Abstract
Arbuscular mycorrhizal (AM) fungi are capable of exploiting organic nitrogen sources, but the molecular mechanisms that control such an uptake are still unknown. Polymerase chain reaction-based approaches, bioinformatic tools, and a heterologous expression system have been used to characterize a sequence coding for an amino acid permease (GmosAAP1) from the AM fungus Glomus mosseae. The GmosAAP1 shows primary and secondary structures that are similar to those of other fungal amino acid permeases. Functional complementation and uptake experiments in a yeast mutant that was defective in the multiple amino acid uptake system demonstrated that GmosAAP1 is able to transport proline through a proton-coupled, pH- and energy-dependent process. A competitive test showed that GmosAAP1 binds nonpolar and hydrophobic amino acids, thus indicating a relatively specific substrate spectrum. GmosAAP1 mRNAs were detected in the extraradical fungal structures. Transcript abundance was increased upon exposure to organic nitrogen, in particular when supplied at 2 mm concentrations. These findings suggest that GmosAAP1 plays a role in the first steps of amino acid acquisition, allowing direct amino acid uptake from the soil and extending the molecular tools by which AM fungi exploit soil resources.
Amino acids reach a considerable quantity in soils of many ecosystems, and this could contribute significantly to the nitrogen (N) nutrition of plants. This is the case in soils in which mineralization processes are low, for example, arctic, boreal (Väre et al., 1997), and heathland soils (Read, 1996), or in poor sites such as wet mires and sand plains (Chen et al., 1999). It is also true for agricultural systems (Scheller, 1996).
Plants, with a few exceptions (Turnbull et al., 1996; Näsholm et al., 1998; Bennett and Prescott, 2004), do not possess the full machinery necessary to exploit such organic sources. Plants have adapted different strategies to access and compete for this key nutrient with the microbial communities of the rhizosphere. One of these is the establishment of symbiotic associations with mycorrhizal fungi (Girlanda et al., 2007).
Apart from the visionary speculation of Frank (1894), who proposed the “organic nitrogen theory” at the end of the 19th century (Read and Perez-Moreno, 2003), the possibility that fungal symbionts might be involved directly in the uptake of organic polymers was largely ignored until the mid-1980s. For a long time, the degradation of organic compounds, the uptake and transfer of organic N to the plant, was considered a prerogative of ectomycorrhizal fungi (Tibbett et al., 2000; Chalot et al., 2002; Tibbett and Sanders, 2002; Sawyer et al., 2003; Guidot et al., 2005; Müller et al., 2007). Unlike ectomycorrhizal fungi, arbuscular mycorrhizal (AM) fungi, which, under natural conditions, colonize the majority of root systems, have mainly been considered for their role in phosphate uptake and translocation (Bucher, 2007; Javot et al., 2007).
There is evidence that AM fungi can also contribute to the increase of N acquisition in host plants, and in recent years advances have been made in the understanding of the movement of N, in particular inorganic N, in AM symbiosis. Experiments based on radioactively labeled N, measurements of the activity of plant enzymes involved in N assimilation, and transcriptional studies of a nitrate reductase in the mycobiont have shown that AM fungi are capable of taking up nitrate and ammonium (Kaldorf et al., 1994, 1998; Johansen et al., 1996; Cliquet et al., 1997; Faure et al., 1998; Subramanian and Charest, 1998; Hawkins et al., 2000; Toussaint et al., 2004). Molecular evidence of ammonium uptake was obtained recently through the characterization of an ammonium transporter that is expressed in the extraradical mycelium of Glomus intraradices (López-Pedrosa et al., 2006). Inorganic N, taken up by the extraradical mycelium, is incorporated into amino acids and translocated to the intraradical mycelium mainly as Arg (Cruz et al., 2007). N is then transferred from the fungus to the plant as ammonium without any loss of carbon skeleton, thanks to the catabolic arm of the urea cycle that converts Arg into ammonium (Govindarajulu et al., 2005; Jin et al., 2005).
AM fungi could also be involved in the acquisition of organic N. Preliminary studies have demonstrated that the development of the extraradical mycelium of AM fungi is stimulated by external organic N sources (St. John et al., 1983a, 1983b; Joner and Jakobsen, 1995; Ravnskov et al., 1999). Näsholm et al. (1998) obtained indirect evidence of amino acid uptake from Deschampsia flexuosa colonized by AM fungi in field conditions. It has been shown that organic N uptake is greatly enhanced by AM colonization (Cliquet et al., 1997) and that AM symbiosis could both enhance the decomposition of N and increase N capture from organic patches (Hodge et al., 2001). More recently, Jin et al. (2005) demonstrated that the extraradical mycelium of AM fungi grown in in vitro cultures can take up and utilize exogenously supplied Arg.
Little is known about the genes involved in organic N metabolism in AM fungi. Until now, only a sequence coding for Gln synthetase has been characterized in Glomus mosseae and G. intraradices (Breuninger et al., 2004). No sequence responsible for organic N uptake has been described to date.
Amino acid transport systems have been studied extensively in higher plants (Okumoto et al., 2002), yeast, and filamentous fungi (Struck et al., 2002; Trip et al., 2002; Wipf et al., 2002b). The transporter classification groups all of the amino acid permeases into the amino acid/polyamine organocation superfamily (Jack et al., 2000). Up to 24 members of the amino acid permease family have been found in the yeast Saccharomyces cerevisiae (Wipf et al., 2002b), most of which have been functionally characterized (Regenberg et al., 1999). As far as mycorrhizal fungi are concerned, to date only two genes have been identified, both from ectomycorrhizal species, Amanita muscaria and Hebeloma cylindrosporum (Nehls et al., 1999; Wipf et al., 2002a).
In this work, we describe and functionally characterize in a yeast mutant an amino acid permease (GmosAAP1) from the AM fungus G. mosseae. GmosAAP1 mRNA was detected in the extraradical mycelium, the fungal structure that explores soil resources. Organic N supplied as the amino acid pool at a concentration of 2 μm or 2 mm determined an increase in the GmosAAP1 transcript levels.
AM fungi are traditionally acknowledged as the microbes that improve mineral supply to a plant, thanks to phosphate uptake, through their external mycelium (Bucher, 2007). Our findings suggest that AM fungi possess other molecular tools to exploit soil resources, since GmosAAP1 may play a role in the first step of amino acid acquisition, allowing direct amino acid uptake from the environment.
