Abstract
The outer integument of the Arabidopsis (Arabidopsis thaliana) ovule develops asymmetrically, with growth and cell division occurring primarily along the region of the ovule facing the base of the gynoecium (gynobasal). This process is altered in the mutants inner no outer (ino) and superman (sup), which lead to absent or symmetrical growth of the outer integument, respectively. INO encodes a member of the YABBY family of putative transcription factors, and its expression is restricted to the gynobasal side of developing ovules via negative regulation by the transcription factor SUP. Other YABBY proteins (e.g. CRABS CLAW [CRC] and YABBY3 [YAB3]) can substitute for INO in promotion of integument growth, but do not respond to SUP regulation. In contrast, YAB5 fails to promote integument growth. To separately investigate the growth-promotive effects of INO and its inhibition by SUP, domain swaps between INO and YAB3, YAB5, or CRC were assembled. The ability of chimeric YABBY proteins to respond to SUP restriction showed a quantitative response proportional to the amount of INO protein and was more dependent on C-terminal regions of INO. A different response was seen when examining growth promotion where the number and identity of regions of INO in chimeric YABBY proteins were not the primary influence on promotion of outer integument growth. Instead, promotion of growth required a coordination of features along the entire length of the INO protein, suggesting that intramolecular interactions between regions of INO may coordinately facilitate the intermolecular interactions necessary to promote formation of the outer integument.
The YABBY family of genes participates in the specification of abaxial identity in plant lateral organs (Siegfried et al., 1999). INNER NO OUTER (INO) is an Arabidopsis (Arabidopsis thaliana) YABBY family member that is specifically expressed in the ovule outer integument and is essential for ovule development (Baker et al., 1997; Villanueva et al., 1999). INO likely modulates the transcription of currently uncharacterized genes involved in the growth of the outer integument in Arabidopsis. While the molecular basis of INO function remains largely unknown, the ability of INO to interact with the putative transcription factor NOZZLE/SPOROCYTELESS (Sieber et al., 2004) further supports the hypothesis that INO modulates expression of genes necessary for outer integument development. Structural domains of YABBY members have been hypothesized based on sequence alignments that show similarities to known protein domain motifs (Siegfried et al., 1999; Villanueva et al., 1999; Bowman, 2000). A region toward the N terminus is similar to Cys2-Cys2 zinc (Zn)-finger domains and has been shown to interact with Zn ions in vitro for the YABBY protein FILAMENTOUS FLOWER (Kanaya et al., 2001). A region (the “YABBY” domain) nearer to the C terminus is predicted to contain two α-helical regions that are thought to participate in DNA binding, in large part due to sequence similarity with the DNA-binding motif of the High Mobility Group transcription factors (Sawa et al., 1999). An understanding of the functions of these regions and a more precise determination of the boundaries of each putative domain would facilitate our understanding of how INO mediates outer integument growth in Arabidopsis.
During Arabidopsis ovule development, outer integument growth is restricted primarily to the side of the ovule that faces the basal region of the gynoecium (gynobasal; Fig. 1). This asymmetry is mediated via SUPERMAN (SUP), a transcription factor that has been shown to restrict INO expression to the gynobasal side of the developing ovule (Meister et al., 2002). While ino mutants failed to initiate or support outer integument growth, it was found that sup mutants resulted in expansion of INO expression to all sides of the developing outer integument. This resulted in outer integument growth on both the gynobasal and gynoapical (toward the stigma) sides of the ovule (Fig. 1). This ectopic growth correlated with gynoapical expansion of expression of INO and of GFP and β-glucuronidase (GUS) coding regions under control of the INO promoter (PROINO), indicating that SUP repression of INO expression was mediated at the transcriptional level (Meister et al., 2002). To further test the nature of INO and SUP antagonism, Meister et al. (2002) placed the coding sequence of CRABS CLAW (CRC), a related YABBY member, under control of PROINO and introduced this into ino-1 plants. Plants expressing PROINO:CRC produced ovules that phenocopied the sup mutant, and reporter analysis indicated expansion of PROINO-driven expression similar to that observed for sup mutant ovules. These results indicated that SUP requires features of the INO protein to repress transcription from PROINO. These features of INO are lacking in CRC. Coupled with evidence that showed that INO participates in positive feedback of its expression, it was concluded that SUP likely inhibits integument growth by disrupting INO autoregulation (Meister et al., 2002).
Figure 1.
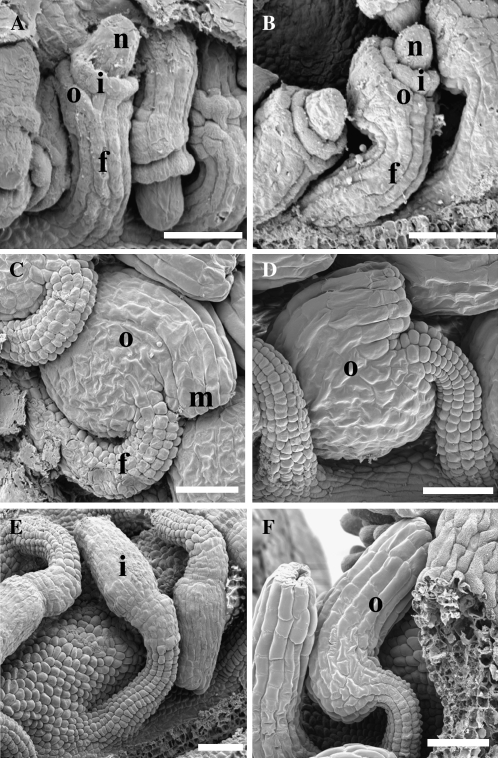
Scanning electron micrographs of wild-type and mutant ovules. During wild-type ovule development, primordia emergence is followed by the appearance of distinct structures, including the funiculus, outer and inner integuments, and the nucellus (A). Asymmetric growth is observed as the ovule develops (B) until the micropyle lies adjacent to the funiculus in mature ovules (C and D). In ino-1 ovules (E), the outer integument fails to initiate and as a result, the symmetrical growth of the inner integument and nucellus are apparent. In sup-5 ovules (F), the outer integument exhibits growth on both sides, leading toward a concentric ring of symmetrical growth. c, Chalaza; i, inner integument; m, micropyle; n, nucellus; o, outer integument. For all panels, the gynobasal side is at left. Scale bars = 25 μm.
