Abstract
Color changes often accompany the onset of ripening, leading to brightly colored fruits that serve as attractants to seed-dispersing organisms. In many fruits, including tomato (Solanum lycopersicum) and pepper (Capsicum annuum), there is a sharp decrease in chlorophyll content and a concomitant increase in the synthesis of carotenoids as a result of the conversion of chloroplasts into chromoplasts. The green-flesh (gf) and chlorophyll retainer (cl) mutations of tomato and pepper, respectively, are inhibited in their ability to degrade chlorophyll during ripening, leading to the production of ripe fruits characterized by both chlorophyll and carotenoid accumulation and are thus brown in color. Using a positional cloning approach, we have identified a point mutation at the gf locus that causes an amino acid substitution in an invariant residue of a tomato homolog of the STAY-GREEN (SGR) protein of rice (Oryza sativa). Similarly, the cl mutation also carries an amino acid substitution at an invariant residue in a pepper homolog of SGR. Both GF and CL expression are highly induced at the onset of fruit ripening, coincident with the ripening-associated decline in chlorophyll. Phylogenetic analysis indicates that there are two distinct groups of SGR proteins in plants. The SGR subfamily is required for chlorophyll degradation and operates through an unknown mechanism. A second subfamily, which we have termed SGR-like, has an as-yet undefined function.
Color change is one of the most dramatic events occurring in fleshy fruits as they begin to ripen, serving as a signal to seed-dispersing fauna that the fruit is ripe, palatable, and nutritious. Fleshy fruits predominantly accumulate carotenoids, anthocyanins, and flavanoids, and the de novo synthesis of these compounds at the onset of ripening is preceded by, or occurs concomitantly with, the degradation of chlorophyll (Seymour et al., 1993). An exception to this generalization occurs in banana (Musa spp.), where the degradation of chlorophyll at the onset of ripening leads to the unmasking of the yellow-pigmented xanthophylls that are already present in immature fruit (Seymour et al., 1993; Drury et al., 1999). Therefore, whereas the degradation of chlorophyll is one of the key events accompanying the reprogramming of cellular metabolism at the onset of fruit ripening, these two events are not necessarily interdependent. While mechanisms of chlorophyll degradation have been elucidated, the regulation and role of chlorophyll degradation in the ripening process and overall quality of fruits remain poorly understood.
Chlorophyll degradation is a highly controlled sequential process that converts the fluorescent chlorophyll molecule into nonfluorescent catabolites that are stored within the vacuole. The basic chlorophyll breakdown pathway is composed of the following steps: chlorophyll b→chlorophyll a→chlorophyllide a→pheophorbide a→red chlorophyll catabolite→fluorescent chlorophyll catabolite→nonfluorescent chlorophyll catabolite (Hortensteiner, 2006). The enzymatic components that catalyze the steps in this pathway and their regulation are not fully understood, but in recent years several have been identified via genetic and biochemical approaches. In higher plants, chlorophyll b is initially converted to chlorophyll a by the action of chlorophyll b reductase. A possible candidate for the enzyme performing this conversion has recently been proposed through the identification of the gene at the non-yellow coloring1 (nyc1) locus of rice (Oryza sativa) that encodes a chloroplast-targeted short-chain dehydrogenase/reductase (Kusaba et al., 2007). The enzyme chlorophyllase catalyzes the conversion of chlorophyll a into chlorophyllide and phytol and is thought to be the rate-limiting step within the chlorophyll breakdown pathway (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999; Harpaz-Saad et al., 2007). Chlorophyllase contains a Ser lipase motif and mutation of conserved residues within this motif reduces enzyme activity (Tsuchiya et al., 2003; Harpaz-Saad et al., 2007). Pheophorbide a oxygenase (PAO) is an Fe-dependent monooxygenase and was identified via the characterization of the lesion mimic mutants accelerated cell death1 (acd1) and lethal leaf spot1 (lls1) of Arabidopsis (Arabidopsis thaliana) and maize (Zea mays), respectively (Gray et al., 1997; Pruzinska et al., 2003). Mutants at these loci exhibit light-dependent cell death phenotypes caused by the generation of reactive oxygen species as a result of photoactivation of accumulated pheophorbide a. PAO converts pheophorbide a into red chlorophyll catabolite, which is in turn converted into fluorescent chlorophyll catabolite by red chlorophyll catabolite reductase (RCCR), a novel enzyme that is distantly related to bilin reductases (Wuthrich et al., 2000). Like PAO, mutations in RCCR, as defined by the acd2 locus, display a lesion mimic phenotype (Mach et al., 2001).
Stay-green mutants have been identified in several plant species and classified on the basis of their chlorophyll retention and general senescence phenotypes (Thomas and Howarth, 2000). Class C stay-green mutants display inhibited chlorophyll degradation, but other aspects of senescence proceed normally, suggesting that these loci specifically affect chlorophyll degradation. One group of class C mutants identified in Festuca pratensis, pea (Pisum sativum), rice, and Arabidopsis has reduced PAO activity and stable pigment-protein complexes within the chloroplast (Thomas and Howarth, 2000; Park et al., 2007; Ren et al., 2007; Sato et al., 2007). Recently, these loci have been shown to encode a family of novel chloroplast-targeted proteins that may promote chlorophyll degradation via destabilization of protein-pigment complexes in the thylakoid membranes through an as-yet undefined mechanism (Armstead et al., 2006, 2007; Jiang et al., 2007; Park et al., 2007; Ren et al., 2007; Sato et al., 2007).
