Abstract
The transition from seed to seedling is mediated by germination, a complex process that starts with imbibition and completes with radicle emergence. To gain insight into the transcriptional program mediating germination, previous studies have compared the transcript profiles of dry, dormant, and germinating after-ripened Arabidopsis (Arabidopsis thaliana) seeds. While informative, these approaches did not distinguish the transcriptional responses due to imbibition, shifts in metabolism, or breaking of dormancy from those triggered by the initiation of germination. In this study, three mechanistically distinct small molecules that inhibit Arabidopsis seed germination (methotrexate, 2, 4-dinitrophenol, and cycloheximide) were identified using a small-molecule screen and used to probe the germination transcriptome. Germination-responsive transcripts were defined as those with significantly altered transcript abundance across all inhibitory treatments with respect to control germinating seeds, using data from ATH1 microarrays. This analysis identified numerous germination regulators as germination responsive, including the DELLA proteins GAI, RGA, and RGL3, the abscisic acid-insensitive proteins ABI4, ABI5, ABI8, and FRY1, and the gibberellin receptor GID1A. To help visualize these and other publicly available seed microarray data, we designed a seed mRNA expression browser using the electronic Fluorescent Pictograph platform. An overall decrease in gene expression and a 5-fold greater number of transcripts identified as statistically down-regulated in drug-inhibited seeds point to a role for mRNA degradation or turnover during seed germination. The genes identified in our study as responsive to germination define potential uncharacterized regulators of this process and provide a refined transcriptional signature for germinating Arabidopsis seeds.
Seeds are essential to the life cycle of most plants and determine when and where plants are established. Strictly speaking, the germination of seeds is defined as the events between the uptake of water by the dry seed and the initiation of elongation of the embryonic root or radicle (Bewley and Black, 1994).
Experiments using whole-genome expression profiling have been performed to gain insight into the transcriptional program of seed germination and its genetic regulation. Nakabayashi et al. (2005) demonstrated that a large percentage of the genome's encoded mRNAs (approximately 55% of genes on the array) can be detected in dry Arabidopsis (Arabidopsis thaliana) seeds, including many seed development-related transcripts. These stored RNAs are degraded within 3 to 6 h after imbibition (HAI), at which point a distinct germination transcriptional program is initiated (Nakabayashi et al., 2005). A variety of approaches have been taken to dissect this shift to the germination transcriptional program in Arabidopsis. Time course experiments have been used to monitor the trends of transcriptional responses and cluster genes into coregulated transcript groups. Using this strategy, Nakabayashi et al. uncovered approximately seven coordinated families of transcriptional patterns during the first 24 HAI. This showed that about one-half of the seed development-associated transcripts are relatively low-abundance RNAs that are not temporally regulated. Approximately 35% of the transcripts analyzed are abundant in dry seeds and quickly lower in abundance in the first 24 HAI; this group contains many late-embryogenesis abundant protein transcripts. Approximately 25% of the transcripts gradually increase over time during the first 24 h; many of these encode proteins involved in metabolism. After 6 h of imbibition, the profile of seed mRNAs changes dramatically as the seed development-associated transcripts are degraded and newly synthesized transcripts appear (Nakabayashi et al., 2005). At this time, the gene expression program in seeds transitions to that characteristic of germination (Carrera et al., 2007).
A study by Carrera et al. (2007) generated a list of genes with differential expression in germinating seeds, referred to as the germination signature. To generate this gene list, the authors compared dormant and nondormant seeds from the Landsberg erecta (Ler) and Cape Verde Islands (Cvi) ecotypes at 24 HAI, enabling the analysis of expression changes between seeds that will, or will not, complete germination due to dormancy. This approach identified genes encoding components of translation, cell wall modification, mobilization of reserves, and the cytoskeleton as germination associated. Dormancy-associated transcripts included dehydrins, heat shock proteins, and storage reserve deposition. These data provided insight into the transcriptional states that differ between germinating and dormant seeds under the same conditions. A seed-specific gene ontology (GO) named TAGGIT was also generated in this study, which facilitates the identification and visualization of the germination signature (Carrera et al., 2007).
Other studies have produced microarray data related to seed germination under different conditions. Experiments using GA-deficient seeds (Ogawa et al., 2003) have described temporal transcript abundance changes in GA-deficient mutant seeds stimulated to germinate by GA treatment or in nongerminating water-imbibed controls. Because both GA treatment effects and germination were studied simultaneously in these experiments, their interpretation in the context of the germination transcriptional program is complicated. Several studies investigating seed dormancy in the Arabidopsis Cvi accession have comprehensively described gene expression during various states of dormancy and several dormancy-breaking treatments (Cadman et al., 2006; Finch-Savage et al., 2007).
In this study, we have taken a different and complementary approach to characterizing the germination signature of Arabidopsis seeds using small-molecule inhibitors of germination. To identify these inhibitors, a chemical genetic screen of approximately 3,200 known bioactive molecules was performed. This strategy enabled the unbiased isolation of small-molecule germination inhibitors from a large set of known bioactive compounds. In general, the chemical genetic approach uses small molecules to probe biological pathways (Stockwell, 2000). This approach has gained popularity in plant biology (Raikhel and Pirrung, 2005) and large screens have identified auxin mimics (Armstrong et al., 2004), protein sorting and vacuole morphology inhibitors (Zouhar et al., 2004; Surpin et al., 2005), cellulose biosynthesis inhibitors (DeBolt et al., 2007), and potential agrichemical lead compounds (Walsh et al., 2006, 2007). Additionally, small molecules can be used to probe natural variation and uncover genes that contribute to pharmacogenetic variation (Cutler and McCourt, 2005; Zhao et al., 2007) The approach can be particularly valuable for probing targets that play essential roles in growth and development because the small-molecule probes act analogously to conditional mutations. In some cases, compounds may bind to a conserved pocket shared by functionally redundant protein family members; therefore, it is expected that small molecules will be advantageous for addressing issues of genetic redundancy. As a result, phenotypes not readily obtained by analysis of single gene mutations may be produced through the action of small molecules.
Herein we describe a screen for inhibitors of Arabidopsis seed germination and the use of these inhibitors to dissect the germination transcriptional program. Global gene expression profiles in the drug-inhibited seeds were compared to those of germinating controls to identify changes in gene expression associated with germination. These data are placed within the context of previously described germination signature (Carrera et al., 2007) and uncover a previously undocumented link between nuclear-encoded plastid transcripts and germination. A seed electronic Fluorescent Pictograph (eFP) browser was also developed and integrated with the TAGGIT GO. This tool facilitates the pictographic visualization of gene expression in seeds using publicly available microarray datasets and aids in hypothesis generation for genes with unknown functions.