Following this study, Meister et al. (2005) tested the ability of other YABBY members to support outer integument growth. When PROINO:YABBY3 (YAB3) was introduced into ino-1 plants, ovules exhibited outer integument growth on all sides of the ovules. This indicated a poor response to SUP repression similar to the response to the presence of PROINO:CRC. In contrast, when PROINO:YAB5 was introduced in ino-1 plants, outer integument growth did not occur, and thus YAB5 was unable to support outer integument growth. To determine the differences between INO and other YABBY members that led to these results, domain swaps between regions of CRC and INO were created. By observing the ability of chimeric transgenes composed of regions 1, 2, or 3 and 4 collectively (Fig. 2A) of CRC or INO to support proper outer integument development, it was determined that multiple regions contributed to INO-specific function in a largely quantitative manner (Meister et al., 2005). While all CRC/INO domain swap lines were able to support outer integument growth, only a subset responded to SUP repression of growth on the gynoapical side of ovules. This suggested that all three regions of INO participate in both the promotion of growth and response to SUP inhibition, although the combination of regions 3 and 4 appeared to play a more significant role (Meister et al., 2005). These results, however, may have been specific to domain swaps between INO and CRC due to the unique role of CRC in gynoecium and nectary specification, in addition to its role in abaxial identity that is common among all YABBY proteins (Alvarez and Smyth, 1999). This is particularly of concern due to the possibility that INO and CRC regions may share some redundancy in function that is not present in other YABBY members, as INO and CRC are the only members with specific reproductive roles.
Figure 2.
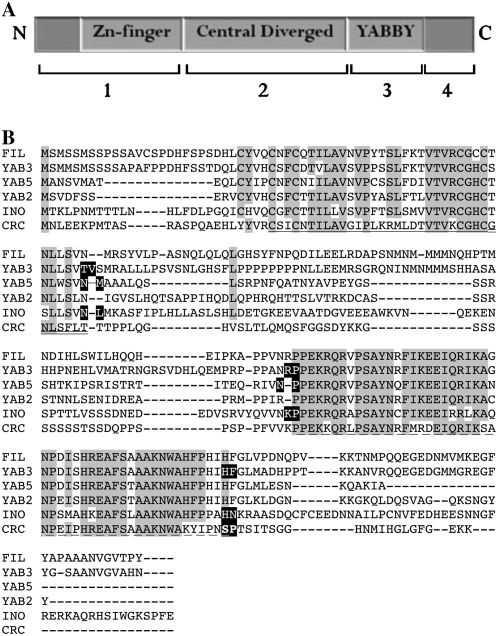
Features of the YABBY family of transcription factors. A, A cartoon representation of the four major regions exchanged in domain swap experiments. The regions designated by the numbers at bottom represent: 1, the N-terminal and Zn-finger regions; 2, the central variable region; 3, the YABBY region; and 4, the C-terminal region. B, An amino acid alignment of YABBY proteins. The Zn-finger and YABBY regions, the borders of which were determined based on previous sequence alignments (Siegfried et al., 1999), are indicated using single solid and broken underlines, respectively. The seven conserved amino acid residues adjacent to the previously defined Zn-finger motif are indicated with a double underline. Residues on either side of protein region boundaries that were used in assembly of chimeric proteins are highlighted in black.
In our current study, exchanges between regions of INO and YAB3 were created to test the hypothesis that domain swap results with INO and CRC resulted from reproductive-specific features of CRC. To separate SUP-responsive defects from growth-promotive effects, domain swap experiments between INO and YAB5 were conducted in parallel, because PROINO:YAB5 did not support outer integument growth in ino-1 plants. To refine our understanding of the importance of the C terminus that was observed in Meister et al. (2005), more defined swaps were made to separate the YABBY region from the more C-terminal portion in domain swap experiments between INO and CRC and INO and YAB3. Through these studies, we show that multiple regions of INO contribute toward the promotion of growth and response to SUP; however, the mechanisms underlying each function are distinct in that a quantitative response was observed only for SUP-related effects.
RESULTS
Nomenclature
In the following sections, representation of the source of each of the exchanged regions in the chimeric cDNAs is indicated by the following: I indicates INO sequence, C indicates CRC sequence, 3 indicates YAB3 sequence, and 5 indicates YAB5 sequence. These designations are listed from left to right starting from the amino terminus and ending with the carboxy terminus. The majority of exchanges focused on three major regions of INO: the N-terminal and Zn-finger regions (1) comprised the first region, the central variable region (2) comprised the second region, and the YABBY (3) and C-terminal (4) regions together comprised the third region (Fig. 2A). However, since the third region possessed greater INO-specific function than the first and second regions in a prior study (Meister et al., 2005), we separately exchanged the YABBY (3) and C-terminal (4) regions in a subset of our experiments. Separate exchange of the C-terminal region 4 is indicated by the following: i indicates INO sequence, 3 indicates YAB3 sequence, and c indicates CRC sequence. Boundaries between regions were selected based on sequence conservation between YABBY members as well as results indicating the importance of seven conserved residues at the carboxyl end of the Zn-finger region (Fig. 2B; Siegfried et al., 1999; Villanueva et al., 1999; Bowman, 2000; Meister et al., 2005).