Fruit of the green-flesh (gf) mutant of tomato (Solanum lycopersicum) ripen to a muddy brown color due to the accumulation of lycopene coupled with a lack of chlorophyll degradation (Kerr, 1956). The lack of chlorophyll degradation in gf is not restricted to fruits because dark- and nutrient-induced chlorophyll loss in leaves is also compromised in the mutant (Akhtar et al., 1999). Together with the retention of chlorophyll in gf, the thylakoid grana, light-harvesting chlorophyll-binding proteins (LHCP), the Rubisco small subunit, and the 33-kD oxygen evolution protein also persist in mutant fruit and leaves (Cheung et al., 1993; Akhtar et al., 1999). However, these phenomena appear not to be caused by a general inhibition of senescence, but rather as a result of inhibition of chlorophyll degradation because senescence-associated marker genes appear to display normal expression patterns in gf (Akhtar et al., 1999). Therefore, gf can be classified as a class C stay-green mutant of tomato. Like gf, fruit of the chlorophyll retainer (cl) mutant of pepper (Capsicum annuum) have ripe fruits that are brown in color due to inhibition of chlorophyll degradation during ripening. Furthermore, the cl locus maps to the long arm of pepper chromosome 1, a region of the pepper genome that is orthologous to the long arm of tomato chromosome 8, where the gf locus is located (Tanksley et al., 1992; Efrati et al., 2005). Together, these data suggest the possibility that GF and CL may represent orthologous loci in tomato and pepper, respectively.
In this article, we describe the molecular identification of GF via the utilization of a positional cloning approach. Through DNA sequence analysis of the gf locus and complementation studies, we demonstrate unequivocally that GF is a member of the STAY-GREEN (SGR) gene family. In addition, we have demonstrated that a mutation in a pepper homolog of GF likely represents the basis for the stay-green phenotype of the pepper cl mutant.
RESULTS
Positional Cloning of GF
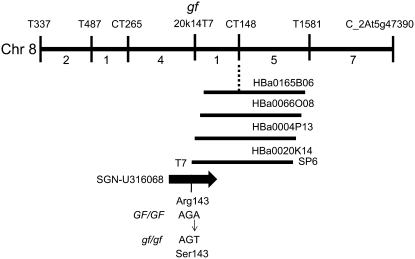
The gf locus has previously been mapped onto the classical genetic map of tomato at a position of approximately 45 cM on the long arm of chromosome 8 (Tanksley et al., 1992). With the aim of refining the map position of the gf locus, we generated two separate F2 populations segregating for tomato and Solanum pennellii alleles on chromosome 8 through crosses between a homozygous gf/gf mutant (LA3534) and two chromosome 8 S. pennellii introgression lines, IL8-2 (GF/GF; LA4074) and IL8-3 (GF/GF; LA4076) that span the long arm of chromosome 8 (Eshed and Zamir, 1994). During the course of this work, we became aware that the map position of the cl locus of pepper had been determined. The cl mutation possesses a phenotype that is very similar to that of the gf mutant in that chlorophyll is retained during fruit ripening, leading to fruit that are brown instead of red. Furthermore, genetic mapping of cl revealed that it is located on the long arm of pepper chromosome 1 between the flanking markers TG510 and CT28 (Efrati et al., 2005). Due to a reciprocal translocation between tomato and pepper chromosomes, this region of pepper corresponds to the long arm of tomato chromosome 8 (Livingstone et al., 1999). The similarity in phenotype and map position of gf and cl suggested that they may represent orthologous loci. We took advantage of the map position of the cl locus and performed a comparative mapping analysis of the gf locus of tomato. We screened 564 F2 plants from the gf/gf X IL8-3 population for recombination between the genetic markers T0337 and C2_At5g47390, which flank the mapping interval of the cl locus. We recovered 20 recombinant individuals and these were grown to maturity and evaluated for the presence or absence of the gf mutant phenotype in ripe fruit and were also genotyped using four additional genetic markers that are located in the interval between T0337 and C2_At5g47390. This analysis revealed that gf was located in a 0.44-cM interval between the markers CT265 and CT148.
As part of the international initiative to sequence the eukaryotic gene space of cultivated tomato, the sequence of numerous bacterial artificial chromosome (BAC) clones residing on chromosome 8 has been determined (http://www.sgn.cornell.edu/about/japan.pl), including the BAC clones LeHBa0165B06 and LeHBa0197J17 that contain the markers CT148 and CT265, respectively. In an effort to generate additional genetic markers between CT148 and CT265 to further our mapping effort, we investigated the structure of these sequenced BAC clones in more detail. The tomato genome-sequencing effort is utilizing a BAC-by-BAC approach and a large number of BAC clones have been genetically fingerprinted and assembled into contigs. Analysis of LeHBA0165B06 revealed that Finger Print Contig analysis had placed this clone within a contig containing three additional BAC clones, LeHBa0004P13, LeHBa0020K14, and LeHBA0066O08 (http://www.genome.arizona.edu/fpc/tomato). We investigated the homologies of the BAC end sequences of these additional clones using the BLASTN tool of the SOL Genomics Network (SGN) to search against the entire collection of Solanaceae unigenes. This analysis revealed that the LeHBa0020K14-T7 BAC end was identical to a region of SGN-U316068. This unigene comprises 26 EST reads and is annotated as the senescence-inducible SGR1 protein of tomato, which is in turn a homolog of the SGR protein of rice (Park et al., 2007). We mapped the LeHBa0020K14-T7 BAC end and found that it cosegregated with the mutant phenotype in our F2 mapping population (Fig. 1).
Figure 1.