RESULTS
Identification of Seed Germination Inhibitors
The commercially available LOPAC library of 1,280 biologically active compounds and the Spectrum library of 2,000 bioactive compounds were screened at 25 μm to identify molecules capable of blocking the germination of wild-type Arabidopsis Columbia (Col) seeds. The LOPAC and Spectrum libraries contain FDA-approved molecules, research inhibitors, natural products, and other bioactive molecules and can be used to identify known bioactive molecules that perturb a process of interest. Our screen for germination inhibitors was part of a larger effort aimed at the identification of both known and novel structure inhibitors of Arabidopsis cell expansion. Details of this larger screening effort will be published elsewhere.
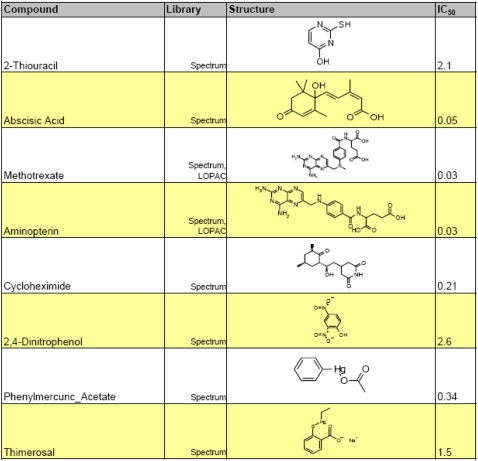
Our screen identified eight compounds capable of inhibiting Arabidopsis germination (Table I), including the plant hormone abscisic acid (ABA), which was present in the Spectrum library. Of the synthetic compounds identified, four stood out as having well-characterized mechanisms of action: methotrexate, aminopterin, cycloheximide, and 2,4-dinitrophenol (2,4-DNP). These compounds are general inhibitors of metabolism, protein synthesis, and energetics and not seed specific in their effects. 2,4-DNP is an uncoupler of mitochondrial membrane transport that blocks ATP synthesis by perturbing the mitochondrial proton gradient (El-Guindy et al., 1981). 2,4-DNP has been shown previously to block lettuce (Lactuca sativa), pea (Pisum sativum), and rice (Oryza sativa) germination (Siegel et al., 1967; Speer, 1973), presumably by blocking ATP biosynthesis. Cycloheximide is a well-characterized inhibitor of translation that acts upon the 60S ribosomal particle and blocks translation elongation. Rajjou et al. (2004) demonstrated that cycloheximide blocks Arabidopsis seed germination. Methotrexate and aminopterin are folic acid analogs that competitively inhibit dihydrofolate reductase (Matthews et al., 1977), which is involved in folate biosynthesis. Folates function as one-carbon donors and play a critical role in thymidine and purine synthesis. As a consequence, these drugs inhibit DNA synthesis, which underlies their use as anticancer agents. Whether methotrexate blocks Arabidopsis germination through the inhibition of DNA replication or other folate-dependent pathways (e.g. Met and formyl-Met synthesis, Ser-Gly interconversion) is not yet clear. Methotrexate, 2,4-DNP, or cycloheximide inhibited the germination of Arabidopsis embryos separated from their endosperms (data not shown), showing that they act on the embryo itself and do act through the maintenance of coat-imposed dormancy (Bewley, 1997). Seeds subjected to cycloheximide, 2,4-DNP, or methotrexate treatment exhibited testa rupture, observed at the time germination was scored (3 d after imbibition; data not shown). Dose response curves for these three inhibitors are provided in Supplemental Figure S1.
Table I.
Compounds from the LOPAC chemical library identified as germination inhibitors at 25 μm
Shown at right are hypocotyl growth inhibition IC50 values, which were conducted to measure the relative potencies of hits.
These well-characterized and mechanistically diverse inhibitors of germination were used to probe the seed germination transcriptional program and complement time course and comparative data of dormant and after-ripened genes in previous studies (Nakabayashi et al., 2005; Carrera et al., 2007). These pharmacological tools were used to define the transcriptional responses that differ between germinating control seeds and germination inhibitor-treated seeds. It is expected that some differences will be drug specific; however, by identifying the responses common across all inhibitory treatments, the drug-specific effects can be reduced.
To identify these germination-selective transcripts, we performed microarray analyses on stratified wild-type seeds imbibed on these drugs and isolated mRNA at 24 h after stratification. At this time point, control (untreated) seeds have shifted from a seed-developmental to a germination transcriptional program (Nakabayashi et al., 2005) and any changes in transcription can be ascribed to this latter process. Duplicate biological replicates were conducted for each drug treatment and triplicate treatments were performed for untreated control samples. The drug concentrations used for these experiments were selected as the lowest concentration of each agent that reproducibly gave 100% inhibition of germination (scored at 3 d after imbibition).
To identify genes that are significantly associated with the germination transcriptional program, we used the Significance Analysis of Microarray (SAM) method (Tusher et al., 2001). This approach indentified transcripts that are significantly up- or down-regulated between all germination inhibitor-treated microarray samples (six microarray datasets total) and controls (triplicate); the large number of replicates enables robust identification of significant responders. This approach identified 207 genes with transcripts that were significantly higher across all nongerminating (drug-inhibited) seed mRNA populations, and therefore lower in the germinating controls. We termed this gene set GermDOWN. We additionally identified 1,008 genes with transcripts that were significantly lower across all nongerminating treatments, and therefore higher in the germinating controls; we refer to this as the GermUP set (Supplemental Table S1).
This set of germination-responsive transcripts was analyzed by GO classification using the The Arabidopsis Information Resource (TAIR) GO annotations tool (complete list in Supplemental Table S2). Table II shows the major ontological differences between the two gene sets. GermUP contains an increased number of nuclear-encoded genes whose products localize to chloroplasts, mitochondria, and plastids relative to GermDOWN. There are also a larger number of genes encoding proteins with hydrolase, transferase, and kinase activities, in addition to the components of electron transport and energy production. These latter data are consistent with the necessity for cell wall modification and increased metabolism prior to the completion of germination (Bewley and Black, 1994). The expression of ribosomal protein-encoding genes is greater in the GermDOWN set than the GermUP set despite the requirement for protein translation for the completion of seed germination (Rajjou et al., 2004).
Table II.