INO/YAB3 Chimera Expression Using PROINO
Chimeric cDNAs composed of INO and YAB3 sequences were expressed using PROINO, and phenotypic effects on outer integument development were scored in an ino-1 mutant background (Table I). Five phenotypic classes were observed among transformed lines containing chimeric transgenes: sup-like, weak-sup, wild type, weak-ino, and ino-like (Fig. 3). Individual transformants sometimes exhibited variation in outer integument development within gynoecia, and these lines were scored based on the phenotype of the majority of ovules of that line. The phenotypic classes observed were based on comparisons with the following control lines: sup-5, ino-1, ino-1 with PROINO:III (INO), and wild type (Table I). In sup-5 plants, ovules exhibited significant growth on both the gynobasal and gynoapical sides, while ino-1 plants exhibited no growth of the outer integument. Most ovules from ino-1 plants containing PROINO:III resembled wild type, indicating rescue of the ino-1 phenotype.
Table I.
Transgenic complementation of ino-1
| Genotypea | PROINO:Transgeneb | Ovule Phenotypec
|
Significantly Different from PROINO:d
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| sup-Like | Weak-sup | Wild Type | Weak-ino | ino-Like | III | 333 | 555 | CCC | ||
| Wild type | –e | – | 25 | – | – | No | Yes | Yes | Yes | |
| ino-1 SUP | – | – | – | – | 25 | Yes | Yes | No | Yes | |
| INO sup-5 | 24 | – | – | – | – | Yes | Yes | Yes | Yes | |
| III | – | – | 28 | 8 | 6 | ∼f | Yes | Yes | Yes | |
| 333 | – | 11 | – | 8 | – | Yes | ∼ | ∼ | ∼ | |
| 555 | – | – | – | 3 | 18 | Yes | ∼ | ∼ | ∼ | |
| CCC | 7 | 13 | – | 4 | 1 | Yes | ∼ | ∼ | ∼ | |
| I33 | 6 | 9 | 6 | 2 | 1 | Yes | Yes | ∼ | ∼ | |
| I3I | 1 | 8 | 10 | 7 | 1 | Yes | Yes | ∼ | ∼ | |
| II3 | 3 | 1 | 13 | 3 | 1 | No | Yes | ∼ | ∼ | |
| 3II | – | 1 | 20 | 2 | – | No | Yes | ∼ | ∼ | |
| 33I | – | 14 | 5 | 1 | – | Yes | Yes | ∼ | ∼ | |
| 3I3 | – | 10 | 18 | – | – | Yes | Yes | ∼ | ∼ | |
| 333i | – | 14 | 3 | 3 | – | Yes | No | ∼ | ∼ | |
| III3 | – | – | 21 | 4 | 1 | No | Yes | ∼ | ∼ | |
| I55 | – | – | 2 | 17 | 8 | Yes | ∼ | Yes | ∼ | |
| I5I | – | – | 3 | 22 | 3 | Yes | ∼ | Yes | ∼ | |
| II5 | 1 | 3 | 9 | 9 | 4 | No | ∼ | Yes | ∼ | |
| 5II | – | – | 3 | 20 | 3 | Yes | ∼ | Yes | ∼ | |
| 55I | – | – | 6 | 14 | 7 | Yes | ∼ | Yes | ∼ | |
| 5I5 | – | – | 1 | 22 | 6 | Yes | ∼ | Yes | ∼ | |
| IIIc | 3 | 8 | 13 | 2 | 0 | Yes | ∼ | ∼ | Yes | |
| CCCi | 3 | 4 | 2 | 18 | 1 | Yes | ∼ | ∼ | Yes | |
Ovules were examined in wild-type, ino-1, or sup-5 mutant plants.
Transgenes were constructed as transcriptional fusions of the listed coding sequence with PROINO and examined in an ino-1 mutant background.
Number of transgenic lines with the corresponding ovule phenotype, as described in the text and illustrated in Figure 3.
Pairwise comparisons of the phenotypic class distribution for each line.
–, No plants of this class observed.
∼, Pairwise comparison not applicable.
Figure 3.