RESULTS
Cloning and Sequence Analysis of GmosAAP1
In order to identify the genes involved in the uptake of N compounds in AM fungi, two oligonucleotides, designed on conserved amino acid domains (Supplemental Fig. S1), were used for PCR on cDNAs obtained from Gigaspora margarita, Gigaspora rosea, G. mosseae, and G. intraradices. A cDNA fragment of approximately 150 bp obtained from G. mosseae extraradical mycelium showed a similarity to previously described fungal amino acid permeases. A full-length cDNA, named GmosAAP1 (accession no. AY882560), was then identified by means of 5′ and 3′ RACE-PCR (Supplemental Fig. S1). Twelve transmembrane domains were predicted for GmosAAP1 (Supplemental Fig. S1) using several programs available on the Web (HMMTOP, TMHMM, SOSUI, and TMPRED). This structure is consistent with that of other amino acid permeases (Van Belle and Andre, 2001).
The overall alignment of GmosAAP1 and vacuolar amino acid transporters described in Schizosaccharomyces pombe (accession no. Q10074) and in S. cerevisiae (accession no. NP_012534) showed a very low similarity level (approximately 12% of identical amino acids). In addition, the weakly conserved motif (T/I/K)LP(L/K/I), which works as a sorting signal for vacuole targeting (Stack et al., 1995), is not present in GmosAAP1. The alignment of a number of plasma membrane amino acid permease sequences from fungi and plants derived from the data bank coupled to a neighbor-joining phylogenetic analysis showed that the fungal transporters are clearly separated from those of the plants. GmosAAP1 clusters with fungal amino acid permease sequences (Supplemental Fig. S2).
In recent years, a number of sequences belonging to ascomycetes that were closely associated to AM spores, grown in pot cultures or from the field, were erroneously assigned to AM fungi (Redecker et al., 1999). For this reason, we wanted to confirm the authenticity of the GmosAAP1 sequence. A couple of specific primers for GmosAAP1 were used on the genomic DNA of a related species, G. intraradices, which was grown in sterile conditions on transformed roots. A PCR product of the expected size was obtained, cloned, and sequenced. The sequence, named GintAAP1, showed a clear similarity to GmosAAP1, with 96.2% identity at the nucleotide level (data not shown).
Functional Characterization of GmosAAP1 in Yeast
The yeast mutant 22Δ8AA, which lacks eight endogenous amino acid transport systems, was transformed with plasmids containing GmosAAP1 cDNA. The growth of 22Δ8AA expressing GmosAAP1 was good on Pro, Asp, Glu, and γ-aminobutyric acid as a single N source (data not shown).
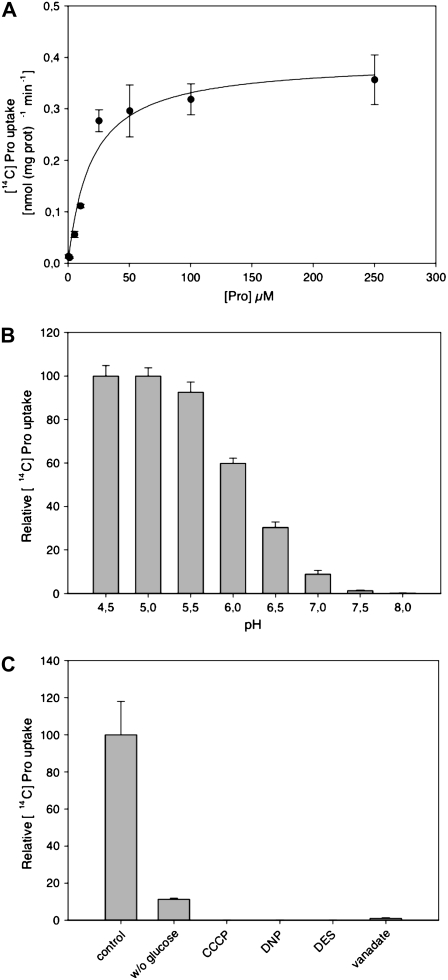
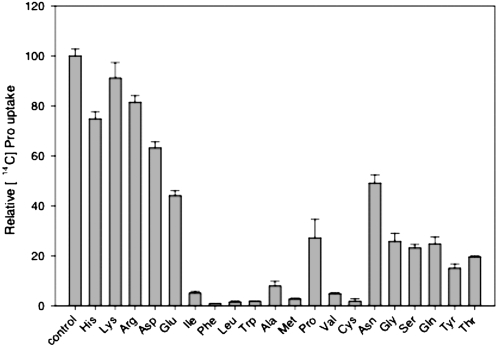
To directly determine the transport properties of GmosAAP1, radiotracer uptake studies were performed using [14C]Pro. Under standard assay conditions, the [14C]Pro uptake was linear for at least 4 min. The uptake rate was concentration dependent and displayed saturation kinetics (Fig. 1A). The Km value for the transport of Pro was 18.8 μm (Fig. 1A). GmosAAP1 activity was clearly pH dependent, with an optimum at approximately pH 4.5 to 5.0 (Fig. 1B). The [14C]Pro uptake depended on the presence of Glc and was sensitive to the protonophores 2,4-dinitrophenol and carbonyl cyanide m-chlorophenylhydrazone and the plasma membrane H+-ATPase inhibitors diethylstilbestrol and vanadate, indicating that energization is required for transport (Fig. 1C). The range of the amino acids that bind to GmosAAP1, and therefore those most probably transported, was determined through their competitive effect on the uptake of the labeled Pro (Fig. 2). Negatively and positively charged amino acids were poorly recognized by GmosAAP1. Neutral, polar, and hydrophobic amino acids were better recognized, with the exception of Cys, which was an excellent competitor. All of the nonpolar, hydrophobic amino acids competed even more efficiently than Pro.
Figure 1.
A, GmosAAP1-mediated [14C]Pro uptake at different substrate concentrations. The experiments were performed at pH 4.5. The values represent means of three independent experiments ± sd. B, pH dependence of the uptake rate of [14C]Pro in the yeast mutant 22Δ8AA expressing GmosAAP1. Yeast expressing GmosAAP1 in pDR196 were measured at different pH values and an 18.8 μm substrate concentration. The values represent means of three independent experiments ± sd. C, Influence of plasma membrane energization on the uptake rate of [14C]Pro in the yeast mutant 22Δ8AA expressing GmosAAP1. The yeast cells were preincubated for 5 min in the presence of 100 mm Glc (control), without Glc, or with Glc and 0.1 mm 2,4-dinitrophenol (DNP), 0.1 mm diethylstilbestrol (DES), 0.1 mm carbonyl cyanide m-chlorophenylhydrazone (CCCP), or 0.1 mm vanadate. The values represent means of three independent experiments ± sd.
Figure 2.
Substrate specificity of GmosAAP1. Inhibition of 18.8 μm [14C]Pro uptake by a 5-fold molar excess of competing amino acids. The data are expressed as percentages of the uptake rate in the presence of 18.8 μm Pro. The values represent means of three independent experiments ± sd.