Structure of the gf locus. Genetic map of the gf locus, located on the long arm of tomato chromosome 8. Genetic markers and the number of recombinant individuals between adjacent markers from a total of 564 F2 plants are shown. The BAC clone HBa0165B06 has been fully sequenced and contains the genetic marker CT148. HBa0165B06 has been placed in a contig with three additional BAC clones. The genetic marker 20k14T7, derived from HBA0020K14, cosegregated with the gf mutant phenotype. 20k14T7 is identical in sequence to the unigene SGN-U316068 that has homology to the SGR protein of rice. Sequence analysis of this gene from wild type (GF/GF) and mutant (gf/gf) revealed a single A→T nucleotide change resulting in an amino acid substitution.
Given the homology of SGN-U316068 and its tight linkage to gf, we reasoned that this would be a strong candidate for GF. Amplification of the corresponding full-length cDNA of SGN-U316068 from GF/GF and gf/gf genetic backgrounds and subsequent sequence analysis of the cloned products revealed a single A→T nucleotide change at position 429 of the predicted coding sequence in the gf/gf mutant. This nucleotide change converts amino acid 143 from an Arg to a Ser in the gf/gf mutant background.
Complementation of the gf Mutant Phenotype
The open reading frame (ORF) of SGN-U316068 amplified from the wild-type (GF/GF) background was inserted downstream of the cauliflower mosaic virus (CaMV) 35S promoter in the binary vector pBI121 that had first been modified to remove the GUS reporter gene. This construct was introduced into the gf/gf mutant background via Agrobacterium-mediated transformation. Seventeen primary transformants were recovered that displayed ripe fruits that were red in color rather than the typical brown color associated with the gf mutation, suggesting that SGN-U316068 was able to complement the mutant phenotype. Genetic complementation was confirmed in segregating T1 progeny. Three independent T1 lines were selected and screened by Southern analysis for the presence or absence of the transgene (data not shown). Fruit from plants that contained the transgene were red, whereas plants that had segregated out the transgene produced brown fruit characteristic of the gf parent (Fig. 2A). Tomato leaves from plants exposed to nutrient stress or detached leaves that have been placed in the dark readily lose chlorophyll and this loss is inhibited in the gf mutant (Akhtar et al., 1999). We utilized the detached leaf assay to examine chlorophyll loss in segregating T1 transgenic lines (Fig. 2B). Leaves from plants that contained the transgene displayed chlorophyll loss following incubation in darkness for 2 weeks, whereas lines that had segregated away the transgene mimicked the gf mutant phenotype. These data provide conclusive evidence that SGN-U316068 encodes the GF protein of tomato.
Figure 2.
Complementation of the gf mutation. A, Fruit phenotype of segregating T1 progeny from three independent transgenic lines expressing a CaMV 35S∷GF construct in the gf/gf mutant background. Fruit in the top image contain the transgene and fruit in the bottom image have segregated out the transgene. Fruit of wild type (AC) and the gf mutant are shown for comparison. Photographs were taken at 10 d postbreaker. B, Leaf degreening assay in segregating T1 progeny described in A. Expanding leaves were detached and floated on water in darkness for 2 weeks.
Sequence Analysis of the cl Locus
Following confirmation that we had cloned the gene responsible for conferring the gf mutation and that it was indeed located within the corresponding genomic interval as the cl locus, we utilized the predicted GF amino acid sequence to search the pepper gene indices (http://compbio.dfci.harvard.edu/tgi/plant.html) for a homologous gene that might serve as a candidate for CL. We identified a single contig of six EST clones (TC4187) that were predicted to encode a pepper protein of 266 amino acids that shared 86% amino acid identity over its entire length to GF. Primers were designed to PCR amplify the entire ORF from cDNA synthesized from RNA isolated from red (CL/CL) and brown (cl/cl) fruited peppers. A single amplification product was obtained from each genotype and these were cloned and sequenced. The nucleotide sequence of the cDNA clones amplified from CL/CL was identical to the sequence of the TC4187 consensus sequence. However, the cDNA clones derived from the cl/cl fruit contained a single T→C change at nucleotide 340 of the protein coding region that resulted in conversion of Trp-114 into an Arg residue. The overlapping map position of gf and cl coupled with their phenotypic similarity and the DNA sequence polymorphism that we detected between red- and brown-fruited peppers strongly suggests that the cl mutation is caused by a mutation in the pepper ortholog of GF.
GF and CL Display a Ripening-Related Expression Pattern
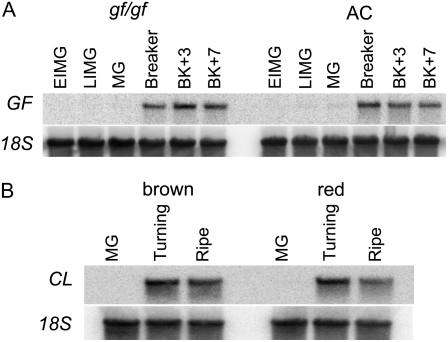
In silico analysis of EST data indicated that the transcripts of both GF and CL were likely to increase in abundance at the onset of fruit ripening in tomato and pepper. The tomato unigene SGN-U316068 is composed of 26 individual EST reads, of which 24 are derived from cDNA libraries prepared from RNA extracted from either ripening fruit or from mixed pools of fruit at different stages of development. The remaining two ESTs are derived from undifferentiated callus and anthesis staged flower cDNA libraries. The consensus sequence TC4187 that corresponds to the pepper CL gene contains only six individual EST reads, but all are derived from RNA isolated from red-ripe pepper fruit. We determined the expression of both GF and CL during fruit development and ripening in both tomato and pepper (Fig. 3). Transcripts were low or undetectable in green fruit, but showed increased abundance at the onset of ripening in both species. In addition, we monitored the expression of both GF and CL in the gf and cl mutant backgrounds to ascertain whether the mutations impacted the expression of either gene. There was no significant difference in transcript abundance detected between wild-type and mutant fruit for either GF or CL, suggesting that the absence of chlorophyll degradation does not influence the expression of these genes.