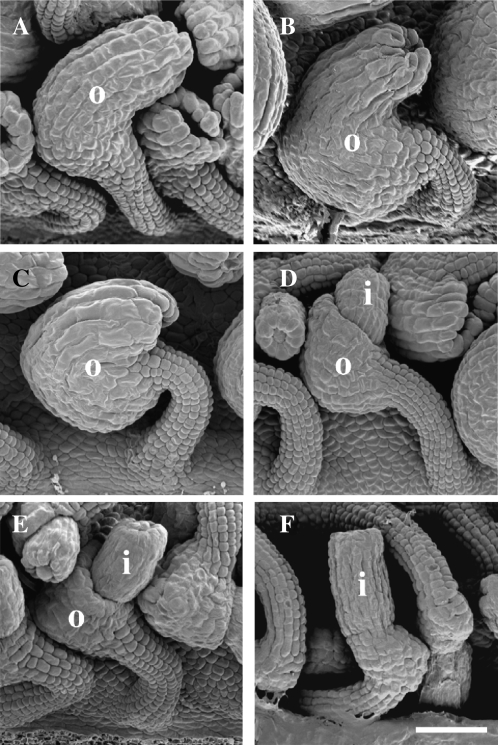
Scanning electron micrographs of representative ovule phenotypic classes observed from INO and YAB3, YAB5, or CRC chimeric cDNAs expressed using PROINO in an ino-1 mutant background. Growth of the outer integument occurred on both the gynobasal and gynoapical sides of ovules of some transformants as shown in A and B, representing sup-like and weak-sup classes, respectively. Other lines showed a wild-type phenotype with growth initiating and proliferating primarily on the gynobasal side of the ovule (C). D and E, A range of outer integument growth was exhibited in lines classified as weak-ino. F, Some transgenic lines showed no outer integument growth and thus exhibited the unaltered ino-1 mutant phenotype. i, Inner integument; o, outer integument. Scale bar = 50 μm.
To determine the significance of differences observed in ovule development for chimeric lines as compared to the control lines, the data were subjected to a Fisher's exact test. A Bonferroni adjustment was used to adjust the α value to account for the increased probability of error when numerous pairwise comparisons are made within a single set of data (Sonnenberg, 1985). Statistical analyses of all domain swap lines in comparison to control lines are reported in Supplemental Tables S1 to S6.
PROINO:III versus PROINO:III/YAB3 Swaps
Ovules of transgenic lines expressing 3II, II3, or III3 under PROINO commonly exhibited wild-type growth of the outer integument and were not significantly different from ovules of lines containing PROINO:III (Table I). In contrast, transgenic lines I3I, 3I3, I33, 33I, and 333i expressed under PROINO were significantly different from PROINO:III plants, with increasing significance as listed from left to right (Table I; Supplemental Table S1). Plants from these lines contained ovules that were more like those induced by PROINO:333 whereby growth of the outer integument frequently occurred on both the gynobasal and the gynoapical sides of the ovules. Based on these results, it appears that the presence of two adjacent regions of INO is sufficient to promote outer integument development like that of the full-length INO protein, with no significant difference between II3 and 3II when compared to INO or each other (Tables I and II). Both I33 and I3I chimeras were significantly different from INO and 3II but were not significantly different from II3. This indicates that adjacent regions 2, 3, and 4 are better able to support growth like that of INO than adjacent regions 1, 2, and 3 (Fig. 2A). C-terminal domain swaps between YAB3 and INO show that the C terminus of INO is not essential for INO function in this context, because PROINO:III3 transgenics were not significantly different from PROINO:III transgenics. Taken together, this indicates that the central variable and YABBY regions contain important information for the unique function of INO in responding to repression by SUP.
Table II.
Pairwise comparisons of phenotypic class distribution
| PROINO:Transgene Comparisona | P Valueb | Significantly Different from Each Other |
|---|---|---|
| I33 versus 3II | 2.31 × 10−05 | Yes |
| I33 versus 3I3 | 1.20 × 10−03 | Yes |
| I3I versus 3II | 3.00 × 10−03 | Yes |
| I3I versus 3I3 | 6.20 × 10−03 | Yes |
| II3 versus 33I | 5.33 × 10−05 | Yes |
| II3 versus 3I3 | 1.30 × 10−03 | Yes |
| 3II versus 33I | 7.84 × 10−06 | Yes |
| 3II versus 3I3 | 5.00 × 10−03 | Yes |
| I33 versus I3I | 1.02 × 10−01 | No |
| I33 versus II3 | 2.13 × 10−02 | No |
| I33 versus 33I | 4.56 × 10−02 | No |
| I3I versus II3 | 6.83 × 10−02 | No |
| I3I versus 33I | 3.77 × 10−02 | No |
| II3 versus 3II | 1.71 × 10−01 | No |
| 33I versus 3I3 | 1.16 × 10−02 | No |
| I55 versus II5 | 8.60 × 10−03 | Yes |
| I5I versus II5 | 5.90 × 10−03 | Yes |
| II5 versus 5I5 | 9.62 × 10−04 | Yes |
| I55 versus I5I | 2.30 × 10−01 | No |
| I55 versus 5II | 2.54 × 10−01 | No |
| I55 versus 55I | 4.10 × 10−01 | No |
| I55 versus 5I5 | 5.73 × 10−01 | No |
| I5I versus 5II | 1.00 × 1000 | No |
| I5I versus 55I | 1.23 × 10−01 | No |
| I5I versus 5I5 | 4.02 × 10−01 | No |
| II5 versus 5II | 1.14 × 10−02 | No |
| II5 versus 55I | 1.55 × 10−01 | No |
| 5II versus 55I | 1.94 × 10−01 | No |
| 5II versus 5I5 | 4.08 × 10−01 | No |
| 55I versus 5I5 | 7.15 × 10−02 | No |
| IIIc versus CCCi | 1.21 × 10−05 | Yes |
Transgenes were constructed as transcriptional fusions of the listed coding sequence with PROINO and examined in an ino-1 mutant background.
The phenotypic class distribution of each set was calculated using Fisher's exact test to determine the P value.
