvYC operon (40,41). What is the basis of isrD regulation by STM1263 and whether it also involves an activation step as seen for the other gene pairs is not yet clear. Taken together, our observations indicate that unlike the majority of the known E. coli sRNAs that act in trans to affect the expression of their target genes, many of the island-encoded genes of S. typhimurium affect the expression of cis-encoded overlapping targets.
Numerous sRNAs were characterized in bacterial pathogens and were shown to affect virulence genes (12,42–55). RNAIII of S. aureus was the first regulatory sRNA shown to be involved in bacterial pathogencity (54). In response to an increase in cell density, it acts to activate the translation of hla mRNA encoding α-hemolysin and represses spa encoding protein A, a main surface adhesin, and rot encoding the transcription factor Rot (45,47,53). Thus, RNAIII controls the switch between colonization that requires surface proteins and pathogenicity along with excretion of toxins. In Pseudomonas, the activity of RsmA, a translational regulator of quorum signal molecules and extracellular products is prevented by the sRNAs, RmsZ in P. aeruginosa and RmsY, RmsZ and RmsX in P. fluorescens (43,46,48,55). The S. typhimurium RsmA-homolog, CsrA, controls genes involved in cell invasion. This activity is counteracted in a redundant manner by two sRNAs, CsrB and CsrC (42,44). Also in S. typhimurium, InvR in SPI-1 regulates the expression of ompD encoding an outer membrane protein while GcvB regulates the expression of multiple ABC transporters of amino acids and peptides (56,57). Seven sRNAs, four related Qrr1-4 and three Csr RNAs, CsrB, CsrC and CsrD regulate the quorum-sensing cascade in Vibrio cholerae (50,51). The four Qrr sRNAs affect hapR mRNA expression, while the three Csr RNAs abolish the activity of CsrA, which in turn, affects the quorum-sensing pathway. Although only a few sRNA genes were shown to affect virulence genes, the observation that mutations in hfq decrease virulence in several pathogens, such as P. aeruginosa, V. cholerae, S. typhimurium, L. monocytogenes and Shigella flexneri strongly indicates that sRNAs play key roles in the control of pathogenesis (58–63).
As a pathogenic bacterium, Salmonella is capable of active invasion of the intestinal epithelium of the host. This process of invasion requires the function of a TTSS encoded within pathogenicity-island 1 (SPI-1), as well as regulators encoded outside of SPI-1. In addition, several environmental conditions such as oxygen limitation, pH, high osmolarity and bacterial growth phase were shown to induce invasion genes (33,64). TTSS forms a needle-like complex that is responsible for the injection of bacterial effector proteins into the cytosol of eukaryotic cells. These effector proteins elicit, in turn, several physiological changes, including actin rearrangement leading to engulfment of the bacterium (65). Intrigued by the expression pattern of IsrP, IsrK and IsrJ sRNAs that are expressed under conditions known to regulate invasion, including pH, low magnesium and low oxygen, we examined the effect of one such sRNA gene, isrJ, on invasion. We found the isrJ mutant strain to exhibit a significant decrease in both the invasion rates and the intracellular accumulation of effector proteins. A recent genome-wide screen for Salmonella genes required for long-term mouse systemic infection, identified a diverse repertoire set of 118 genes that contribute to a long-lasting infection (66). Among these, ∼30% correspond to horizontally acquired regions such as pathogenicity islands (SPI 1-5), prophages (Gifsy 1 and 2) and the pSLT virulence plasmid. Interestingly, apart from the known virulence functions, several SPI-1 encoded TTSS functions were shown to be required for establishment and maintenance of persistent infections. Our results, showing that the isrJ-deficient strain exhibits a significant decrease in the levels of translocation and invasion, indicate that the island-encoded sRNA, IsrJ, plays an important role in the process of infection of the host by Salmonella.
The sRNA RyhB, initially discovered in E. coli, has been shown to down-regulate a large number of transcripts encoding iron-using proteins, modulating the usage of intracellular iron. (21,67–69). ryhB-like genes were also characterized in P. aeruginosa, V. cholera and Shigella dysenteriae (28,29,70). These ryhB genes were shown to affect growth under iron starvation conditions as well as virulence functions such as biofilm formation, chemotaxis and acid resistance (28,71,72).