Key differences in the ontological classification of the GermUP and GermDOWN datasets
| GO Classification | GermDOWN | GermUP | |
|---|---|---|---|
| GO descriptor | |||
| Cellular component | Chloroplast | 14 | 192 |
| Mitochondria | 13 | 69 | |
| Plastid | 5 | 90 | |
| Ribosome | 22 | 6 | |
| Activity | Kinase | 7 | 91 |
| Hydrolase | 20 | 182 | |
| Transferase | 16 | 182 | |
| Process | Electron transport or energy | 3 | 62 |
| TAGGIT Ontology Classification
|
|||
| TAGGIT category | |||
| Cell cycle | 1 | 6 | |
| Cytoskeleton | 0 | 8 | |
| Glycolysis and gluconeogeneis | 4 | 21 | |
| Krebs cycle | 0 | 14 | |
| Photosynthesis | 0 | 8 | |
These gene sets were also classified according to the seed-specific GO called TAGGIT (Carrera et al., 2007). This ontology groups genes according to their function in seeds and provides a more meaningful classification of genes within the context of germination. Included in the TAGGIT ontology were all the GO annotated genes in Arabidopsis related to protein translation. These can be found in the TAGGIT gene list used in this study (Supplemental Table S2). TAGGIT classification of the gene sets reveals there are a greater number of genes corresponding to the cell cycle, cytoskeleton, glycolysis and gluconeogenesis, Krebs cycle, and photosynthesis in the GermUP set than the GermDOWN set (Table II). As observed using GO classification, there were more genes associated with protein translation in the GermDOWN gene set than the GermUP set. This suggests these transcripts are inhibited in response to these germination inhibitors despite their induction during the normal germination process (Nakabayashi et al., 2005; Holdsworth et al., 2008). With the exception of the genes associated with protein translation, these ontological classifications are consistent with the physiological process known to be involved in the germination process (Bewley and Black, 1994).
Visualization of Gene Expression in Dormant and Germinating Seeds Using the Seed eFP Browser
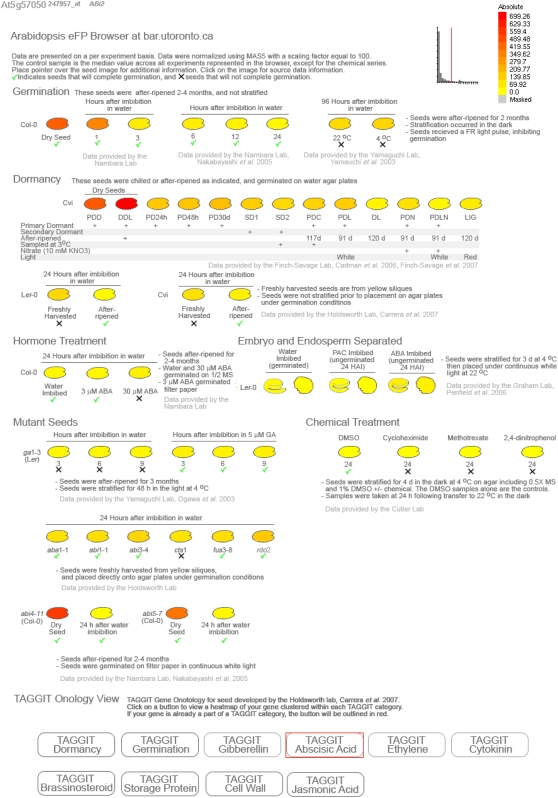
The majority of the genes indentified by our SAM approach lacked an ascribed function so we developed an online tool to visualize gene expression patterns in seeds using our data and publicly available microarray datasets. Winter et al. (2007) describe the eFP browser where experimental gene expression data are painted onto pictographic representations of the tissues and conditions from which the RNA samples were obtained. A seed eFP browser was created and included as part of the Bio-Array Resource (BAR) online bioinformatics tools package (www.bar.utoronto.ca; Toufighi et al., 2005). Datasets displayed include an Arabidopsis seed germination time course (Nakabayashi et al., 2005), cold treatment of seeds (Yamauchi et al., 2003), seed dormancy in the Arabidopsis accession Cvi (Cadman et al., 2006; Finch-Savage et al., 2007), freshly harvested dormant and after-ripened germinating Col and Cvi seeds and freshly harvested mutant seeds (Carrera et al., 2007), ABA treatment of seeds (provided by the lab of E. Namabara), GA treatment of seeds (Ogawa et al., 2003), isolated embryo and endosperm tissue (Penfield et al., 2006), and the data described in this work. Information related to the conditions in which the experiments were done is included on the output graphic, including a green checkmark for seeds that will ultimately germinate, and a black X for those that will not complete germination (see Fig. 1).
Figure 1.
Seed eFP browser output. eFP output is illustrated using ABI2 (At5g57050) as an example. Transcript abundance is high in dry seeds of both Col and Cvi accessions. These transcripts rapidly decline by 3 HAI in Col seeds, whereas a low level of expression is maintained in dormant Cvi seeds. The TAGGIT button belonging to the ABA category is highlighted at the bottom with a red box, indicating ABI2 is a part of this group of genes.
To extract information from the seed eFP browser, a user enters an Arabidopsis Genome Initiative (AGI) ID for a given gene of interest, and the transcript expression pattern and intensity across the available experiments are plotted pictographically. This visualization facilitates the interpretation of microarray data and serves as a tool for hypothesis generation as to gene function. For example, the expression pattern of the gene encoding ABI2 (At5g57050) is shown in the seed eFP output format (Fig. 1). Transcript abundance of this dormancy-mediating gene strongly correlates with seed samples representing the dormant state of Arabidopsis seeds.
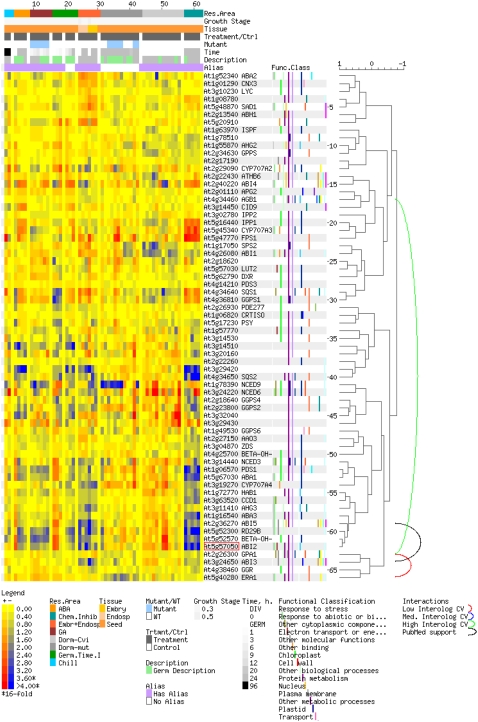
The TAGGIT seed-specific GO (Carrera et al., 2007) was integrated into the seed eFP browser. A series of buttons representing the TAGGIT gene categories is present at the bottom of the seed eFP browser output screen. These buttons may be clicked at any time to produce a hierarchically clustered heat map (Toufighi et al., 2005) for each of the TAGGIT genes in the category across all seed microarray experiments. If the queried AGI ID in the seed eFP browser belongs to one of the TAGGIT categories, the corresponding button is highlighted with a red box at the bottom of the screen (Fig. 1). Upon viewing the highlighted TAGGIT category, the AGI ID entered by the user is highlighted by a red box within the heat map gene list, allowing the user to see where their gene is situated (Fig. 2). We present these datasets as a new view in the eFP browser tool in addition to the hundreds of other seed datasets, all of which may be queried using the tools on the BAR.
Figure 2.