PROINO:333 versus PROINO:INO/YAB3 Swaps
When comparing PROINO:INO/YAB3 domain swap lines with lines expressing PROINO:333 (in an ino-1 background), all lines except I3I and 333i transgenics were significantly different from PROINO:333 (Table I). Although INO/YAB3 domain swap lines were similar to PROINO:333 transgenics (all swaps resulted in a proportion of individuals with weak-sup ovules), many domain swap lines possessed wild-type ovules, in contrast with PROINO:333 transgenics. This is notable with PROINO:I3I, a line that did not meet our most stringent criterion for statistically significant difference from PROINO:333. However, this construct led to a total of 10 wild-type individuals, while PROINO:333 did not result in any wild-type individuals and in fact was different from PROINO:333 if the significance level was raised to a mere 1.04% cutoff. We therefore consider this line to show a real difference from PROINO:333. Although all lines were able to support growth of the outer integument, the extent of growth varied among lines (Table I). Based on our results, the central variable region of YAB3 contains sufficient ability to promote some growth of the outer integument on the gynoapical side of ovules, as evidenced by PROINO:I33, I3I, and 33I transgenic lines. The presence of adjacent regions from YAB3 did not enhance the ability of chimeric proteins to stimulate outer integument growth, because PROINO:I33 and 33I were not significantly different from PROINO:I3I (Table II). The results for PROINO:333i indicate that the C terminus of YAB3 is not necessary for the promotion of growth nor escape from SUP repression when expressed in ovules, as these transgenics were not significantly different from PROINO:333 plants. This also shows that all three primary regions of INO contribute to its ability to respond to SUP, with the central variable region providing the smallest increment of this activity.
INO/YAB3 Chimera Expression Using the Cauliflower Mosaic Virus 35S Promoter
In parallel with phenotypic analysis of chimeric YABBY expression using PROINO, ectopic expression using the generally active cauliflower mosaic virus 35S promoter (PRO35SCaMV) was pursued to address the functionality of the chimeric cDNAs. When expressed ectopically, YABBY family members, including both YAB3 and INO, have been shown to alter leaf morphology, leading to narrowed and curled leaves, likely due to the abaxialization of adaxial tissue types (Eshed et al., 1999; Siegfried et al., 1999; Meister et al., 2005). Detection of similar effects from ectopic expression of the chimeric YABBY/INO genes would show that the genes produced functional YABBY-type proteins. With examples shown in Figure 4, all chimeric lines created with the PRO35SCaMV expression vectors resulted in leaf abnormalities consistent with ectopic YABBY effects. Thus, all of the chimeric proteins functioned as YABBYs, as was observed in prior domain swap experiments with CRC and INO (Meister et al., 2005).
Figure 4.
Ectopic expression of wild-type or chimeric YABBY proteins produced alterations in vegetative and reproductive development. Expression using PRO35SCaMV often resulted in gynoecia and cauline leaves that were misshapen and had irregular surfaces (A) when compared to wild-type Arabidopsis (B). Leaves of the transgenic plants were often narrow and epinastic (C) as compared to those of wild-type plants (D). Scale bar = 10 mm.
INO/YAB5 Chimera Expression Using PROINO
The majority of ino-1 mutant plants harboring the PROINO:555 (YAB5) construct exhibited no growth of the outer integument, and when any growth was observed, it was very limited (Table I). Following the strategy described for domain swaps between YAB3 and INO, chimeric YABBY proteins composed of YAB5 and INO were created and expressed using PROINO. As with the YAB3 and INO domain swaps, a range of phenotypes was observed and compared to the control lines (Fig. 3; Table I). The data were analyzed as for the YAB3/INO domain swaps.
PROINO:III versus PROINO:INO/YAB5 Swaps
Although the ability to support outer integument growth was markedly less than that seen for INO/YAB3 domain swaps, all INO/YAB5 chimeras were able to support some degree of growth (Table I). The transgenic line PROINO:II5 was the only line that did not significantly differ from PROINO:III (Table I), because a majority of ovules from this line exhibited wild-type growth of the outer integument. This line, however, was not significantly different from PROINO:5II or 55I lines that most often resulted in ovules with weak ino phenotypes (Tables I and II), indicating that some degradation of activity relative to the wild-type INO protein is likely. Overall, the data demonstrate that any one region from INO is sufficient to promote outer integument growth, albeit weakly in many INO/YAB5 chimeric lines. A quantitative effect was not observed, as PROINO:5II did not support growth to a greater extent than PROINO:55I.
PROINO:555 versus PROINO:INO/YAB5 Swaps
All INO/YAB5 domain swap lines expressed using PROINO (in an ino-1 background) were significantly different from ino-1 plants expressing PROINO:555 (Table I). This is largely because a YAB5/INO chimeric gene containing any one region of INO was sufficient to support some growth of the outer integument for all chimeric lines observed. As expected, ino-1 plants were not significantly different from PROINO:555 plants, because YAB5 was rarely able to support any growth of the outer integument (Table I).
INO-YAB5 Chimera Expression Using PRO35SCaMV
As with ectopic expression analysis of YAB3 and INO domain swaps, all chimeric lines of domain swaps between YAB5 and INO resulted in the characteristic YABBY overexpression phenotypes observed previously (Fig. 4; Eshed et al., 1999; Siegfried et al., 1999; Meister et al., 2005). Thus, the weak or absent ability of the chimeric proteins to support integument growth results from specific properties of the proteins, rather than being an indication of a generally inactive protein.