Figure 3.
Ripening-related expression of GF and CL in tomato and pepper. A, GF expression during fruit development and ripening in gf/gf and wild-type (AC) genetic backgrounds. Northern-blot analysis of RNA extracted from tomato fruit harvested at the early immature green (EIMG), late immature green (LIMG), mature green (MG), breaker, breaker + 3 d (BK+3) and breaker +7 d (BK+7) stages of development. The term breaker reflects the onset of fruit ripening in tomato. B, CL expression during fruit ripening in brown (cl/cl) and red (CL/CL) genetic backgrounds. Northern-blot analysis of RNA extracted from pepper fruit harvested at the MG, turning, and ripe stages of development. Turning represents the onset of fruit ripening in pepper.
GF and CL Represent New Members of the SGR Gene Family
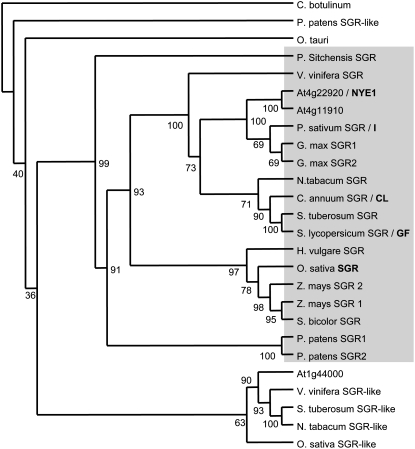
Through a combination of positional cloning and candidate gene analysis, we have identified single-nucleotide changes that define the mutations at the gf and cl loci of tomato and pepper. These mutations arise in orthologs of the rice SGR gene. The SGR family is conserved in plants and distant homologs are present in algae and, unexpectedly, in species of Bacillus and Clostridium, but not in other bacterial genomes. To investigate the relationship of this family, we collected predicted full-length sequences from GenBank and other family and organism-specific databases and performed a phylogenetic analysis using the predicted protein sequences (Fig. 4). The data clearly indicate that there are two clades that represent distinct subfamilies of these genes in plants. The family highlighted by the gray box most likely represents genes that are functionally equivalent to SGR and contains several members that have been shown through mutational analysis to be involved in chlorophyll degradation, including family members from Arabidopsis, rice, and pea, as well as GF and CL. These proteins range from approximately 260 to 295 amino acids in length. They have a highly conserved central core that displays between 70% and 95% amino acid identity to the GF protein, but are fairly divergent at both their N and C termini. This divergence is particularly apparent between proteins from different plant families. Within this SGR subfamily, some species, such as Arabidopsis, soybean (Glycine max), corn, and Physcomitrella patens contain paralogous genes, although we have not uncovered any evidence of this within the Solanaceae. In addition, a second distinct subfamily is evident in plants that contains members of several plant families and includes homologs from rice and Arabidopsis. The proteins within this subfamily are between 251 and 260 amino acids and are more distantly related to GF, sharing approximately 50% amino acid identity within the central core of the protein. To date, no mutant phenotypes have been assigned to any of the members of this subfamily, but, like the SGR subfamily, using both the TargetP (http://www.cbs.dtu.dk/services/TargetP) and Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html) prediction servers, all are predicted to encode proteins that are targeted to the chloroplast.
Figure 4.
Phylogenetic analysis of the SGR family. Protein alignments based using N terminus deleted amino acid sequences were performed using ClustalX. Phylogenetic relationships between the proteins were analyzed using the PHYLIP 3.67 suite of programs (http://evolution.genetics.washington.edu/phylip.html). The maximum parsimony, distance matrix, and likelihood methods of the Protpars and Seqboot programs were utilized to estimate phylogenies. A nonrooted tree phylogenetic tree was generated using Consense and the Treeview package using the Clostridium botulinum protein as the outgroup. The single most parsimonious tree obtained in a heuristic search following 100 random sequence addition replicates is shown. Bootstrap percentage supports are indicated at the branches of the tree. Sequence identifiers and accession numbers are described in “Materials and Methods.” Proteins shown in bold indicate where a mutant phenotype has been described.
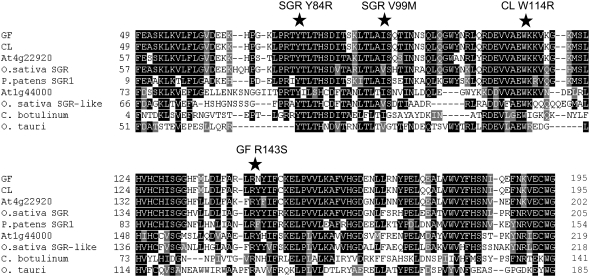
The mutations at the gf and cl loci result in the amino acid substitutions R143S and W114R, respectively. An amino acid alignment of GF and CL, together with representative proteins from each family subgroup, indicated that Arg-143 and Trp-114 represent invariant residues within the SGR family (Fig. 5). This observation was found to hold true even within a group of nine proteins from various Clostridium and Bacillus species (data not shown). From the various mutant loci described to date within the SGR family, two mutant alleles in rice SGR also result in amino acid substitutions that convert Y84C and V99M (Jiang et al., 2007; Park et al., 2007). Our alignments showed that Tyr-84 also represents a completely conserved residue in all the protein sequences that we recovered from database searches and that Val-99 is substituted in about 50% of the proteins for an Ile residue, but that no other amino acid occurs at this position (Fig. 5). These conserved residues are all located within the central core of this protein family, toward the N terminus of the mature protein, suggesting that this region of the protein is important for function.
Figure 5.