ABA TAGGIT ontology. TAGGIT ontology categories showing expression data for these genes across all seed microarray experiments are shown. The ABI2 gene is highlighted with a red box. Hierarchical clustering of expression data is represented by the tree to the right, and the curved lines demonstrate confirmed and predicted protein-protein interactions (Geisler-Lee et al., 2007).
Examination and Visualization of Germination-Regulated Transcripts
The GermDOWN dataset contained numerous genes previously identified as negative regulators of germination potential. These included the ABA-insensitive genes (fold change is in brackets after each gene) ABI4 (1.7 times) and ABI5 (2.7 times) whose corresponding mutants germinate on ABA concentrations inhibitory to the wild type (Koornneef et al., 2002). Genes encoding negative regulators of GA action were also found in this group of genes, including the DELLA genes RGA (1.3 times), GAI (1.7 times), and RGL3 (2.0 times). The protein products of these genes act to repress germination through the inhibition of GA action.
Various phosphatases that act in the ABA signaling pathway have been described, such as ABI1, ABI2, HAB1, HAB2, AHG1, and AHG3 (Leung et al., 1997; Robert et al., 2006; Yoshida et al., 2006; Nishimura et al., 2007). Numerous uncharacterized protein phosphatase genes were identified using our method, for example, the protein phosphatase 2C (PP2C; At1g67820; 1.8 times down-regulated), which follows a similar expression pattern during seed germination as ABI2 (Supplemental Table S3; Fig. 1). Both of these genes are expressed at a high level in the dry seed and decline by 6 HAI. Other uncharacterized phosphatases that were identified include the PP2C genes At2g20630, At5g53140, At3g02750, and At4g38520, the PP2A gene At1g69960, type 1 PP2A (At3g46820), acid phosphatase (At4g29270), and a purple acid phosphatase (At2g27190). Phosphatase regulatory proteins were also identified, including two regulatory subunits of PP2A encoded by At1g13460 and At1g17720.
Numerous genes encoding kinases were also identified in the gene sets. Notably, a number of mitogen-activated protein (MAP) kinase genes are germination responsive, including MPK9 (At3g18040), MPK20 (At2g42880), MKK2 (At4g29810), MAPKKK12 (At3g06030), and MAPKKK20 (At3g50310). The possibility that a MAP kinase cascade mediates the completion of seed germination remains to be investigated. The expression pattern of MAPKKK20 suggests this gene plays a role in this process. Expression of this gene is induced following imbibition, likely through the action of GA, and is expressed primarily in the endosperm (Supplemental Table S3).
A homolog of the dormancy quantitative trait locus DOG1 (Bentsink et al., 2006) was also present in the GermDOWN set. Expression of this gene is exclusive to seeds (Schmid et al., 2005) and follows the same pattern of transcript abundance as the uncharacterized PP2C mentioned above (Supplemental Table S3). A putative endo-β-mannanase (At5g66460; 1.8 times down-regulated) is also present in this gene set. Transcripts of this gene are strongly induced in germinating seeds by 12 HAI and are present exclusively in the endosperm (Supplemental Table S3). This induction is dependent on the presence of GA, an expression pattern similar to the tomato (Lycopersicon esculentum) mannanase LeMAN2, which is proposed to play a role in the release from coat dormancy (Bewley, 1997).
The polyamine spermidine inhibits Arabidopsis seed germination when applied exogenously (Mirza and Bagni, 1991). Spermidine is synthesized from S-adenosyl-Met, a substrate shared with ethylene biosynthesis. Several genes involved in the synthesis of these compounds are germination responsive, including S-adenosyl-Met synthase 1 (At1g02500), S-adenosyl-Met transporter 1 (At4g39460), and spermidine synthetase 1 (At1g23820). This observation suggests the synthesis of polyamines is transcriptionally regulated with respect to seed germination. Transcript abundance of these genes increases prior to the completion of germination and following dormancy-breaking treatments (Supplemental Table S3). Spermidine synthase 1 is highly expressed in both the embryo and endosperm, whereas the S-adenosyl-Met synthase 1 gene is predominantly expressed in the endosperm, while its transporter is more highly expressed in the embryo. The other S-adenosyl-Met synthase gene (At4g01850) is highly expressed in both the embryo and endosperm and is not germination responsive according to our analysis. Based on the ethylene TAGGIT category output from the seed eFP browser, the ethylene synthesis enzyme 1-aminocyclopropane-1-carboxylic acid oxidase 2 (At1g62380) is predominantly expressed in the embryo (Supplemental Table S3). Collectively, these observations suggest that, during germination, S-adenosyl-Met is synthesized in the endosperm in response to conditions favorable for germination and transported to the embryo for the purpose of producing spermidine and ethylene.
Nitrate is capable of breaking seed dormancy in Arabidopsis (Hilhorst and Karssen, 1988; Alboresi et al., 2005), and evidence suggests this occurs by interacting with the ABA signaling pathway (E. Nambara, personal communication). The nitrate reductase 1 gene (At1g77760) was present in the GermDOWN gene set, consistent with this proposed role in countering the inhibitory effects of ABA during germination.
The soluble GA receptor GID1A was also present in the GermDOWN list (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Nakajima et al., 2006). This protein acts as a positive regulator of seed germination (Iuchi et al., 2007) and is strongly down-regulated by 3 HAI (Supplemental Table S3). This observation is consistent with the finding that GA response occurs within the first 24 HAI in Arabidopsis seed germination (Nambara et al., 1991).
Within the GermUP list were the negative regulators of ABA response ABI8, which encodes a protein of unknown function (Brocard-Gifford et al., 2004), and the FIERY1 inositol phosphatase (Xiong et al., 2001). When mutated, these genes each confer ABA-resistant seed germination. The ENHANCED RESPONSE TO ABA1 (ERA1) encodes a farnesyl transferase enzyme whose activity attenuates ABA response. The substrate for this enzyme is farnesyl diphosphate, and both farnesyl diphosphate synthase genes were present in the GermUP gene set. Whether the abundance of farnesyl diphosphate acts to regulate seed germination remains to be established because farnesyl diphosphate is the precursor of a large number of secondary metabolites.
A number of genes mediating light responses in seeds were included in the GermUP list, including PHOT1, NPH3, PAP1, LHY, and PIF3 (Parks and Quail, 1991; Whitelam et al., 1993). The DFL1 gene also mediates light responses and was present in the GermDOWN gene list.
Drug-Inhibited Germination Signatures
Carrera et al. (2007) characterized the germination transcriptional signature by identifying genes differentially regulated between dormant and germinating after-ripened seeds at 24 HAI using the Col and Cvi ecotypes. To place our data within the context of this work, we compared our germination-responsive genes with the lists of genes generated by Carrera et al. as being up-regulated specifically in after-ripened (germinating) seeds and dormant (nongerminating) seeds.