C-Terminal Domain Swaps Using PROINO
As mentioned previously, domain swap experiments that exchanged regions 1, 2, or 3 and 4 from CRC and INO suggested that regions 3 and 4 provided more functional information than regions 1 and 2 (Fig. 2A; Meister et al., 2005). To address whether this specific information was contained within region 3 or 4, these regions were swapped to individually evaluate their roles. When the C-terminal domain swaps were expressed using PROINO, the majority of lines showed an ability to support outer integument growth (Fig. 3). The strategy for classification described above was used to evaluate phenotypes and statistical significance.
Transgenic lines expressing IIIc or CCCi under PROINO were significantly different from PROINO:III, with a larger degree of significance for the latter line (Table I; Supplemental Table S5). These results are due to a significant proportion of PROINO:IIIc plants that contained ovules with weak-sup or sup-like phenotypes. This suggests that the C terminus of CRC is sufficient to overcome the repressive action of SUP. In contrast, the majority of transgenic lines containing PROINO:CCCi were only able to support outer integument growth weakly (Table I). This result implies that the ability of CRC domain swaps to support outer integument growth like that of PROINO:CCC (CRC) transgenic plants was dependent on the presence of the C-terminal region of CRC. As expected, IIIc and CCCi were significantly different from each other (Table II) due to the ability of IIIc to produce wild-type ovules, whereas CCCi provided only limited support for growth of the outer integument.
PROINO:IIIc and CCCi transgenic lines were significantly different from PROINO:CCC plants (Table I). Although several lines with PROINO:IIIc had weak-sup and sup-like phenotypes, several plants also had wild-type ovules unlike plants containing PROINO:CCC. PROINO:CCCi plants rarely supported outer integument growth more than that observed for a weak-ino phenotype, indicating the importance of the C-terminal portion of CRC for CRC-like function when expressed in ovules.
C-Terminal Domain Swaps Using PRO35SCaMV
As for the other tested constructs, the C-terminal domain swaps were tested for YABBY function through expression from PRO35SCaMV. The resulting chimeric lines exhibited leaf abnormalities consistent with ectopic YABBY effects. Thus, the limited function of CCCi in supporting integument growth does not appear to result from a general loss of all protein activity.
DISCUSSION
Previous work using INO and CRC domain swaps by Meister et al. (2005) addressed the particular properties of these two reproductive-specific YABBY members in the context of growth promotion and response to SUP inhibition. The unique role of CRC during nectary development (Bowman and Smyth, 1999; Baum et al., 2001), however, indicates that the effects observed in INO/CRC domain swaps may have been due to features not shared by other YABBY members. Such a specialized divergence for CRC is consistent with the observation that other YABBY proteins cannot substitute for CRC in carpel or nectary development (Meister et al., 2005). To address this issue, our current study expands on this work by examining domain swaps between INO and YAB3, a YABBY protein produced in both vegetative and reproductive organs that also fails to respond to SUP, and YAB5, the only Arabidopsis YABBY member that failed to support outer integument growth when expressed under PROINO (Meister et al., 2005).
YAB3 and INO Domain Swaps
Our results using YAB3/INO domain swaps were largely similar to those for CRC and INO, indicating that the reproductive role of CRC does not confer a special ability to function in INO/CRC chimeras. In INO/YAB3 domain swap lines, the presence of adjacent regions 2, 3, and 4 from INO supported outer integument growth more like that of the complete INO protein than chimeras that possessed only one region from INO or two, nonadjacent regions from INO. This observation parallels that of Meister et al. (2005), who found that INO/CRC domain swap lines were most similar to wild type when regions toward the C-terminal portion of INO were included. Furthermore, it was found that inclusion of the central variable region of YAB3 was sufficient to overcome SUP repression in most cases. This suggests that there are responsive elements to SUP repression within the central variable region of INO that are lacking in YAB3, as the differential response to SUP is the only difference observed between the expression of YAB3 or INO using PROINO.
Although the presence of the central variable region of YAB3 in I3I did confer some resistance to SUP repression, the central region of INO in 3I3 was not sufficient for the proper response to SUP repression (Table I). This implies that there are sequences outside of the central variable region that are also important for proper SUP repression. In contrast, it appears that there is little information necessary for INO function within the C terminus, because plants expressing INO with an exchanged C terminus (PROINO:IIIc and PROINO:III3) possessed wild-type ovules (Table I).
YAB5 and INO Domains Swaps
PROINO:555 plants rarely supported any outer integument growth, but plants possessing YAB5/INO chimeras of any combination were capable of supporting outer integument growth at least to a limited extent (Table I). This indicates that all regions contribute to INO-specific function. While PROINO:II5 resulted in ovule phenotypes that could not be statistically differentiated from PROINO:III, no other chimeric combinations between INO and YAB5 were able to support growth as well as INO itself. Interestingly, the other chimeric YAB5/INO transgenes with two regions from INO did not exhibit any greater rescue of the ino-1 phenotype than did YAB5/INO chimeric proteins with only one region from INO. Therefore, a combination of differences between INO and YAB5 distributed over at least three regions of the protein is required to render the protein incapable of supporting outer integument growth in ino-1 plants.
In contrast with domain swaps between INO and CRC or YAB3, quantitative effects due to increase in the fraction of INO content were not observed for YAB5/INO chimeras. Based on our hypothesis that INO makes multiple contacts with proteins and/or DNA targets to elicit INO-specific function, these results for YAB5/INO domain swaps were surprising. Although we do not present direct evidence, it seems likely that the YAB5 protein is capable of binding to INO DNA targets but fails to drive proper expression of these genes. This is supported by our observation that 5I5 chimeras, which contain DNA-binding domains exclusively of YAB5 origin, supported outer integument growth to a limited extent, although we cannot rule out the potential role of the central variable region in this interaction.