Amino acid alignment of SGR homologs. Protein alignments based on the highly conserved central core of the SGR proteins were performed using ClustalW. Conserved amino acids are indicated by shaded squares. Highly conserved amino acids that, when mutated, give rise to mutant phenotypes (rice, Y84C and V99M; pepper, W114R; and tomato, R143S) are indicated by stars. Accession numbers are as follows (in parentheses): GF (EU414632); CL (EU414631); rice SGR (EAZ09856); P. patens SGR1 (EDQ70701); rice SGR-like (NP_001054370); Clostridium botulinum (YP_001391480); Oryza tauri (CAL56489). The Arabidopsis sequences At1g44000 and At4g22920 are based on TAIR annotations (http://www.arabidopsis.org).
DISCUSSION
Stay-green mutants have been identified in an array of plant species. The gf and cl mutants of tomato and pepper display a stay-green phenotype that leads to the production of brown fruits as the result of carotenoid accumulation and lack of chlorophyll degradation. Through a combination of genetic mapping and candidate gene analysis, we have provided strong evidence indicating that the stay-green phenotypes of the gf and cl loci are the result of single amino acid substitutions in orthologs of the recently identified SGR gene of rice (Park et al., 2007). The SGR gene family is required for chlorophyll degradation and, in addition to rice SGR, mutant alleles that display stay-green phenotypes have been identified in Arabidopsis, pea, and F. pratensis (Armstead et al., 2006, 2007; Ren et al., 2007; Sato et al., 2007).
Variation in Structure and Function within the SGR Gene Family
Mutations in the SGR gene family have been designated as class C stay-green mutants that are deficient in their ability to break down chlorophyll, but have an otherwise normal pattern of senescence (Thomas and Howarth, 2000). One feature of the class C stay-green mutants is that the LHCP complexes persist in leaves and fruits, whereas other senescence processes continue as normal, suggesting that destabilization of the pigment protein complexes is required for chlorophyll degradation to proceed (Cheung et al., 1993; Akhtar et al., 1999; Thomas and Howarth, 2000; Park et al., 2007). Rice SGR has been shown to bind the LHCP II protein both in vivo and in vitro, suggesting that it may act to destabilize the pigment-protein interactions, therefore allowing the action of proteases and catabolic enzymes to degrade LHCPs and chlorophyll, respectively (Park et al., 2007). The availability of single amino acid substitutions in SGR will facilitate studies on the structure-function of this protein in more detail. Four amino acid substitutions that lead to strong phenotypic effects have now been identified in SGR orthologs in rice, tomato, and pepper (Jiang et al., 2007; Park et al., 2007). Three of these substitutions occur in amino acid residues that are invariant within all the SGR family members across diverse species boundaries and the fourth, SGR V99M, occurs at a residue that features either a Val or an Ile with approximately equal frequency (Fig. 5). The V99M substitution in rice SGR did not disrupt binding to LHCP II, indicating that this amino acid may be important for some other aspect of SGR function, such as an unknown catalytic activity or tertiary structure (Park et al., 2007). The rice mutant alleles together with the gf and cl alleles identified during the course of this study provide important tools for defining the function of the SGR family.
Our phylogenetic analysis indicates that there are two subfamilies of SGR-like proteins in plants (Fig. 4A). This conclusion is similar to that obtained in a recently published study of the SGR family (Aubry et al., 2008). The function of the subfamily that contains the Arabidopsis gene At1g44000 is currently unknown, but, like the SGR subfamily, these proteins are also predicted to be targeted to the chloroplasts and it will be of interest to determine whether they also function in chlorophyll degradation. In silico analysis of the expression pattern of At1g44000 using the Genevestigator suite of analysis programs (https://www.genevestigator.ethz.ch) revealed that this gene is expressed in a variety of tissues at different stages of development and is induced and repressed by different stimuli. However, At1g44000 does appear to be more highly expressed in cotyledons and developing leaves and has lower expression in senescent leaves. This is in contrast to At4g22920/AtNYE1, which shows increased expression in senescent organs (Ren et al., 2007). Functional analysis of this distinct subfamily is required to elucidate whether members also function in an aspect of chlorophyll turnover.
Overexpression of SGR in rice and Arabidopsis yielded plants that produced leaves with pale phenotypes or plants that died following transfer from tissue culture (Jiang et al., 2007; Park et al., 2007; Ren et al., 2007). We complemented the gf mutation using a construct driven by the CaMV 35S promoter; however, of the 17 primary transgenic lines that showed complementation of the gf mutant phenotype, we did not recover any plants that had a constitutively pale phenotype. The underlying reason for this difference between our experiments and those performed in rice and Arabidopsis remains unknown, but could simply be reflected by a difference in transgene expression levels or an as-yet unexplained difference in biology between these species. Low-level overexpression of Arabidopsis NYE1 (At4g22920) resulted in normal plant phenotypes, whereas high-level expression caused yellowing of leaves, suggesting that there is a direct correlation between NYE1/SGR expression levels and chlorophyll loss (Ren et al., 2007). Nevertheless, when we placed detached leaves in the dark, we saw a consistently greater chlorophyll loss in our transgenic lines than in our wild-type controls, suggesting that constitutive expression of GF can promote chlorophyll degradation (Fig. 2B).
Chlorophyll Degradation during Fruit Ripening
Chlorophyll degradation and the transition of chloroplasts into chromoplasts represent a dramatic change in metabolism at the onset of fruit ripening. However, little is known about the role of chlorophyll degradation and the regulation of this pathway during the ripening process, nor the overall contribution that it makes to fruit quality. In both the gf and cl mutants, the thylakoid membranes persist during fruit ripening, leading to the occurrence of both thylakoids and plastoglobuli within the plastids (Cheung et al., 1993; Roca et al., 2006). The consequences of this abnormal plastid transition have not been extensively investigated, although both gf and cl mutants show reduced rates of carotenoid accumulation during ripening (Ramirez and Tomes, 1964; Roca et al., 2006). Given the reduced rates of thylakoid breakdown and carotenoid accumulation in gf and cl, one might predict that other fruit quality characteristics may be altered. For example, several important aroma volatiles are derived from either fatty acid metabolism or carotenoid breakdown and these may well be altered in mutants as an indirect consequence of inhibited chlorophyll breakdown (Goff and Klee, 2006).