GmosAAP1 Gene Expression Profiles
Gene expression analysis was performed by reverse transcription (RT)-PCR assays on different stages of the G. mosseae life cycle: sporocarps germinated in water, extraradical mycelium, and mycorrhizal roots from which the external hyphae were removed. The last two samples were collected from pot cultures: Cucumis sativus mycorrhizal roots watered with a Long Ashton solution containing 1 mm nitrate. Three months after inoculation, the roots presented the typical structures of AM symbiosis and a good mycorrhization level according to Trouvelot et al. (1986): F = 87% (frequency of mycorrhization of the root fragments), M = 52% (intensity of root cortex colonization), a = 49% (average presence of arbuscules within the infected areas), and A = 26% (arbuscule abundance in the root system).
In order to exclude cross-hybridization with the plant material, oligonucleotides for the G. mosseae 28S ribosomal gene (van Tuinen et al., 1998) and for GmosAAP1 (G1/G2) were first tested on the C. sativus genomic DNA. No amplification product was obtained (data not shown).
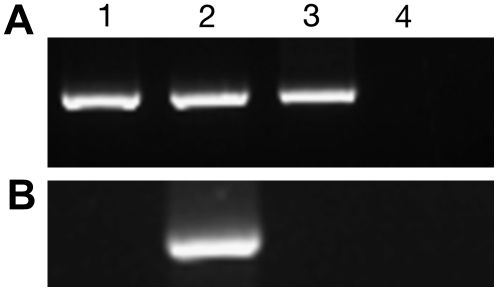
The RT-PCR experiments with G. mosseae 28S ribosomal primers gave an amplified fragment of the expected size (380 bp) from the germinated sporocarps, extraradical mycelium, and intraradical fungal structure cDNAs (Fig. 3A). Amplifications with GmosAAP1 primers generated a PCR product (780 bp), but only in the sample corresponding to extraradical mycelium cDNA (Fig. 3B).
Figure 3.
Gel electrophoresis of RT-PCR products obtained with oligonucleotides specific for the G. mosseae 28S rDNA (A) or GmosAAP1 (B) on the following samples: lane 1, germinating spores; lane 2, extraradical mycelium; lane 3, intraradical mycelium; lane 4, no template.
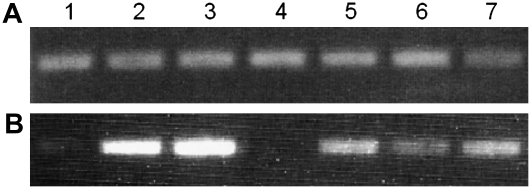
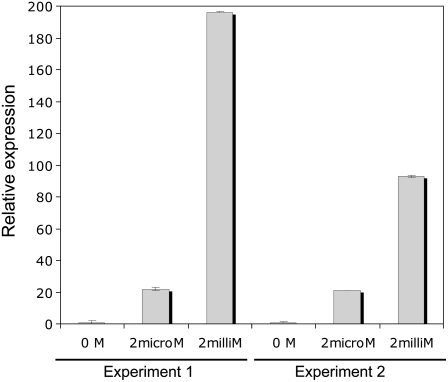
To investigate whether GmosAAP1 expression was modulated by organic N in the surrounding medium, semiquantitative and real-time RT-PCR assays were performed on pot culture extraradical mycelium treated for 72 h with a modified Long Ashton solution without N (0 m) or with a Long Ashton solution containing as N source a pool of amino acids (Leu, Ala, Asn, Lys, and Tyr) or NH4+ or NO3− at two different concentrations (2 μm or 2 mm). The amount of mRNA obtained from different samples was first calibrated using fungus-specific 28S rRNA primers. The mRNA samples were then amplified with GmosAAP1-specific oligonucleotides. The PCR product corresponding to GmosAAP1 was considerably more abundant for the two samples treated with the amino acid pool at 2 μm and 2 mm (Fig. 4). Real-time RT-PCR assays were performed to obtain a quantitative measurement of this induction. In fact, an increase of the GmosAAP1 transcripts level was observed on two independent biological samples after exposure to organic N. In spite of a certain variability in the independent samples, the GmosAAP1 transcript levels were particularly abundant after the 2 mm treatment (Fig. 5).
Figure 4.
Gel electrophoresis of RT-PCR products obtained with oligonucleotides specific for the G. mosseae 28S rDNA (A) or GmosAAP1 (B) on external mycelium treated as follows: lane 1, no N (0 m); lane 2, 2 μm of the amino acid pool (Leu, Ala, Asn, Lys, and Tyr); lane 3, 2 mm of the amino acid pool; lane 4, 2 μm KNO3; lane 5, 2 mm KNO3; lane 6, 2 μm (NH4)2SO4; lane 7, 2 mm (NH4)2SO4.
Figure 5.
Real-time RT-PCR analysis of the GmosAAP1 mRNA in extraradical mycelium treated with 0 m N or 2 μm or 2 mm of the amino acid pool. Relative expression levels were obtained with the comparative threshold cycle method (see “Materials and Methods” for details) and were normalized with respect to the GmosAAP1 levels in the 0 m treatment.
DISCUSSION
The role of AM fungi in litter degradation and in the uptake of organic nutrients from the soil is an ecologically important issue that has particular relevance in plant nutrition (Hodge et al., 2001). As stated by Alexander (2007), “In view of the worldwide distribution of arbuscular mycorrhizas, sometimes in highly organic soils, this is a topic ripe for further exploration.”
As a first step toward the identification of the molecular machinery that allows amino acid uptake and organic N transport in AM symbiosis, we have identified a cDNA sequence (GmosAAP1) from the AM fungus G. mosseae that shows a remarkable similarity to amino acid transporters. According to the transporter classification, GmosAAP1 belongs to the amino acid/polyamine organocation superfamily. Most of these transporters exhibit a uniform topology, with 12 putative α-helical transmembrane domains and cytoplasmically located N- and C-terminal hydrophilic regions (Wipf et al., 2002b). The number and distribution of the transmembrane domains of GmosAAP1 mirror an identical topology. The GmosAAP1 protein shows a limited similarity, in terms of primary sequence and structure, to vacuolar amino acid transporters described in yeast (Russnak et al., 2001). Together, the data suggest that GmosAAP1 is a plasma membrane protein. The identification of a partial sequence showing high similarity to GmosAAP1 from in vitro-grown G. intraradices clearly supports the authenticity of the sequence. Since in yeast and filamentous fungi AAPs usually belong to a multigene family (Wipf et al., 2002b), it is likely that G. mosseae also possesses additional AAPs.