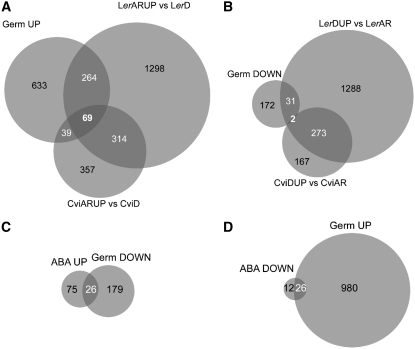
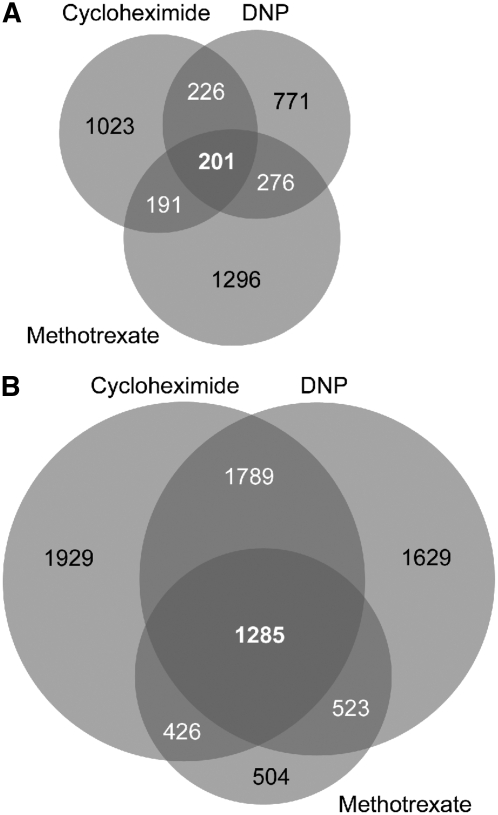
Our GermUP gene set shared one-third of its genes with LerARUP genes and one-tenth with the CviARUP gene set (Fig. 3A; Supplemental Table S4), indicating similarity between the transcripts from our analysis and those induced during germination in the analysis by Carrera et al. A total of 69 genes were shared between all three treatments, including the tubulin genes TUA4 (At1g04820), TUB1 (At1g75780), and TUB7 (At2g29550), and the homeobox gene ATHB31 (At1g14440).
Figure 3.
Overlap of drug-defined transcript profiles and prior germination data. Proportional Venn diagrams illustrating overlap between previously described germination signatures (Carrera et al., 2007) and gene expression in ABA-inhibited seeds are shown. A, Venn diagram showing genes up-regulated in germinating Ler and Cvi seeds in relation to the GermUP dataset. B, Genes up-regulated in dormant Ler and Cvi seeds compared with the GermDOWN dataset. C, GermDOWN and ABAUP gene sets as determined using the SAM method. D, GermUP and ABADOWN gene sets.
The GermDOWN dataset was less similar to genes up-regulated in dormant seeds of Ler and Cvi (Fig. 3B; Supplemental Table S4). Only 33 of 205 GermDOWN genes were shared with LerDUP, and two shared with CviDUP, being a zinc finger (At5g66730) and a late embryogenesis abundant protein (At1g72100). This is consistent with the fact that the seeds used in the study were after-ripened and stratified prior to being placed under germination conditions. Their transcriptional profile consequently reflects a loss of dormancy in these seeds. Collectively, these data suggest that the chemical inhibition of seed germination is not inducing a state of dormancy in seeds, but rather is blocking the execution of germination.
The effect of the germination inhibitors was also compared to the transcript profiles obtained by inhibiting germination with the hormone ABA under these same conditions. SAM was performed on microarray data generated from mRNA isolated from ABA-inhibited seeds and germinating control seeds. This analysis identified 101 genes up-regulated in ABA (ABAUP) and 38 genes down-regulated (ABADOWN). Shared between ABAUP and GermDOWN were 26 genes, including the putative endo-β-mannanase (At5g66460; Fig. 3C; Supplemental Table S4). ABADOWN shared 26 of 38 genes with GermUP (Fig. 3D). As expected, there are common changes in gene expression between ABA and drug-mediated inhibition of seed germination because both inhibit germination. The large number of ABA-specific transcriptional responses likely reflects ABA-responsive transcripts that are not normally responsive to germination.
To evaluate the quality of our microarray data, quantitative reverse transcription (qRT)-PCR experiments were conducted using a small set of genes (ABI4, ABI5, β-mannanase) that showed modest, but significant, changes during germination (as determined using the SAM method). RNA samples for biological duplicate replicates for each of the chemical treatments were subjected to qRT-PCR along with triplicate germinating RNA controls using two different control primer pairs. We observed good concordance between the microarray data and the qRT-PCR data (Supplemental Fig. S1). The quality of our microarray data likely stems in part from our use of six mRNA populations (two biological replicates for each of three inhibitors) in generating the nongerminating seed mRNA populations.
Effects of the Individual Drug Treatments on Gene Expression
The influence of each drug treatment on gene expression was examined. Given that each inhibitor treatment was done in duplicate, the SAM method could not be utilized and we focused on characterizing genes with greater than 2-fold change relative to the controls.
The total number of genes down-regulated by drugs was 2.6 times greater than those up-regulated (Supplemental Table S5), a similar trend to that obtained by using the SAM method (Supplemental Table S1). TAGGIT ontological analysis of these gene sets was performed (Supplemental Table S5). Cycloheximide up-regulated a greater number of genes related to ABA than the other two treatments, and methotrexate induced a proportionally larger number of genes related to cell wall modification. The union of gene sets 2-fold or more up-regulated by each of the treatments (Fig. 4A) included the hormone synthesis and response genes GA2-β-dioxygenase (At1g30040), NCED9 (At1g78390), GA 20-oxidase-3 (At5g07200), and ABI5. Cycloheximide treatment increases the transcript abundance of several hormone synthesis genes dramatically, up-regulating NCED6 (At3g2422) 50-fold, GA 2-oxidase-8 49-fold, and GA2-β-dioxygenase 25-fold. These data suggest a factor produced through de novo protein synthesis during seed germination may be critical for the regulation of some hormone biosynthetic enzyme gene expression.
Figure 4.
Drug-specific transcriptional responses. Effect of the individual drug treatments on gene expression in Arabidopsis seeds is shown. Gene sets were defined as genes up- or down-regulated 2-fold or more by each of the treatments relative the control. A, Up-regulated genes in each of the three drugs. B, Genes down-regulated.
Within the down-regulated gene sets from the individual treatments, no striking trends are apparent when taking into account the difference in the size of the gene sets (Fig. 4B). Cycloheximide does not have a strong effect on the abundance of translation-related transcripts, whereas methotrexate inhibits transcripts related to DNA repair relative to the other treatments. The DNP treatment specifically down-regulated the GA synthesis gene GA4 (At1g4120) and ABA synthesis gene ABA2 (At2g42350), in addition to the ABA response regulator HAB1 (At3g54400). Cycloheximide uniquely inhibited the expression of numerous GA and ABA synthesis genes, including NCED3 (At1g52060), NCED4 (At3g60440), GA 3-oxidase (At4g10290), GA5 (At4g08020), and the ABA-degrading enzyme CYP707A1 (At3g60630).