With the exception of CHLOROPHYLLASE, few expression data are available for genes involved in the chlorophyll degradation pathway during the ripening of fleshy fruits. CHLOROPHYLLASE transcripts were found to be of low abundance and constitutively expressed during natural ripening in citrus, but expression and enzyme activity could be stimulated by ethylene treatment (Jacob-Wilk et al., 1999; Fujii et al., 2007). This finding led the authors to conclude that chlorophyllase may not be the regulator of chlorophyll breakdown during the natural ripening of citrus fruits, although recent evidence has suggested that chlorophyllase represents the rate-limiting step in chlorophyll degradation following posttranslational modification of the enzyme (Harpaz-Saad et al., 2007). The prevalence of ESTs for both GF and CL in cDNA libraries prepared from fruit tissues suggested that the expression of these genes might be ripening regulated and we confirmed this by northern analysis (Fig. 3). We also searched the tomato and pepper EST collections using NYC1-related, chlorophyllase, PAO, and RCCR protein sequences from Arabidopsis. We identified homologs of all of these genes in the tomato EST collection, except for chlorophyllase, suggesting that this may represent a low expressed transcript in tomato, which would be in agreement with expression levels determined in citrus (Jacob-Wilk et al., 1999). Tomato homologs of NYC1-related, PAO, and RCCR were defined by the following SGN unigenes; SGN-U320581, SGN-U313134, and SGN-U319240, respectively. Based upon their occurrence in ESTs sequenced from fruit cDNA libraries, all three of the genes corresponding to these tomato unigenes are expressed in tomato fruit. However, the distribution of ESTs derived from fruit libraries compared to the total number of ESTs in the unigene ranged from between 20.5 (eight of 39 ESTs)—44.4% (four of nine; PAO, eight of 39 ESTs; RCCR, three of nine ESTs; and NYC1, four of nine ESTs) compared to 92% for GF (24 of 26 ESTs), suggesting that other components of the chlorophyll degradation pathway are not as highly expressed in tomato fruit as GF.
The gf and cl mutations are used in commercial breeding lines of tomato and pepper and the gf mutation likely forms the genetic basis for several heirloom tomato varieties that have names typically associated with the colors black and purple, including, among others, Carbon, Black Krim and Purple Passion. Defining the molecular basis of both the gf and cl mutations provides genetic markers that can be utilized to select these traits early during breeding programs and to clearly define the genetic basis of the multitude of heirloom tomato varieties that display phenotypic similarity to the gf mutation. The chlorophyll degradation pathway has been extensively studied in leaves, where it forms an integral component of the senescence process because the nutrients are recycled at a particular stage in the plant's life cycle or in response to environmental challenges. In contrast, in fleshy fruits, such as tomato and pepper, chlorophyll degradation accompanies the onset of fruit ripening and the associated conversion of chloroplasts to chromoplasts marks a brief period of high metabolic activity leading to the synthesis of a range of compounds that serve as attractants for seed-dispersing animals. Identification of the molecular basis of the gf and cl mutations has revealed novel alleles in SGR orthologs that will aid functional studies of this protein family and provides tools for comparing the regulation of chlorophyll degradation between vegetative and reproductive tissues together with the influence of this process on fruit quality.
MATERIALS AND METHODS
Plant Material and Growth Treatments
Tomato (Solanum lycopersicum) seeds homozygous for the gf mutation (LA3534) and the Solanum pennellii introgression lines IL8-2 (LA4074) and IL8-3 (LA4076) were obtained from the Tomato Genetics Resource Center, University of California, Davis. The parental cultivar Ailsa Craig was originally obtained from the Glasshouse Crops Research Institute (Littlehampton, Sussex, UK). Pepper (Capsicum annuum) seeds of the Ancho type that were either red fruited (CL/CL) or brown fruited (cl/cl) were provided by Dr. S. Moore and Dr. M. Jahn, Department of Plant Breeding, Cornell University. Plants were grown in peat-based compost supplemented with fertilizer in greenhouses equipped with heating and cooling systems and supplemental lighting either at Cornell University (Ithaca, NY) or Michigan State University (East Lansing, MI). Fruits for RNA extraction were harvested at the indicated stage of maturity with the term breaker being used to determine the onset of fruit ripening in both tomato and pepper. Seeds and locular gel were removed and the pericarp frozen in liquid nitrogen and stored at −80°C until use. The leaf degreening assay was performed by harvesting expanding terminal leaflets from the fourth true leaf of young tomato plants prior to the onset of flowering. Leaves were excised and floated on water in sealed petri dishes. The dishes were wrapped in aluminum foil and placed in darkness for 2 weeks at room temperature.
Molecular Markers and Genetic Mapping
Genomic DNA isolation was performed as described previously (Barry et al., 2005). Details of tomato genetic maps and the chromosome 8 molecular markers can be accessed through the SGN (http://www.sgn.cornell.edu). Many of the genetic markers utilized in this study were originally RFLP based and we have converted these into cleaved amplified polymorphic sequence (CAPS)-based markers for the purpose of this study. Briefly, primers designed from the sequence of the genetic marker were utilized to amplify the corresponding tomato and S. pennellii alleles from gf/gf (LA3534) and IL8-3 (LA4076), respectively. These fragments were purified using the Charge Switch PCR purification kit and cloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen). The sequence of these marker alleles was determined and the CAPS designer tool (http://www.sgn.cornell.edu/tools/caps_designer/caps_input.pl) was used to determine a restriction enzyme suitable for yielding polymorphisms. Markers were tested on the parents of the mapping population and the F1 progeny of their cross to confirm their utility prior to mapping in the F2 population. The primers and restriction enzymes used to generate these markers are available in Supplemental Table S1.