GmosAAP1 Encodes a Functional Amino Acid Transporter
Yeast has provided a genuine heterologous expression system for the characterization of many nutrient and metabolite transporters from animals, plants, and nonyeast fungi. This instrument is particularly valuable for organisms that are currently recalcitrant to genetic transformation, such as AM fungi. In fact, the few transporters described in this group of fungi to date have been functionally characterized using yeast mutants (Harrison and van Buuren, 1995; Gonzalez-Guerrero et al., 2005; López Pedrosa et al., 2006).
The 22Δ8AA mutant strain expressing GmosAAP1 was able to grow using some amino acids as a single N source, thus confirming the amino acid transport capability of GmosAAP1. The uptake rate for Pro was concentration dependent and displayed saturation kinetics, with a Km value (18.8 μm) that is comparable with amino acid concentrations found in the soil (Scheller, 1996). The GmosAAP1 activity has an optimum at approximately pH 4.5 to 5.0, which is consistent with the pH optimum described for the uptake of Glu and Gln by mycelia of the ectomycorrhizal fungus Paxillus involutus (Chalot et al., 1995). The strong dependence on the presence of Glc and a proton gradient indicates that GmosAAP1-mediated transport requires a secondary active transport mechanism that is similar to those of its yeast homologs (Opekarová et al., 1993). Competition experiments indicate a preferential affinity of GmosAAP1 for all nonpolar and hydrophobic amino acids, suggesting a relatively narrow substrate specificity.
GmosAAP1 Is Expressed in Extraradical Structures
Qualitative RT-PCR assays indicated that GmosAAP1 was expressed in the external mycelium but not in the intraradical fungal structures of plants treated with millimolar nitrate concentrations. GmosAAP1 transcripts were not detected in sporocarps germinated in a water/agar medium; however, for a more comprehensive view, the expression in asymbiotic stages should be studied considering other growth conditions (e.g. exposure to different N sources).
GmosAAP1 expression in extraradical hyphae responded to the presence of different concentrations of organic N. In spite of a certain variability in the two independent biological samples, an induction was constantly observed after the two amino acid pool treatments in comparison with the 0 m treatment. In particular, GmosAAP1 was strongly up-regulated after the 2 mm treatment. It is worth noting that the amino acid pool contained three amino acids, Leu, Ala, and Tyr, which, from the competition studies obtained in the yeast mutant (Fig. 2), are likely to be substrates of GmosAAP1. This might reflect a common mechanism of AAP regulation, that is, the transcriptional induction by the substrate (Grauslund et al., 1995).
The hypothesis that organic N acts as a signaling molecule in AM fungi is also supported by the observation that limiting organic N conditions induce a specific response at the transcriptional level in extraradical structures of G. intraradices (Cappellazzo et al., 2007).
The main function of GmosAAP1, as indicated by the putative localization on the plasma membrane, its expression in extraradical hyphae, and the biochemical properties in terms of Km, may be the uptake of amino acids from the soil solution. A similar role has also been suggested for the gene identified in H. cylindrosporum, although detailed expression studies were not performed (Wipf et al., 2002a). AmAAP1 gene expression has only been studied in A. muscaria mycelium grown in pure culture, and no data are available concerning its expression during its interaction with host plants (Nehls et al., 1999).
CONCLUSION
N nutrition of AM plants, in particular with regard to organic N sources, remains a largely unexplored area. With the identification of a gene that encodes a functional amino acid transporter, we offer experimental evidence that the AM fungus G. mosseae, which is extensively present in agricultural systems and often used as a component of commercial inocula, possesses molecular tools for the uptake not only of phosphate (Benedetto et al., 2005) but also of organic N from the soil. This finding could contribute to a better understanding of the organic N metabolism in AM fungi and lead to new important questions on its impact on host plant nutrition.
MATERIALS AND METHODS
Biological Material
The Glomus mosseae ‘BEG 12’ (International Bank for the Glomeromycota; http://www.kent.ac.uk/bio/beg/) inoculum (sporocarps and mycorrhizal roots) was obtained from Biorize. For germination, the sporocarps were collected with forceps, surface sterilized with 3% (w/v) chloramine-T, and placed in water-agar (1.5%, w/v) at 25°C in the dark. The inoculum was also used in pot culture to obtain mycorrhizal plants. Cucumis sativus ‘Marketmore’ seeds were previously surface sterilized for 30 s in 98% sulfuric acid and then rinsed several times with distilled sterile water. The seeds were left to germinate for 7 d on water-agar plates at 24°C in the dark. The seedlings were then transferred to 0.3-L plastic pots containing heat-sterilized (3 h at 180°C) quartz sand and G. mosseae inoculum (1:10, v/v) and kept in a growth chamber for a 13-h photoperiod at 20°C/24°C dark/light. The plants were watered every second day with water and the other day with a Long Ashton solution (Hewitt, 1966) containing a low phosphorus concentration (3.2 μm NaHPO4·12H2O). After 3 months, when the mycorrhization was fully established, the roots were carefully washed and then submerged for 72 h in a modified Long Ashton solution. According to the treatment, this solution was used directly (no N sample) or two different concentrations (2 μm or 2 mm) of KNO3, (NH4)2SO4, or a pool of amino acids with different biochemical properties (Leu, Ala, Asn, Lys, and Tyr) were added.
Two sets of independent mycorrhizal plants were treated. The extraradical mycelium and mycorrhizal root pieces, devoid of external hyphae, were collected with forceps under the stereomicroscope lens and immediately frozen in liquid N.
Root organ cultures of Agrobacterium rhizogenes (Ri T-DNA)-transformed carrot (Daucus carota) roots were used for monoxenic cultivation of the AM fungus Glomus intraradices ‘MUCL 43194’ (Declerck et al., 2005) obtained from GINCO (http://www.mbla.ucl.ac.be/ginco-bel). The carrot roots inoculated with G. intraradices were grown in two-compartment petri dishes on Strullu-Romand medium (Declerck et al., 1998) and solidified with 3 g L−1 Phytagel. The petri dishes were incubated horizontally in an inverted position at 27°C in the dark for 3 to 4 weeks. The G. intraradices extraradical structures were recovered by solubilizing the solid Strullu-Romand medium with sterile 50 mm Tris-HCl, pH 7. The extraradical structures were collected with forceps, rinsed with sterilized water, recovered by vacuum filtration on a sterilized polyvinylidene fluoride membrane (Durapore GVWP; Millipore), and immediately frozen in liquid N and stored at −80°C until used.
DNA and RNA Extractions
Genomic DNA was extracted from approximately 100 mg of roots using the hexadecyl-trimethyl-ammonium bromide protocol (Henrion et al., 1994). The roots were ground in liquid N to a fine powder, a hexadecyl-trimethyl-ammonium bromide extraction buffer was immediately added, and the samples were incubated at 65°C for 1 h. Crude lysates were extracted once with phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v) and once with chloroform. Aqueous phases were precipitated with 1.5 volume of isopropanol (−20°C overnight). The DNA pellet was washed with 70% ethanol, dried, and then resuspended in 20 μL of water.