The three-way intersection of the genes down-regulated by the individual drug treatments includes numerous genes related to photosynthesis, including LHCA6 (At1g19150), LHCA3 (At1g61520), LHCB3 (At5g54270), LHCB5 (At4g10340), and PSAH2 (At1g52230). Many genes related to photosynthesis were also identified as down-regulated in the presence of germination inhibitors (within the GermUP gene set) by SAM. These include PSI subunit D-1 (At4g02770), LHCA5 (At1g45474), PSII BY (At1g67740), chlorophyll synthetase (At3g51820), Rubisco activase (At2g39730), and the chlorophyll synthesis enzymes PORA (At5g54190) and PORB (At4g27440).
Nuclear-Encoded Transcripts for Plastid-Localized Proteins Are Induced During Germination and Inhibited by ABA
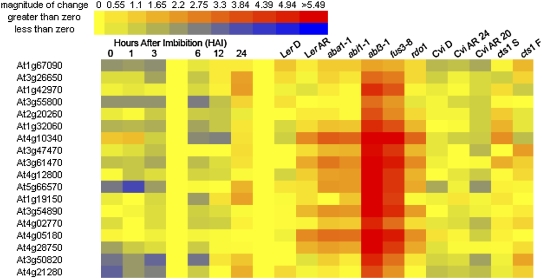
Given the germination-responsive nature of the nuclear-encoded components of photosynthesis, we examined the expression pattern of nuclear-encoded genes involved in photosynthesis and localized to this organelle. A group of 17 genes were tightly coregulated during seed germination (Supplemental Table S6) and are induced between 6 and 12 HAI in wild-type seeds (Fig. 5). These genes are more highly expressed in the ABA-deficient genotype aba1-1, the ABA-insensitive mutant abi1-1. This is seen more strongly in the abi3-4 and fus3-8 mutants (Carrera et al., 2008), which result in the premature loss of embryonic identity and a green seed phenotype. Nuclear-encoded photosynthetic genes are also more highly expressed in the rdo or reduced dormancy mutant. These data suggest that ABA regulates the expression of nuclear-encoded photosynthetic genes during germination, perhaps through the maintenance of dormancy and embryonic cellular identity. These observations are consistent with the finding that the light-responsive G-box motif is in close association with the ABA response element in the promoters of genes that are subject to modulation by retrograde signaling (Koussevitzky et al., 2007).
Figure 5.
Photosynthetic nuclear-encoded transcript responses in germinating seeds. Transcript abundance of genes encoding the protein components of photosynthesis in wild-type and various mutant Arabidopsis seeds during germination are shown. Log2-transformed data are relative to the mean of all seed samples in the BAR, with red indicating transcript abundance above the mean and blue indicating abundance below the mean. Arabidopsis mutants were freshly harvested from their mother plants and placed to germinate on agar-water for 24 h (Carrera et al., 2008). Heat maps were generated at the BAR (www.bar.utoronto.ca).
DISCUSSION
In this study, we induced a nongerminating seed phenotype using chemicals. The use of three mechanistically distinct inhibitors enabled germination to be probed without focusing on the effects of a single pathway, such as GA biosynthesis or ABA signaling, as has been done previously. By pooling the data of transcriptional responses triggered in seeds inhibited by these diverse mechanisms, we were able to isolate the transcriptional signature that defines germinating seeds. SAM identified numerous genes previously demonstrated to play potentially repressive roles in seed germination as differentially regulated between our germinating and nongerminating mRNA populations. These genes include GAI, RGA, RGL3, GID1A, ABI4, ABI5, ABI8, FRY1, and the light response-regulating genes PHOT1, NPH3, PAP1, LHY, PIF3, and DFL1. These findings illuminate the strength of the approach we took for uncovering genes related to the regulation of seed germination. A smaller ratio of genes was differentially up-regulated in germinating seeds (GermUP compared with after-ripened) relative those in dormant (GermDOWN compared with dormant; Fig. 3, A and B). These observations are consistent with the loss of dormancy in the seed samples used and suggest that chemical inhibition of germination does not reinitiate dormancy.
Regulation of Germination by Transcription
The GermDOWN gene set was significantly smaller than that of GermUP (207 and 1,008, respectively). In spite of its small size, this set contains many important factors that affect hormone sensitivity during germination (ABI4, ABI5, RGL3, RGA, GAI). The ABI loci are positive ABA response factors and the DELLA/RGL proteins negative GA response factors. It therefore appears that, during germination, there is a generalized reduction in the transcript levels of several factors that provide negative regulatory inputs into germination. Thus, one hypothesis to emerge from our observations is that the degradation or turnover of regulatory transcripts could be an important mechanism in the control of germination. The utilization of an mRNA degradation/turnover mechanism to promote germination is consistent with observations by Rajjou et al. (2004) that de novo transcription is not required for the completion of Arabidopsis seed germination. New inhibitors of RNA degradation with good seed permeability would be valuable tools for probing this hypothesis further. Additionally, if mutants defective in RNA metabolism factors, such as SAD1 and ABH1 (Kuhn and Schroeder, 2003), are defective in this degradation/turnover process, this could explain why these mutants are hypersensitive to ABA during germination. Last, our data suggest that the postimbibition down-regulation of regulatory transcripts is likely to be a regulated event during germination because imbibitions alone are not sufficient to trigger the response. Thus, a postimbibition step associated with germination appears to operate in this process.
Regulation of the Plastid Transcriptome during Germination
Transcription in the plastid has been implicated in the regulation of germination potential (Demarsy et al., 2006). Seeds imbibed in Tagetin, an inhibitor of the plastid-encoded RNA polymerase, show a delay in seed germination. In addition, the plastid-encoded RNA polymerase protein is present in germinating seeds and is likely active.
The plastid may be regulating germination potential through the production of energy in some capacity. The up-regulation of photosynthetic machinery may also be a reflection of the seed's commitment to germinate in anticipation of autotrophic growth. The detection of nuclear-encoded plastid transcripts in the drug-inhibited seeds may reflect this inhibition of the transition to the vegetative state and the maintenance of the nonphotosynthetic embryonic state. The inhibition of nuclear-encoded photosynthetic transcripts by ABA and abi1, abi3, and fus3 suggests that this phenomenon is related to the maintenance of embryonic identity and, possibly, genome-uncoupled (GUN) signaling (Koussevitzky et al., 2007; Fig. 5). The embryo may maintain the GUN signal repressing the production of the photosynthetic machinery until it is committed to complete germination. The up-regulation of photosynthetic genes in LerAR over LerD supports this hypothesis. Genes encoding photosynthetic machinery may therefore serve as molecular markers for seeds that are to complete germination.