It is likely that the binding of YAB5 to INO DNA targets, or its interaction with trans-factors needed for expression of these targets, is inefficient due to the absence or misorientation of critical residues that facilitate these interactions. Reduction in these interactions could be produced by disruption of intramolecular contacts necessary for formation of the most active protein structure due to the presence of the YAB5 sequences. If the presence of any region of YAB5 could cause such disruption, then this could explain the lack of additivity observed for multiple segments of INO. Thus, any level of contribution from INO to YAB5/INO chimeras promotes growth, but the ability to support growth like that of full-length INO requires interactions between all three regions of the chimeric protein. Alternatively, the nonquantitative behavior of INO in these chimeras may simply be a result of greater structural divergence of YAB5 from other YABBY members, disrupting intermolecular interactions no matter the amount of INO identity within each chimera. We hypothesize that YAB3 and CRC at least partially conserve the inter- and intramolecular contact sites and so behave more like INO regions in the chimeric proteins.
C-Terminal Domain Swaps
YAB3/INO C-terminal domain swaps had little effect on the ability of the chimeric proteins to influence ovule development (Table I). These results suggest that the C-terminal portion of INO does not include information that can overcome the sup-like response observed when the YAB3 protein is expressed in ovules.
In contrast with the C-terminal domain swaps between YAB3 and INO, the swaps between CRC and INO show that the C-terminal region of CRC does possess unique information (Table I). The distribution of phenotypes among transgenics with CCCi spans the entire range of phenotypic classes; however, the majority of plants possessed weak-ino ovules. This suggests that the ability of the CRC protein to both support outer integument growth and overcome SUP repression is at least partially dependent on certain residues contained within the C-terminal region. This could reflect the need for specific intermolecular interactions between the C-terminal region and the rest of the CRC protein for formation of an active structure. In further support of the importance of the C-terminal region, while most plants containing PROINO:IIIc exhibited a wild-type ovule phenotype, several transgenic plants possessed weak-sup ovules and sup-like ovules (Table I). Thus, the presence of the C-terminal portion of CRC was sufficient for the IIIc protein to overcome SUP repression in some plants. This ability, however, is not exclusively found within the C-terminal region, as PROINO:IIIc plants did not support the sup-like or weak-sup phenotype as frequently as PROINO:CCC. Based on the results of YAB3/INO domain swaps, it appears unlikely that the C-terminal portion of INO played a role in the phenotypes observed for PROINO:IIIc and PROINO:CCCi; rather, the presence or lack of the C-terminal region of CRC was the critical factor in determining the extent and nature of outer integument growth.
CONCLUSION
We have found that no particular region of INO contained the specific information responsible for either the differential promotion of outer integument growth or differential response to SUP. This parallels results described for INO/CRC domain swaps in Meister et al. (2005), indicating that reproductive-specific features of CRC do not contribute to the effects observed for INO/CRC domains swaps. Furthermore, we present novel evidence that shows that the C-terminal region of CRC is necessary for chimeric proteins to support outer integument growth and escape SUP suppression in ovules, while, in contrast, the C-terminal region of INO does not appear to possess unique information necessary for INO function. We have further shown that while the central variable region of YAB3 is sufficient to overcome SUP growth repression, the central variable region of INO is not sufficient to mediate suppression by SUP. This novel finding suggests that while there are SUP-responsive elements in the central variable region of INO that have diverged in YAB3, there exist elements outside of the central variable region of INO that participate in the response to SUP. Lastly, we have shown that a single region derived from INO is sufficient to confer growth-promoting activity of YAB5/INO chimeras; however, in contrast with YAB3 and CRC domains swaps with INO, features of INO are required along the entire length of YAB5/INO chimeras for wild-type outer integument growth.
The positive quantitative effect of INO observed in domain swaps with YAB3 and CRC may reflect multiple unique adaptations of the INO protein for the specialized role it plays in reproductive development. Such specialization is supported by the observation that YAB5 cannot substitute for INO but ectopic expression of YAB5 or INO results in similar vegetative phenotypes. Thus, features of YABBY proteins necessary for interactions that lead to the vegetative effects of ectopic expression appear to be conserved among all Arabidopsis YABBY paralogs, and intramolecular interactions necessary for establishing a structure that can function in this capacity must also be preserved.
From a mechanistic standpoint, our data indicate that the INO protein makes multiple contacts with proteins or DNA sequences that are required for the proper temporal and spatial expression of genes necessary for the establishment of the outer integument. Different sets of intermolecular contacts may be disrupted by particular differences between INO, CRC, YAB3, and YAB5, explaining why different regions of these proteins have the greatest effects in domain swaps. In support of this observation, in vitro studies using pull-down assays demonstrated an ability of the Zn-finger and YABBY regions of the INO protein to independently bind the transcription factor NOZZLE/SPOROCYTELESS, a protein that is necessary for the establishment of the nucellus and pollen sacs in Arabidopsis (Sieber et al., 2004). This supports our findings that domains with discrete function cannot be readily identified in INO and that the protein instead acts as an integrated unit that likely requires multiple intermolecular interactions on a variety of surfaces of INO to form active complexes capable of driving proper outer integument growth.