We identified IsrE, an island-encoded paralog of RyhB. Comparison between the expression patterns of the two paralogs revealed that their regulation differs considerably. Both genes are induced in response to iron limitation, oxidative stress and at stationary phase. However, in exponential phase cells, under conditions of iron limitation or oxidative stress, RyhB levels exceed those of IsrE, whereas at stationary phase, IsrE levels are much higher than those of RyhB. RNA analysis of these genes in fur-deficient strains demonstrated that the activity of the isrE promoter in response to iron metabolism is minor in comparison to its activity during stationary phase. In contrast, RyhB promoter exhibits the opposite. The different expression pattern observed for isrE may stem from its genetic origin and from it being encoded by a foreign DNA segment that was acquired by horizontal transfer. Alternatively, it is possible that the binding of these sRNAs to different targets affect their steady state levels via their degradation.
Also intriguing is the observation that under conditions of iron depletion, the growth of wild type S. typhimurium was inhibited, while the double mutant of ryhB and isrE grew well. A similar phenotype was detected for the ryhB mutant in V. cholera (28). Possibly, the redistribution of intracellular iron requires a temporary growth arrest, or it leads to it. Consistent with the observation of the differential regulation, we found that the two sRNAs are not functionally redundant, as the growth arrest phenotype of the wild type strain is abolished with the disruption of either one of the genes. Together the data suggest that the two genes ryhB and isrE may each have individual targets.
Numerous horizontally acquired virulence factors such as the ones involved in invasion are regulated by regulatory circuits some of which may have existed in the organism prior to the acquisition of the pathogenicity island (33). The presence and effects of the island-encoded gene, IsrE, points to the opposite process, in which genes acquired by horizontal transfer were adapted to control the expression of genes of the genome core.
In this study we show that island encoded sRNA genes play an important role within the network that regulates bacterial adaptation to environmental changes and stress conditions and thus controls virulence. Deciphering the roles of sRNAs within this network and fully understanding the ways in which these sRNAs are involved in virulence is an exciting direction for further studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Michael McClelland for generously providing the sequences of the islands of S. typhimurium and to Hilla Giladi for useful comments on the manuscript. We thank Yaniv Baum for technical assistance. Supported by The Israel Science Foundation founded by The Israel Academy of Sciences and Humanities, grant number 663/02, The United States-Israel Binational Science Foundation, grant number 2001032, The Israel Science Foundation-Bikura Program, grant number 1342/05, The Israeli Ministry of Science, grant number 3/2559 and by BACRNAs, a Specific Targeted Research Project supported by European Union's FP6 Life Science, Genomics and Biotechnology for Health, LSHM-CT-2005-018618. (SA). Funding to pay the Open Access publication charges for this article was provided by BACRNAs, a Specific Targeted Research Project supported by European Union's FP6 Life Science, Genomics and Biotechnology for Health, LSHM-CT-2005-018618.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gerdes K, Wagner EG. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Masse E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Valentin-Hansen P, Johansen J, Rasmussen AA. Small RNAs controlling outer membrane porins. Curr. Opin. Microbiol. 2007;10:152–155. doi: 10.1016/j.mib.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Vanderpool CK. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 2007;10:146–151. doi: 10.1016/j.mib.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 7.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Sogaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Sansonetti PJ, Kopecko DJ, Formal SB. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 1982;35:852–860. doi: 10.1128/iai.35.3.852-860.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groisman EA, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 16.Keusch GT. Salmonellosis. Harrisons Principles of Internal Medicine. 1994. McGraw-Hill, Inc., New York, pp. 671–676. [Google Scholar]
- 17.Donnenberg MS. Pathogenic strategies of enteric bacteria. Nature. 2000;406:768–774. doi: 10.1038/35021212. [DOI] [PubMed] [Google Scholar]
- 18.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 19.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 20.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 21.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones BD, Lee CA, Falkow S. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 1992;60:2475–2480. doi: 10.1128/iai.60.6.2475-2480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CA, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl Acad. Sci. USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills E, Baruch K, Charpantier X, Kobi S, Rosenshine I. Cell Host Microbe. Real time analysis of effector translocation by the type III secretion system of enteropathogenic E. coli. doi:10.1016/j.chom.2007.11.007 (in press). [DOI] [PubMed] [Google Scholar]