The genomic DNA from G. intraradices extraradical mycelium from in vitro cultures was obtained with the DNAeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions.
RNA was extracted from approximately 100 sporocarps germinated in water-agar, 100 mg of mycorrhizal roots, 100 mg of nonmycorrhizal roots, and 0.2 to 0.3 mg of G. mosseae extraradical mycelium using the SV Total RNA Isolation System Kit (Promega). The RNA was precipitated with 6 m LiCl and resuspended in 20 μL of sterile water. The RNA samples were routinely checked for DNA contamination by RT-PCR analyses conducted using the 28S rRNA universal primers NS1/NS2 (White et al., 1990) and the One-Step RT-PCR Kit (Qiagen) according to Cappellazzo et al. (2007). The first strand of cDNA was synthesized using Sensiscript reverse transcriptase (Qiagen) and random primers according to the manufacturer's instructions.
PCR and RT-PCR
The PCR experiments were carried out with the oligonucleotides NITPLUS (5′-GCCCTGCGCTTCTTCATCGG-3′) and NITMINUS (5′-AAATGGCCGGCATGACGAAG-3′). The primers were designed by Dr. E. Soragni (University of Parma, Italy). cDNAs from spores, germinated spores, or external mycelium of Gigaspora margarita ‘BEG 34’, Gigaspora rosea ‘BEG 9’, G. mosseae ‘BEG 12’, and G. intraradices ‘DAOM181602’, kindly provided by Prof. P. Franken (Institute for Vegetable and Ornamental Crops, Grossbeeren, Germany), were used as templates. PCR was carried out in a final volume of 30 μL containing 10 mm Tris-HCl, pH 8.3, 50 mm KCl, 1.1 mm MgCl2, 0.01% gelatin, 200 μm of each dNTP, 1 μm of each primer, 50 to 100 ng of cDNA, and 1 unit of REDTaqTM DNA polymerase (Sigma). A PCR program was conducted in a Perkin-Elmer GeneAmp 9700 thermal cycler according to these parameters: 95°C for 5 min (one cycle), 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min (35 cycles), and 72°C for 5 min (one cycle). The negative controls for all PCR experiments consisted of reaction mixtures from which template DNA was omitted. The PCR products were separated on a 1.2% Tris-acetate EDTA/0.5× agarose gel and visualized by ethidium bromide staining.
Specific primers for G. mosseae 28S rRNA (5.21/NDL22; van Tuinen et al., 1998) and GmosAAP1 G1 (5′-CTGGAGAGAAGATATCAAC-3′) and G2 (5′-CATGCCCTGAGGAGCAGCG-3′) were tested on DNA and cDNA samples. The PCR program was conducted in a Perkin-Elmer GeneAmp 9700 thermal cycler according to these parameters: 95°C for 3 min (one cycle), 92°C for 45 s, annealing temperature (55°C for NS1/NS2, 64°C for 5.21/NDL22, and 54°C for G1/G2) for 45 s, 72°C for 45 s (35 cycles), and 72°C for 5 min (one cycle).
PCR was carried out on G. intraradices genomic DNA with APH1 (5′-GTCGTCGCTGCTTTCTCCTTCGG-3′) and APEF1 (5′-CAAGGAGACCAAAGGCGATCTG-3′) primers using an annealing temperature of 48°C.
The PCR products were purified from agarose gels using the QIAEX II gel extraction kit (Qiagen) and directly cloned in the pGEM-T vector (Promega). Plasmid DNA was extracted using the Qiagen Mini kit and sequenced by Genelab (Enea).
RACE-PCR
The amplification of GmosAAP1 5′ and 3′ cDNA ends was performed by RACE using the SMART RACE cDNA amplification kit (BD Biosciences) utilizing a combination of specific forward primers, AT5 (5′-CCCTGCGCTTCTTCATCGGCTATCTC-3′) and AT7 (5′-GCACACTGCTTGGCCCTCGCTGAGATGG-3′), and reverse primers, AP1 (5′-GACCGACCATCTCAGTACCACCG-3′) and APH2 (5′-CAATTCGCCAGGAGACCTGCTTG-3′), in nested PCR. The reactions were carried out in a GeneAmp 9700 thermal cycler according to the manufacturer's instructions.
Semiquantitative and Real-Time RT-PCR
Semiquantitative RT-PCR was performed on two independent biological samples. RNA samples were calibrated using ribosomal primers (5.21/NDL22); specific AAP primers (G1/G2) were then used to evaluate the GmosAAP1 mRNA level in each treatment. PCR was allowed to proceed for a different number of cycles to determine the exponential amplification phase. Reactions were carried out in a final volume of 50 μL using the previously described conditions. RT-PCR experiments were conducted using two technical replicates.
Individual real-time reactions were assembled in a final volume of 20 μL with 0.15 μm of each oligonucleotide, 10 μL of 2× iQ SYBR Green Supermix (Bio-Rad), plus an appropriate volume of each cDNA preparation. The following primers were used: 5.21 and 28S1 (5′-CACTTCAGTACGAGATCGAAG-3′) for the fungal 28S ribosomal gene, Tef1 (5′-GCAGAACGTGAGCGTGGTAT-3′) and Tef2 (5′-ACCAGTACCGGCAGCAATAA-3′) for the fungal elongation factor gene, and AAP1 (5′-TACTCCTCCCACCGATTACG-3′) and AAP2 (5′-CCGATGATGAGATAGCCGAT-3′) for the GmosAAP1 gene.
The PCR cycling program (15 s at 95°C followed by 30 s at 62°C for 28S rRNA and Tef genes and at 64°C for the GmosAAP1 gene) included a heating step (3 min at 95°C) at the beginning of each run. Real-time RT-PCR was carried out with an ICycler apparatus (Bio-Rad). A melting curve (55°C–95°C, with a heating rate of 0.5°C per 10 s and continuous fluorescence measurement) was recorded at the end of each run to assess amplification product specificity (Ririe et al., 1997). All of the reactions were performed with at least two technical replicates, and only comparative threshold cycle values with a sd that did not exceed 0.3 were considered. The comparative threshold cycle method (Rasmussen, 2001) was used to calculate the GmosAAP relative expression level.