Cloning of GF and CL cDNA and Expression Analysis
Full-length cDNAs corresponding to the tomato unigene SGN-U316068 were amplified from breaker fruit cDNA synthesized from either the GF/GF or the gf/gf background using the primers U316068F (5′-GGACTTTTATCAAACAGCTAACTTGCA-3′) and U316068R (5′-GGCACAACCCAACTTACAATAATTGTA-3′). Fragments were amplified using the Pfu Ultra DNA polymerase (Stratagene) and cloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen). Full-length cDNA fragments, corresponding to the pepper homolog of SGR, were amplified from cDNA synthesized from pepper fruit RNA extracted at the breaker stage of development. The primer sequences PEPGF-F (5′-CACAACTCCTCTTTAAGTTTCTACTC-3′) and PEPGF-R (5′-TCTTGCTTCCACAAACCCTATAATGA-3′) were designed from the consensus contig TC4187 assembled in the pepper gene indices (http://compbio.dfci.harvard.edu/tgi/plant.html). Fragments were amplified from both red (CL/CL) and brown (cl/cl) fruited peppers using the FastStart high-fidelity PCR system (Roche Applied Science) and ligated into the pGEM-T Easy vector system (Promega). In both the tomato and pepper reverse transcription-PCR experiments, four clones were sequenced from each genetic background to confirm the existence of the genetic polymorphisms. Gene expression was determined using northern-blot analysis on total RNA extracted from fruit samples with full-length GF and CL as previously described (Griffiths et al., 1999; Barry et al., 2005).
DNA Constructs and Plant Transformation
The full-length cDNA sequence of GF was reamplified from a cloned template using the primers GFCOMPF and GFCOMPR, which are identical to U316068F and U316068R, respectively, but with the addition of a BamHI linker on the forward primer and a SmaI restriction site on the reverse primer. This fragment was cloned using TOPO cloning as described above. The fragment was excised using the restriction enzyme sites in the linkers and ligated downstream of the CaMV 35S promoter in the binary vector pBI121, previously modified by removal of the UidA coding region by digestion with SacI followed by polishing with T4 DNA polymerase and subsequent BamHI digestion. Construct fidelity was confirmed by DNA sequencing. Transgenic tomato plants were generated through cotyledon-derived explants via Agrobacterium tumefaciens-mediated transformation (strain LBA4404), using previously described methods (Fillatti et al., 1987).
DNA Sequence Analysis and Bioinformatics Resources
DNA sequences were assembled using Sequencher Version 4.7 (Genecodes Corporation). Amino acid sequences were deduced from cDNA clones using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Sequences used for comparisons and phylogenetic analysis were downloaded from organism-specific databases or from GenBank (http://www.ncbi.nlm.nih.gov/Genbank). The Arabidopsis (Arabidopsis thaliana) sequences At1g44000, At4g11910, and At4g22920 are based on The Arabidopsis Information Resource (TAIR) annotations (http://www.arabidopsis.org). The following sequences were obtained using their GenBank accession numbers: rice (Oryza sativa) SGR, EAZ09856; rice SGR-like (Os04g0692600), NP_001054370; soybean (Glycine max) SGR1, AAW82959; soybean SGR2, AAW82960; maize (Zea mays) SGR1, NP_001105770; maize SGR2, NP_001105771; sorghum (Sorghum bicolor) SGR, AAW82958; oat (Avena sativa) SGR, AAW82955; pea (Pisum sativum) SGR, BAF76351; tobacco (Nicotiana tabacum) SGR, ABY19382; Physcomitrella patens SGR1, EDQ70701; P. patens SGR2, EDQ62217; P. patens SGR-like, EDQ81746; Physcomitrella sitchensis SGR ABK22344; Ostreococcus tauri, CAL56489; and Clostridium botulinum, YP_001391480. The grape (Vitis vinifera) sequences are based upon annotation derived from a grape genome database (http://www.plantgdb.org/VvGDB/index.php) from the following sequence identifiers: grape SGR, PUT-157a-Vitis_vinifera-4592; and grape SGR-like, PUT-157a-Vitis_vinifera-43151296. Solanaceae SGR homologs were obtained from sequences deposited at SGN (http://www.sgn.cornell.edu) with the following unigene identifiers: potato (Solanum tuberosum) SGR, SGN-U272740; potato SGR-like, SGN-U274726; and tobacco SGR-like, SGN-U369489. Amino acid alignments were generated using either ClustalW or ClustalX and were decorated using the Boxshade server (Version 3.2.1; http://www.ch.embnet.org). Phylogenetic trees were constructed using the PHYLIP Version 3.67 suite of programs (http://evolution.genetics.washington.edu/phylip.html) and visualized using Treeview software.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU414631 (CL) and EU414632 (GF).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Development of PCR-based markers flanking the gf locus.