CONCLUSION
Our approach of using mechanistically diverse chemical inhibitors combined with pooled transcriptomic data enabled us to uncouple the imbibition and germination transcriptional response. Notably, a large number of known regulators of hormone sensitivity were identified as germination responsive using this approach. Given that a multitude of genes with unknown function are also observed to be germination responsive, our data suggest that new factors in germination may potentially be uncovered by using our data to guide future genetic studies. Attempts to test this hypothesis are currently under way using T-DNA insertion lines. Collectively, our data provide a refined picture of the germination transcriptional signature and demonstrate the utility of small-molecule screens for seed biology.
MATERIALS AND METHODS
Small-Molecule Screens
To identify germination inhibitors, the LOPAC library (1,280 compounds) obtained from Sigma-Aldrich and the Spectrum library (Microsource Discovery Systems) were screened. Ninety-six-well microtiter plates (Greiner) were used for bioassays, prepared with 25 μm compound, 0.8% agar (w/v), and 0.34× Murashige and Skoog salts (Sigma-Aldrich). Compounds were diluted from 2.5 mm stock solutions in dimethyl sulfoxide (DMSO) such that all wells contained 1% DMSO final concentration. Arabidopsis (Arabidopsis thaliana) seeds of the Col ecotype were harvested from plants grown under 24 h artificial light and after-ripened for 1 year at room temperature and ambient humidity. The seeds were then surface sterilized using 10% bleach and 0.01% Tween-20 in double deionized water, followed by four washes with double deionized water. They were then suspended in 0.1% agar (w/v) and sown into wells containing compound (approximately 15 seeds per well in 20 mL suspension), and then stratified for 4 d at 4°C. After 4 d of growth in the dark at 22°C, the plates were examined to identify germination and cell expansion inhibitors. Candidate hits were then retested to validate bioactivity. To compare relative potencies of hits, dose curves were conducted and used to infer hypocotyl growth inhibition IC50 values. The approximately 250 hits identified that affect cell expansion, but not germination, will be described elsewhere. Hypocotyl effects are our preferred comparator because most bioactive compounds do not inhibit germination, but do inhibit hypocotyl growth.
Tissue Preparation and RNA Extraction
Col wild-type seeds were sown on 0.5× Murashige and Skoog medium (approximately 2,500 seeds per 150-mm plate) containing either 25 μm 2,4-DNP, 1 μm cycloheximide, 2 μm methotrexate, 1 μm ABA, or 1% DMSO (DMSO is the carrier solvent and all treatments contain 1% DMSO). Chemical concentrations were chosen from dose response curves as doses that yield robust inhibition of germination. 2,4-DNP, cycloheximide, methotrexate, and ABA (± isomers) were purchased from Sigma-Aldrich. Seeds were stratified on drug, hormone, or control plates for 4 d and then incubated in the dark at 22°C for 24 h. Seeds were collected, frozen in liquid nitrogen, then ground to fine powder form with frozen mortar and pestle, after which total RNA was extracted using the RNAqueous kit (Ambion) or using the phenol-chloroform extraction protocol, as described by Suzuki et al. (2004). All replicates for microarrays are biological replicates, not technical replicates. RNA purity of each sample was assessed using OD260:OD280 ratios, and only samples possessing ratios between 1.7 and 2.2 were utilized further. RNA was additionally analyzed by gel electrophoresis to confirm integrity prior to cRNA labeling. For each sample analyzed, 5 μg of total RNA was converted to biotin-labeled cRNA using oligo(dT) priming as described by the manufacturer (Enzo kit; Affymetrix) and hybridized to 22K ATH1 Affymetrix microarrays at the Affymetrix Genechip facility (University of Toronto). Duplicate samples were hybridized for cycloheximide, methotrexate, and 2,4-DNP treatments, and triplicate samples were hybridized for control and ABA treatments. Probe sets with expression signals called present or marginal by the statistical algorithms (GCOS) were used in our analyses. SAM (Tusher et al., 2001) was used to identify probe sets that are significantly regulated by treatments using a false discovery rate of 5%. Microarray data are available from the BAR (http://bar.utoronto.ca) under Project 29 of BAR's project browser (http://bar.utoronto.ca/affydb/cgi-bin/affy_db_proj_browser.cgi), using the following identifiers: control DMSO samples, bot0142, bot0314, bot0394; 2,4-DNP samples, bot0246, bot0247; cycloheximide samples, bot0256, bot0257; methotrexate samples, bot0293, bot0294. Affymetrix CEL files are available upon request.
GO Analysis of Microarray Data
Pseudoproportional Venn diagrams were generated using Adobe Photoshop. The GO analysis was done on the TAIR Web site (www.arabidopsis.org), and TAGGIT analysis was done using a macro provided by Dr. Michael Holdsworth (University of Nottingham, UK).
Feeding and Embryo Experiments
Arabidopsis seeds were imbibed in water for 6 h on moistened filter paper in a 90-mm petri plate before their embryos were dissected from their endosperm. The isolated embryo was then placed on a 0.8% (w/v) agar-water plates containing a final concentration of 25 μm of the drug and 1% (v/v) final concentration of DMSO. Wild-type embryos germinated within 24 h, whereas the drug-treated embryos failed to germinate after 1 week.
qRT-PCR Experiments
Total RNA samples were quantified using a NanoDrop Spectrophotometer (Nanodrop Technologies). RT reactions were performed using the Fermentas first-strand cDNA synthesis kit following the manufacturer's directions with 100 ng total RNA input. qPCRs were performed using a Chromo 4 real-time PCR detector (Bio-Rad) controlled by Opticon Monitor 3 software. Standard curves were generated using five concentrations in triplicate in a dilution series, followed by the test sample run with two biological samples per treatment (each analyzed with triplicate technical replicates). qPCR results were quantified by the Pfaffl method as described in the real-time PCR applications guide (Bio-Rad). The DMSO-treated samples (i.e. germinating samples) were used as calibrators and the test samples were either cycloheximide-, 2,4-DNP-, or methotrexate-treated samples (i.e. nongerminating samples). The target genes were normalized against the reference β-tubulin gene (TUB4) or against a Zn-finger control (At5g18650), which was chosen because we noticed that many typical housekeeping genes exhibit significantly altered expression levels in the germination inhibitor datasets (see Supplemental Table S1 for examples). Primers utilized and PCR conditions are presented in Supplemental Table S7.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Dose response curves for inhibitors utilized in microarray experiments.
Supplemental Figure S2. qRT-PCR validation of microarray results.
Supplemental Table S1. Gene lists from SAM, GO ontology analysis, TAGGIT ontology analysis, and selected output from the seed eFP Browser.
Supplemental Table S2. GO and TAGGIT ontology analyses of SAM selected germination-responsive transcripts.