Sequence Analyses
Sequence analyses were performed with Sequencher (Gene Codes Corporation), BLASTX software available from the National Center for Biotechnology Information (Altschul et al., 1997), and ClustalW (Thompson et al., 1994). The secondary structure was predicted using HMMTOP (Tusnady and Simon, 2001), TMHMM (Sonnhammer et al., 1998), SOUSI (Hirokawa et al., 1998), and TMPRED (Hofmann and Stoffel, 1993). The protein family, domain, and functional sites were searched using the InterProScan program (Mulder et al., 2003).
Phylogenetic analyses were performed using version 3.1 of the MEGA (Molecular Evolutionary Genetic Analysis) program available on the Web (http://www.megasoftware.net/mega.html) and analyzed by the neighbor-joining algorithm (Kumar et al., 2004).
Yeast Growth and Transformation
The yeast strain used was a mutant lacking multiple amino acid uptake systems, 22Δ8AA (Matα gap1-1 put4-1 uga4-1 Δcan1 Δapl1 Δlyp1 Δhip1 Δdip5 ura3-1; Fischer et al., 2002). The full-length cDNA of GmosAAP1 was cloned in the yeast expression vector pDR196 (Wipf et al., 2003), exploiting the Saccharomyces cerevisiae homologous recombination process. Approximately 100 ng of XhoI-linearized pDR196 vector was mixed with 100 ng of the PCR product of GmosAAP1 obtained using specific primers (For, 5′-GCTGCAGGAATTCGATATCAAGCTTATCGATACCGTCGACCATGTACCACCGGGGAACCAAGAG-3′, and Rev, 5′-TACGACTCACTATAGGGCGAATTGGGTACCGGGCCCCCCGTACATGCTTAGTAAAAGC-3′) containing an extension of 40 bp of sequence homology with the vector sequence flanking the XhoI restriction site. The mixture was used to transform the ΔYAP yeast (Wu et al., 1993). Colonies carrying recombinant plasmids (pDR196-GmosAAP1) were screened using a selective ura− medium.
The pDR196-GmosAAP1 construct was used to transform the 22Δ8AA yeast strain. Transformants were selected on solid minimal synthetic defined medium supplemented with 10 mm (NH4)2SO4. The plasmid DNA was isolated and reintroduced into the mutant strain 22Δ8AA. The cDNA clone GmosAAP1 was able to restore the growth of the mutant under selective conditions. The empty vector pDR196 was used as a negative control.
Transport Measurements
S. cerevisiae cells were grown to a logarithmic phase for uptake studies. The cells were harvested at an optical density at 600 nm of 0.5, washed twice in water, and resuspended in buffer A (0.6 m sorbitol and 50 mm potassium phosphate at the desired pH) to a final optical density at 600 nm of 5. Prior to the uptake measurements, the cells were supplemented with 100 mm Glc and incubated for 5 min at 30°C. To start the reaction, 100 μL of this cell suspension was added to 100 μL of the same buffer containing at least 0.46 kBq [14C]Pro with a specific activity of 8.58 GBq/mmol (Amersham) and unlabeled amino acid to the concentrations used in the experiments. Sample aliquots of 50 μL were removed after 30, 60, 120, and 240 s, transferred to 4 mL of ice-cold buffer A, filtered on glass fiber filters, and washed twice with 4 mL of buffer A. The uptake of carbon-14 was determined by liquid scintillation spectrometry. Competition for Pro uptake was performed by adding a 5-fold molar excess of the respective competitors to 18.8 μm Pro.
For analysis of the pH dependence, incubations were performed in 100 mm potassium phosphate buffer adjusted to the different pH values, 100 mm Glc, and 150 μm [14C]Pro. The influence of plasma membrane energization on the uptake rate of [14C]Pro was analyzed by incubating the yeast cells for 5 min in the presence of 100 mm Glc (control), without Glc, or with Glc and 0.1 mm 2,4-dinitrophenol, 0.1 mm diethylstilbestrol, 0.1 mm carbonyl cyanide m-chlorophenylhydrazone, or 0.1 mm vanadate. The transport measurements were repeated independently and represent means of at least three experiments.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY882560 (GmosAAP1) and AM940008 (GintAAP1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ClustalW alignment and predicted hydrophobicity profile of GmosAAP1.
Supplemental Figure S2. Unrooted phylogram obtained using the neighbor-joining algorithm.
Supplementary Material
Acknowledgments
We thank Dr. P. Franken for the cDNAs and for the G. mosseae Tef sequence and Elodie Oger for providing the G. intraradices in vitro cultures.
This work was supported by the Italian MIUR Projects (Prin 2006, Soil Sink) and IPP-Consiglio Nazionale delle Ricerche (Biodiversity National Project) to P.B. and the University of Torino (60% Project, 2007) to L.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Luisa Lanfranco (luisa.lanfranco@unito.it).
The online version of this article contains Web-only data.
References
- Alexander IJ (2007) A knight of symbiosis. New Phytol 176 499–501 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetto A, Magurno F, Bonfante P, Lanfranco L (2005) Expression profiles of a phosphate transporter gene (GmosPT) from the endomycorrhizal fungus Glomus mosseae. Mycorrhiza 15 620–627 [DOI] [PubMed] [Google Scholar]
- Bennett JN, Prescott CE (2004) Organic and inorganic nitrogen nutrition of western red cedar, western hemlock and salal in mineral N-limited cedar-hemlock forests. Oecologia 141 468–476 [DOI] [PubMed] [Google Scholar]
- Breuninger M, Trujillo CG, Serrano E, Fischer R, Requena N (2004) Different nitrogen sources modulate activity but not expression of glutamine synthetase in arbuscular mycorrhizal fungi. Fungal Genet Biol 41 542–552 [DOI] [PubMed] [Google Scholar]
- Bucher M (2007) Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol 173 11–26 [DOI] [PubMed] [Google Scholar]
- Cappellazzo G, Lanfranco L, Bonfante P (2007) A limiting source of organic nitrogen induces specific transcriptional responses in the extraradical structures of the endomycorrhizal fungus Glomus intraradices. Curr Genet 51 59–70 [DOI] [PubMed] [Google Scholar]
- Chalot M, Javelle A, Blaudez D, Lambilliote R, Cooke R, Sentenac H, Wipf D, Botton B (2002) An update on nutrient transport processes in ectomycorrhizas. Plant Soil 244 165–175 [Google Scholar]
- Chalot M, Kytoviita M, Brun A, Finlay RD, Söderstrom B (1995) Factors affecting amino acid uptake by the ectomycorrhizal fungus Paxillus involutus. Mycol Res 99 1131–1138 [Google Scholar]