Supplementary Material
This work was supported by Michigan State University and the Michigan Agricultural Experiment Station (grant to C.B.), and National Science Foundation Plant Genome grants (grant nos. 05–01778 and 06–06595 to J.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Cornelius S. Barry (barrycs@msu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Akhtar MS, Goldschmidt EE, John I, Rodoni S, Matile P, Grierson D (1999) Altered patterns of senescence and ripening in gf, a stay-green mutant of tomato (Lycopersicon esculentum Mill.). J Exp Bot 50 1115–1122 [Google Scholar]
- Armstead I, Donnison I, Aubry S, Harper J, Hortensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al (2006) From crop to model to crop: identifying the genetic basis of the staygreen mutation in the Lolium/Festuca forage and amenity grasses. New Phytol 172 592–597 [DOI] [PubMed] [Google Scholar]
- Armstead I, Donnison I, Aubry S, Harper J, Hortensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al (2007) Cross-species identification of Mendel's/locus. Science 315 73. [DOI] [PubMed] [Google Scholar]
- Aubry S, Mani J, Hörtensteiner S (2008) Stay-green protein, defective in Mendel's green cotyledon mutant, acts independent and upstream of pheophorbide a oxygenase in the chlorophyll catabolic pathway. Plant Mol Biol (in press) [DOI] [PubMed]
- Barry CS, McQuinn RP, Thompson AJ, Seymour GB, Grierson D, Giovannoni JJ (2005) Ethylene insensitivity conferred by the Green-ripe and Never-ripe 2 ripening mutants of tomato. Plant Physiol 138 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, McNellis T, Piekos B (1993) Maintenance of chloroplast components during chromoplast differentiation in the tomato mutant green flesh. Plant Physiol 101 1223–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury R, Hortensteiner S, Donnison I, Bird CR, Seymour GB (1999) Chlorophyll catabolism and gene expression in the peel of ripening banana fruits. Physiol Plant 107 32–38 [Google Scholar]
- Efrati A, Eyal Y, Paran I (2005) Molecular mapping of the chlorophyll retainer (cl) mutation in pepper (Capsicum spp.) and screening for candidate genes using tomato ESTs homologous to structural genes of the chlorophyll catabolism pathway. Genome 48 347–351 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D (1994) A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79 175–179 [Google Scholar]
- Fillatti JJ, Kiser J, Rose R, Comai L (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium-tumefaciens vector. Biotechnology 5 726–730 [Google Scholar]
- Fujii H, Shimada T, Sugiyama A, Nishikawa F, Endo T, Nakano M, Ikoma Y, Shimizu T, Omura M (2007) Profiling ethylene-responsive genes in mature mandarin fruit using a citrus 22K oligoarray. Plant Sci 173 340–348 [Google Scholar]
- Goff SA, Klee HJ (2006) Plant volatile compounds: sensory cues for health and nutritional value? Science 311 815–819 [DOI] [PubMed] [Google Scholar]
- Gray J, Close PS, Briggs SP, Johal GS (1997) A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89 25–31 [DOI] [PubMed] [Google Scholar]
- Griffiths A, Barry C, Alpuche-Solis A-G, Grierson D (1999) Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. J Exp Bot 50 793–798 [Google Scholar]
- Harpaz-Saad S, Azoulay T, Arazi T, Ben-Yaakov E, Mett A, Shiboleth YM, Hortensteiner S, Gidoni D, Gal-On A, Goldschmidt EE, et al (2007) Chlorophyllase is a rate-limiting enzyme in chlorophyll catabolism and is posttranslationally regulated. Plant Cell 19 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortensteiner S (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57 55–77 [DOI] [PubMed] [Google Scholar]
- Jacob-Wilk D, Holland D, Goldschmidt EE, Riov J, Eyal Y (1999) Chlorophyll breakdown by chlorophyllase: isolation and functional expression of the Chlase1 gene from ethylene-treated citrus fruit and its regulation during development. Plant J 20 653–661 [DOI] [PubMed] [Google Scholar]
- Jiang HW, Li MR, Liang NB, Yan HB, Wei YL, Xu X, Liu JF, Xu Z, Chen F, Wu GJ (2007) Molecular cloning and function analysis of the stay green gene in rice. Plant J 52 197–209 [DOI] [PubMed] [Google Scholar]
- Kerr EA (1956) Green flesh, gf. Rpt Tomato Genet Coop 6 17 [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone KD, Lackney VK, Blauth JR, van Wijk R, Jahn MK (1999) Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics 152 1183–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT (2001) The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al (2007) The senescence-induced stay-green protein regulates chlorophyll degradation. Plant Cell 19 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinska A, Tanner G, Anders I, Roca M, Hortensteiner S (2003) Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA 100 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez DA, Tomes ML (1964) Relationship between chlorophyll and carotenoid biosynthesis in dirty-red (green-flesh) mutant in tomato. Bot Gaz 125 221–226 [Google Scholar]
- Ren GD, An K, Liao Y, Zhou X, Cao YJ, Zhao HF, Ge XC, Kuai BK (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca M, Hornero-Mendez D, Gandul-Rojas B, Minguez-Mosquera MI (2006) Stay-green phenotype slows the carotenogenic process in Capsicum annuum (L.) fruits. J Agric Food Chem 54 8782–8787 [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Nishimura M, Yamaguchi H, Kusaba M (2007) Mendel's green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc Natl Acad Sci USA 104 14169–14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Taylor JE, Tucker GA, editors (1993) Biochemistry of Fruit Ripening. Chapman & Hall, London
- Tanksley SD, Ganal MW, Prince JP, De-Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132 1141–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H, Howarth CJ (2000) Five ways to stay green. J Exp Bot 51 329–337 [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimada H, Masuda T, Takamiya K (1999) Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA 96 15362–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Suzuki T, Yamada T, Shimada H, Masuda T, Ohta H, Takamiya K (2003) Chlorophyllase as a serine hydrolase: identification of a putative catalytic triad. Plant Cell Physiol 44 96–101 [DOI] [PubMed] [Google Scholar]
- Wuthrich KL, Bovet L, Hunziker PE, Donnison IS, Hortensteiner S (2000) Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. Plant J 21 189–198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.