Supplemental Table S3. Selected output from the seed eFP Browser. eFP output for genes mentioned in the text are presented in this file.
Supplemental Table S4. Germination signature of Arabidopsis seeds.
Supplemental Table S5. Effects of the individual drug treatments based on genes showing a 2-fold change or more in transcript abundance.
Supplemental Table S6. Gene lists of nuclear-encoded components of photosynthesis that show an increase in transcript abundance during seed germination.
Supplemental Table S7. qRT-PCR primers and reaction conditions.
Supplementary Material
Acknowledgments
We thank Dr. Eiji Nambara (RIKEN, Japan), Dr. Michael Holdsworth (Nottingham, UK), Dr. Bill Finch-Savage (Warwick, UK), Dr. Shinjiro Yamaguchi (RIKEN, Japan), and Dr. Ian Graham (York, UK) for providing seed microarray data. We also thank Hardeep Nahal for assisting with the creation of the seed eFP Browser and Thanh Nguyen for processing RNA samples for Affymetrix hybridizations. We are grateful to Peter McCourt for helpful suggestions on the manuscript prior to publication.
This work was supported by National Science and Engineering Research Council Discovery grants (to S.R.C. and N.J.P.), by a Canadian Research Chair in Plant Functional Genomics (to S.R.C.), and by the University of California, Riverside (startup funds to S.R.C.). Microarray experiments were conducted on equipment funded by Genome Canada/Ontario Genomics Institute.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sean R. Cutler (sean.cutler@ucr.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN (2005) Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ 28 500–512 [DOI] [PubMed] [Google Scholar]
- Armstrong JI, Yuan S, Dale JM, Tanner VN, Theologis A (2004) Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc Natl Acad Sci USA 101 14978–14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD (1997) Breaking down the walls—a role for endo-beta-mannanase in release from seed dormancy? Trends Plant Sci 2 464–469 [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination, Ed 2. Plenum Press, New York
- Brocard-Gifford I, Lynch TJ, Garcia ME, Malhotra B, Finkelstein RR (2004) The Arabidopsis thaliana ABSCISIC ACID-INSENSITIVE8 encodes a novel protein mediating abscisic acid and sugar responses essential for growth. Plant Cell 16 406–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46 805–822 [DOI] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Dietrich D, Footitt S, Theodoulou FL, Holdsworth MJ (2008) Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J 53 214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Peer W, Schmuths H, Footitt S, Theodoulou FL, Holdsworth MJ (2007) Gene expression profiling reveals defined functions of the ATP-binding cassette transporter COMATOSE late in phase II of germination. Plant Physiol 143 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S, McCourt P (2005) Dude, where's my phenotype? Dealing with redundancy in signaling networks. Plant Physiol 138 558–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S, Gutierrez R, Ehrhardt DW, Melo CV, Ross L, Cutler SR, Somerville C, Bonetta D (2007) Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc Natl Acad Sci USA 104 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Mache S (2006) Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol 142 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Guindy MM, Neder AC, Gomes CB (1981) 2,4-Dinitrophenol—mechanism of action. Cell Mol Biol Incl Cyto Enzymol 27 399–402 [PubMed] [Google Scholar]
- Finch-Savage WE, Cadman CSC, Toorop PE, Lynn JR, Hilhorst HWM (2007) Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J 51 60–78 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O'Toole N, Ammar R, Provart NJ, Millar AH, Geisler M (2007) A predicted interactome for Arabidopsis. Plant Physiol 145 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst HW, Karssen CM (1988) Dual effect of light on the gibberellin- and nitrate-stimulated seed germination of Sisymbrium officinale and Arabidopsis thaliana. Plant Physiol 86 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D (2008) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13 7–13 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Suzuki H, Kim YC, Iuchi A, Kuromori T, Ueguchi-Tanaka M, Asami T, Yamaguchi I, Matsuoka M, Kobayashi M (2007) Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J 50 958–966 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5 33–36 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316 715–719 [PubMed] [Google Scholar]
- Kuhn JM, Schroeder JI (2003) Impacts of altered RNA metabolism on abscisic acid signaling. Curr Opin Plant Biol 6 463–469 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DA, Alden RA, Bolin JT, Freer ST, Hamlin R, Xuong N, Kraut J, Poe M, Williams M, Hoogsteen K (1977) Dihydrofolate reductase: x-ray structure of the binary complex with methotrexate. Science 197 452–455 [DOI] [PubMed] [Google Scholar]
- Mirza JI, Bagni N (1991) Effects of exogenous polyamines and difluoromethylornithine on seed germination and root growth of Arabidopsis thaliana. Plant Growth Regul 10 163–168 [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41 697–709 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46 880–889 [DOI] [PubMed] [Google Scholar]
- Nambara E, Akazawa T, McCourt P (1991) Effects of the gibberellin biosynthetic inhibitor uniconazol on mutants of Arabidopsis. Plant Physiol 97 736–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50 935–949 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwalhara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH (1991) Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikhel N, Pirrung M (2005) Adding precision tools to the plant biologist's toolbox with chemical genomics. Plant Physiol 138 563–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert N, Merlot S, N'Guyen V, Boisson-Dernier A, Schroeder JI (2006) A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett 580 4691–4696 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Siegel SM, Lederman M, Daly O, Roberts K (1967) Effects of metabolic poisons on rice—comparative sensitivity of aerobic and anaerobic modes of germination. Plant Physiol 42 1489–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer HL (1973) Effect of arsenate and other inhibitors on early events during germination of lettuce seeds (Lactuca sativa L.). Plant Physiol 52 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell BR (2000) Chemical genetics: ligand-based discovery of gene function. Nat Rev Genet 1 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M, Rojas-Pierce M, Carter C, Hicks GR, Vasquez J, Raikhel NV (2005) The power of chemical genomics to study the link between endomembrane system components and the gravitropic response. Proc Natl Acad Sci USA 102 4902–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Kawazu T, Koyama H (2004) RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 37 542–544 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J 43 153–163 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow T, Hsing YC, Kitano H, Yamaguchi I (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698 [DOI] [PubMed] [Google Scholar]
- Walsh TA, Bauer T, Neal R, Merlo AO, Schmitzer PR, Hicks GR, Honma M, Matsumura W, Wolff K, Davies JP (2007) Chemical genetic identification of glutamine phosphoribosylpyrophosphate amidotransferase as the target for a novel bleaching herbicide in Arabidopsis. Plant Physiol 144 1292–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, Matsumura W, Davies JP (2006) Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol 142 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK (2001) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S (2003) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chow TF, Puckrin RS, Alfred SE, Korir AK, Larive CK, Cutler SR (2007) Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nat Chem Biol 3 716–721 [DOI] [PubMed] [Google Scholar]
- Zouhar J, Hicks GR, Raikhel NV (2004) Sorting inhibitors (Sortins): chemical compounds to study vacuolar sorting in Arabidopsis. Proc Natl Acad Sci USA 101 9497–9501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.