- Chen A, Chambers SM, Cairney JWG (1999) Utilisation of organic nitrogen and phosphorus sources by mycorrhizal endophytes of Woollsia pungens (Cav.) F. Muell. (Epacridaceae). Mycorrhiza 8 181–187 [Google Scholar]
- Cliquet JB, Murray PJ, Boucaud J (1997) Effect of the arbuscular mycorrhizal fungus Glomus fasciculatum on the uptake of amino nitrogen by Lolium perenne. New Phytol 137 345–349 [DOI] [PubMed] [Google Scholar]
- Cruz C, Egsgaard H, Trujillo C, Ambus P, Requena N, Martins-Loucao MA, Jacobsen I (2007) Enzymatic evidence for the key role of arginine in nitrogen translocation by arbuscular mycorrhizal fungi. Plant Physiol 144 782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declerck S, Strullu DG, Fortin JA, editors (2005) In Vitro Culture of Mycorrhizas. Springer-Verlag, Heidelberg
- Declerck S, Strullu DG, Plenchette C (1998) Monoxenic culture of the intraradical forms of Glomus sp. isolated from a tropical ecosystem: a proposed methodology for germplasm collection. Mycologia 90 579–585 [Google Scholar]
- Faure S, Cliquet JB, Thephany G, Boucaud J (1998) Nitrogen assimilation in Lolium perenne colonized by the arbuscular mycorrhizal fungus Glomus fasciculatum. New Phytol 138 411–417 [DOI] [PubMed] [Google Scholar]
- Fischer WN, Loo DDF, Koch W, Ludewig U, Boorer KJ, Tegeder M, Rentsch D, Wright EM, Frommer WB (2002) Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J 29 717–731 [DOI] [PubMed] [Google Scholar]
- Frank AB (1894) Die Bedeutung der Mykorrhizapilze für die gemeine Kiefer. Forstwissenschaftliche Centralblat 16 1852–1890 [Google Scholar]
- Girlanda M, Perotto S, Bonfante P (2007) Mycorrhizal fungi: their habitats and nutritional strategies. In CP Kubicek, IS Druzhinina, eds, The Mycota. IV. Environmental and Microbial Relationships, Ed 2. Springer-Verlag, Berlin, pp 229–256
- Gonzalez-Guerrero M, Azcon-Aguilar C, Mooney M, Valderas A, MacDiarmid CW, Eide DJ, Ferrol N (2005) Characterization of a Glomus intraradices gene encoding a putative Zn transporter of the cation diffusion facilitator family. Fungal Genet Biol 42 130–140 [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Pfeffer PE, Jin H, Abubaker J, Douds DD, Allen JW, Bucking H, Lammers PJ, Shachar-Hill Y (2005) Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435 819–823 [DOI] [PubMed] [Google Scholar]
- Grauslund M, Didion T, Kielland-Brandt MC, Andersen HA (1995) BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim Biophys Acta 1269 275–280 [DOI] [PubMed] [Google Scholar]
- Guidot A, Verner MC, Debaud JC, Marmeisse R (2005) Intraspecific variation in use of different organic nitrogen sources by the ectomycorrhizal fungus Hebeloma cylindrosporum. Mycorrhiza 15 167–177 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, van Buuren ML (1995) A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature 378 626–629 [DOI] [PubMed] [Google Scholar]
- Hawkins H, Johansen A, George E (2000) Uptake and transport of organic and inorganic nitrogen by arbuscular mycorrhizal fungi. Plant Soil 226 275–285 [Google Scholar]
- Henrion B, Chevalier G, Martin F (1994) Typing truffle species by PCR amplification of the ribosomal DNA spacers. Mycol Res 98 37–43 [Google Scholar]
- Hewitt EJ (1966) Sand and Water Culture Methods Used in the Study of Plant Nutrition. Technical Communication No. 22. Commonwealth Agriculture Bureau, East Malling, UK, pp 431–432
- Hirokawa T, Seah BC, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14 378–379 [DOI] [PubMed] [Google Scholar]
- Hodge A, Campbell CD, Fitter AH (2001) An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature 413 297–299 [DOI] [PubMed] [Google Scholar]
- Hofmann H, Stoffel W (1993) TMbase: a database of membrane spanning protein segments. Biol Chem 374 166 [Google Scholar]
- Jack DL, Paulsenã TN, Saier MH Jr (2000) The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146 1797–1814 [DOI] [PubMed] [Google Scholar]
- Javot H, Pumplin N, Harrison MJ (2007) Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ 30 310–322 [DOI] [PubMed] [Google Scholar]
- Jin H, Pferrer PE, Douds DD, Piotrowski E, Lammers PJ, Shachar-Hill Y (2005) The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. New Phytol 168 687–696 [DOI] [PubMed] [Google Scholar]
- Johansen A, Finlay RD, Olsson PA (1996) Nitrogen metabolism of the external hyphae of the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 133 705–712 [Google Scholar]
- Joner EJ, Jakobsen I (1995) Growth and extracellular phosphatase activity of arbuscular mycorrhizal hyphae is influenced by soil organic matter. Soil Biol Biochem 27 1153–1159 [Google Scholar]
- Kaldorf M, Schmelzer E, Bothe H (1998) Expression of maize and fungal nitrate reductase genes in arbuscular mycorrhiza. Mol Plant Microbe Interact 11 439–448 [DOI] [PubMed] [Google Scholar]
- Kaldorf M, Zimmer W, Bothe H (1994) Genetic evidence for the occurrence of assimilatory nitrate reductase in arbuscular mycorrhizal and other fungi. Mycorrhiza 5 23–28 [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5 150–163 [DOI] [PubMed] [Google Scholar]
- López-Pedrosa A, Gonzalez-Guerrero M, Valderas A, Azcon-Aguilar C, Ferrol N (2006) GintAMT1 encodes a functional high-affinity ammonium transporter that is expressed in the extraradical mycelium of Glomus intraradices. Fungal Genet Biol 43 102–110 [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res 31 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T, Avolio M, Olivi M, Benjdia M, Rikirsch E, Kasaras A, Fitz M, Chalot M, Wipf D (2007) Nitrogen transport in the ectomycorrhiza association: the Hebeloma cylindrosporum-Pinus pinaster model. Phytochemistry 68 41–51 [DOI] [PubMed] [Google Scholar]
- Näsholm T, Ekblad A, Nordin A, Giesler R, Högberg M, Högberg P (1998) Boreal forest plants take up organic nitrogen. Nature 392 914–916 [Google Scholar]
- Nehls U, Kleber R, Wiese J, Hampp R (1999) Isolation and characterization of a general amino acid permease from the ectomycorrhizal fungus Amanita muscaria. New Phytol 144 343–349 [Google Scholar]
- Okumoto S, Schmidt R, Tegeder M, Fischer WN, Rentsch D, Frommer WB, Koch W (2002) High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J Biol Chem 277 45338–45346 [DOI] [PubMed] [Google Scholar]