- 26.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, Segura-Salazar J, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 2005;187:4005–4014. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl Acad. Sci. USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchmeier NA, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect. Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kingsley RA, Baumler AJ. Salmonella interactions with professional phagocytes. Subcell. Biochem. 2000;33:321–342. doi: 10.1007/978-1-4757-4580-1_13. [DOI] [PubMed] [Google Scholar]
- 32.Schlosser-Silverman E, Elgrably-Weiss M, Rosenshine I, Kohen R, Altuvia S. Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J. Bacteriol. 2000;182:5225–5230. doi: 10.1128/jb.182.18.5225-5230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altier C. Genetic and environmental control of Salmonella invasion. J. Microbiol. 2005;43:85–92. [PubMed] [Google Scholar]
- 34.Charpentier X, Oswald E. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 2004;186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hershberger PA, deHaseth PL. RNA polymerase bound to the PR promoter of bacteriophage lambda inhibits open complex formation at the divergently transcribed PRM promoter. Implications for an indirect mechanism of transcriptional activation by lambda repressor. J. Mol. Biol. 1991;222:479–494. doi: 10.1016/0022-2836(91)90491-n. [DOI] [PubMed] [Google Scholar]
- 36.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 40.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhee KY, Opel M, Ito E, Hung S, Arfin SM, Hatfield GW. Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc. Natl Acad. Sci. USA. 1999;96:14294–14299. doi: 10.1073/pnas.96.25.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altier C, Suyemoto M, Lawhon SD. Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA. Infect. Immun. 2000;68:6790–6797. doi: 10.1128/iai.68.12.6790-6797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burrowes E, Abbas A, O’Neill A, Adams C, O’Gara F. Characterisation of the regulatory RNA RsmB from Pseudomonas aeruginosa PAO1. Res. Microbiol. 2005;156:7–16. doi: 10.1016/j.resmic.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Fortune DR, Suyemoto M, Altier C. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect. Immun. 2006;74:331–339. doi: 10.1128/IAI.74.1.331-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 2006;61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 46.Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, Camara M, Williams P, Haas D. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2004;186:2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl Acad. Sci. USA. 2005;102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 50.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 51.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Mangold M, Siller M, Roppenser B, Vlaminckx BJ, Penfound TA, Klein R, Novak R, Novick RP, Charpentier E. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol. Microbiol. 2004;53:1515–1527. doi: 10.1111/j.1365-2958.2004.04222.x. [DOI] [PubMed] [Google Scholar]
- 53.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valverde C, Lindell M, Wagner EG, Haas D. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J. Biol. Chem. 2004;279:25066–25074. doi: 10.1074/jbc.M401870200. [DOI] [PubMed] [Google Scholar]
- 56.Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 57.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 60.Figueroa-Bossi N, Lemire S, Maloriol D, Balbontin R, Casadesus J, Bossi L. Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 2006;62:838–852. doi: 10.1111/j.1365-2958.2006.05413.x. [DOI] [PubMed] [Google Scholar]
- 61.Sharma AK, Payne SM. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 2006;62:469–479. doi: 10.1111/j.1365-2958.2006.05376.x. [DOI] [PubMed] [Google Scholar]
- 62.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jager KE, Blasi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 64.Jones BD. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 2005;43:110–117. [PubMed] [Google Scholar]
- 65.Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 2001;3:1281–1291. doi: 10.1016/s1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- 66.Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacques JF, Jang S, Prevost K, Desnoyers G, Desmarais M, Imlay J, Masse E. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 2006;62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- 68.Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 70.Murphy ER, Payne SM. RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect. Immun. 2007;75:3470–3477. doi: 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mey AR, Craig SA, Payne SM. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 2005;73:5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oglesby AG, Murphy ER, Iyer VR, Payne SM. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Mol. Microbiol. 2005;58:1354–1367. doi: 10.1111/j.1365-2958.2005.04920.x. [DOI] [PubMed] [Google Scholar]