- Opekarová M, Caspari T, Tanner W (1993) Unidirectional arginine transport in reconstituted plasma-membrane vesicles from yeast overexpressing CAN1. Eur J Biochem 211 683–688 [DOI] [PubMed] [Google Scholar]
- Rasmussen R (2001) Quantification on the LightCycler instrument. In S Meuer, C Wittwer, K Nakagawara, eds, Rapid Cycle Real-Time PCR: Methods and Applications. Springer-Verlag, Heidelberg, pp 21–34
- Ravnskov S, Larsen J, Olsson PA, Jakobsen I (1999) Effects of various organic compounds growth and phosphorus uptake of an arbuscular mycorrhizal fungus. New Phytol 141 517–524 [Google Scholar]
- Read DJ (1996) The structure and function of the ericoid mycorrhizal root. Ann Bot (Lond) 77 365–374 [Google Scholar]
- Read DJ, Perez-Moreno J (2003) Mycorrhizas and nutrient cycling in ecosystems: a journey towards relevance? New Phytol 157 475–492 [DOI] [PubMed] [Google Scholar]
- Redecker D, Hijri M, Dulieu H, Sanders IR (1999) Phylogenetic analysis of a dataset of fungal 5.8S rDNA sequences shows that highly divergent copies of internal transcribed spacers reported from Scutellospora castanea are of ascomycete origin. Fungal Genet Biol 28 238–244 [DOI] [PubMed] [Google Scholar]
- Regenberg B, During-Olsen L, Kielland-Brandt MC, Holmberg S (1999) Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr Genet 36 317–328 [DOI] [PubMed] [Google Scholar]
- Ririe KM, Rasmussen RT, Wittwer CT (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem 245 154–160 [DOI] [PubMed] [Google Scholar]
- Russnak R, Konczal D, McIntire SL (2001) A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J Biol Chem 276 23849–23857 [DOI] [PubMed] [Google Scholar]
- Sawyer NA, Chambers SM, Cairney JW (2003) Utilisation of inorganic and organic nitrogen sources by Amanita species native to temperate eastern Australia. Mycol Res 107 413–420 [DOI] [PubMed] [Google Scholar]
- Scheller E (1996) Aminosäuregehalte von Ap- und Ah-Horizonten verschiedener Böden und deren Huminsäuren- und Fulvosäuren-Fraktionen. Mitteilungen Deutsche Bodenkundliche Gesellschaft 81 417–424 [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6 175–182 [PubMed] [Google Scholar]
- St. John TV, Coleman DC, Reid CPP (1983. a) Association of vesicular-arbuscular mycorrhizal hyphae with soil organic particles. Ecology 64 957–959 [Google Scholar]
- St. John TV, Coleman DC, Reid CPP (1983. b) Growth and spatial distribution of nutrient-absorbing organs: selective exploitation of soil heterogeneity. Plant Soil 71 487–493 [Google Scholar]
- Stack JH, DeWald DB, Takegawa K, Emr SD (1995) Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol 129 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck C, Ernst M, Hahn M (2002) Characterization of a developmentally regulated amino acid transporter (AAT1p) of the rust fungus Uromyces fabae. Mol Plant Pathol 3 23–30 [DOI] [PubMed] [Google Scholar]
- Subramanian KS, Charest C (1998) Arbuscular mycorrhizae and nitrogen assimilation in maize after drought and recovery. Plant Physiol 102 285–296 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbett M, Hartley M, Hartley S (2000) Comparative growth of ectomycorrhizal basidiomycetes (Hebeloma spp.) on organic and inorganic nitrogen. J Basic Microbiol 40 393–395 [PubMed] [Google Scholar]
- Tibbett M, Sanders FE (2002) Ectomycorrhizal symbiosis can enhance plant nutrition through improved access to discrete organic nutrient patches of high resource quality. Ann Bot (Lond) 89 783–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint JP, St-Arnaud M, Charest C (2004) Nitrogen transfer and assimilation between the arbuscular mycorrhizal fungus Glomus intraradices Schenck & Smith and Ri T-DNA roots of Daucus carota L. in an in vitro compartmented system. Can J Microbiol 50 251–260 [DOI] [PubMed] [Google Scholar]
- Trip H, Evers ME, Konings WN, Driessen AJ (2002) Cloning and characterization of an aromatic amino acid and leucine permease of Penicillium chrysogenum. Biochim Biophys Acta 1565 73–80 [DOI] [PubMed] [Google Scholar]
- Trouvelot A, Kough JL, Gianinazzi-Pearson V (1986) Mesure du taux de mycorhization VA d'un système radiculaire: recherche de méthodes d'estimation ayant une signification fontionelle. In V Gianinazzi-Pearson, S Gianinazzi, eds, Physiological and Genetic Aspects of Mycorrhiza. INRA Press, Paris, pp 217–221
- Turnbull MH, Schmidt S, Erskine PD, Richards S, Stewart GR (1996) Root adaptation and nitrogen source acquisition in natural ecosystems. Tree Physiol 16 941–948 [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17 849–850 [DOI] [PubMed] [Google Scholar]
- Van Belle D, Andre B (2001) A genomic view of yeast membrane transporters. Curr Opin Cell Biol 13 389–398 [DOI] [PubMed] [Google Scholar]
- van Tuinen D, Jacquot E, Zhao B, Gollotte A, Gianinazzi-Pearson V (1998) Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol Ecol 7 879–887 [DOI] [PubMed] [Google Scholar]
- Väre H, Vestberg M, Ohtonen R (1997) Shifts in mycorrhiza and microbial activity along an oroartic altitudinal gradient in northern Fennoscandia. Arct Alp Res 29 93–104 [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In MA Innis, DH Gelfand, JJ Sninsky, TJ White, eds, PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, pp 315–322
- Wipf D, Benjdia M, Rikirsch E, Zimmermann S, Tegeder M, Frommer WB (2003) An expression cDNA library for suppression cloning in yeast mutants, complementation of a yeast his4 mutant, and EST analysis from the symbiotic basidiomycete Hebeloma cylindrosporum. Genome 46 177–181 [DOI] [PubMed] [Google Scholar]
- Wipf D, Benjdia M, Tegeder M, Frommer WB (2002. a) Characterization of a general amino acid permease from Hebeloma cylindrosporum. FEBS Lett 528 119–124 [DOI] [PubMed] [Google Scholar]
- Wipf D, Ludewig U, Tegeder M, Rentsch D, Koch W, Frommer WB (2002. b) Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem Sci 27 139–147 [DOI] [PubMed] [Google Scholar]
- Wu A, Wemmie JA, Edgington NP, Goebl M, Guevara JL, Moye-Rowley WS (1993) Yeast bZIP proteins mediate pleiotropic drug and metal resistance. J Biol Chem 268 18850–18858 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.