Overall, our results suggest that INO's function during ovule development involves a coordination of molecular events that are distinct from those events that are shared among the YABBY family. These reproductive-specific events, dependent upon all three regions of INO, are necessary for the promotion of integument growth and the response to modulation by SUP. While YAB3 and CRC can participate in all the intermolecular interactions necessary for the promotion of growth, they fail to participate in interactions necessary for repression by SUP. Biochemical identification of factors that associate with INO and gene targets of INO-mediated transcriptional regulation will elucidate the mechanisms by which INO promotes the development of the outer integument and reveal the particular contacts made by each region of the protein. Because we have not detected physical interactions between INO and SUP, or INO and PROINO using yeast two- and one-hybrid studies, respectively (data not shown), the identification of additional factors involved in this process is key to understanding the molecular events that coordinate ovule development.
MATERIALS AND METHODS
Construct Assembly
Chimeric Coding Sequences
Chimeric coding sequences were created using overlap extension PCR (Horton et al., 1990) to create fusions between regions of YAB3, YAB5, INO, and CRC. Using the same strategy employed by Meister et al. (2005), chimeric cDNAs were synthesized and inserted into pLITMUS28 (New England Biolabs) using the restriction sites BamHI/XbaI or EcoRI/XbaI depending on the source of template being used (data not shown). The sequence of the chimeric cDNAs was confirmed on an Applied Biosystems 3730 sequencing system.
PROINO Expression Constructs of Chimeric cDNAs
Chimeric cDNAs were introduced into pRJM42 that contained the regions 5′ and 3′ of the genomic INO coding sequence necessary for the proper temporal and spatial expression pattern of endogenous INO (Meister et al., 2004). Assembly of these constructs was performed using the restriction sites BamHI/XbaI or BglII/XbaI according to the particular chimeric cDNA used (data not shown). This cloning strategy replaced the CRC coding sequence originally incorporated in pRJM42.
PRO35SCaMV Expression Constructs of Chimeric cDNAs
Chimeric cDNAs digested with BglII/XbaI were introduced into pMON999 digested with the same enzymes. pMON999 contained a modified PRO35SCaMV (Kay et al., 1987) with a polyadenylation signal sequence of nopaline synthase 3′.
Plant Growth and Transformation
Fragments including the chimeric cDNAs in the appropriate expression cassettes listed above were excised with NotI, the chimeric genes were inserted into pMLBART using the same site, and the resulting plasmids were transferred into the Agrobacterium strain ASE via triparental matings (Figurski and Helinski, 1979; Fraley et al., 1985; Gleave, 1992). Wild-type Landsberg erecta (Ler) and INO (Columbia/ino-1 [Ler]) plants (Arabidopsis [Arabidopsis thaliana]) were grown on soil in constant light at 20°C for transformation using the floral dip method (Kranz and Kirchheim, 1987; Clough and Bent, 1998). T1 lines were selected following germination on soil by treatment with the herbicide glufosinate ammonium (Finale; Farnham Companies), which selects for the phosphinothricin acetyl transferase gene. Homozygous ino-1 plants were identified using a detectable polymorphism between Columbia and Ler ecotypes at the INO locus as described previously (Meister et al., 2002).
Microscopy
Determination of phenotype class for transformants containing chimeric cDNAs expressed using PROINO was initially performed using dark-field microscopy. Representative lines of each phenotypic class were then fixed and prepared for analysis using scanning electron microscopy following methods described previously (Broadhvest et al., 2000).
Statistical Analysis
Statistical analysis of significant differences between data sets of each chimera tested and against control lines was performed using Fisher's exact test at the UC Davis Statistical Laboratory. Significant differences between phenotypic effects of the transgenes were calculated with the modified Bonferroni adjustment (α value/number of pairwise comparisons) to reduce the rate of false-positives while conducting simultaneous pairwise comparisons. This method has been shown to be advantageous when multiple statistical analyses are conducted simultaneously (Wright, 1992; Morikawa et al., 1996).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Pairwise comparison of ovule phenotypes between PROINO:INO/YAB3 and PROINO:III.
Supplemental Table S2. Pairwise comparison of ovule phenotypes between PROINO:INO/YAB3 and PROINO:333.
Supplemental Table S3. Pairwise comparison of ovule phenotypes between PROINO:INO/YAB5 and PROINO:III.
Supplemental Table S4. Pairwise comparison of ovule phenotypes between PROINO:INO/YAB5 and PROINO:555.
Supplemental Table S5. Pairwise comparison of ovule phenotypes between PROINO:INO/CCC and PROINO:III.
Supplemental Table S6. Pairwise comparison of ovule phenotypes between PROINO:INO/CCC and PROINO:CCC.
Supplementary Material
Acknowledgments
We thank Sangman Kim, Kristina Passerini, and Abel Unzueta for technical help during the analysis of transgenic lines, the UC Davis School of Medicine and the MCB Imaging Facility for use of scanning electron microscopes, Kay Robinson-Beers for some wild-type scanning electron microscope images, Debra Skinner, Robert Meister, and Ryan Brown for technical advice, Neil Willits for help with statistical analysis, and Stacey Harmer for helpful support.
This work was supported by the National Science Foundation (grant no. IOB–0419531 to C.S.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Charles S. Gasser (csgasser@ucdavis.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alvarez J, Smyth DR (1999) CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126 2377–2386 [DOI] [PubMed] [Google Scholar]
- Baker SC, Robinson-Beers K, Villanueva JM, Gaiser JC, Gasser CS (1997) Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145 1109–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum SF, Eshed Y, Bowman JL (2001) The Arabidopsis nectary is an ABC-independent floral structure. Development 128 4657–4667 [DOI] [PubMed] [Google Scholar]
- Bowman JL (2000) The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol 3 17–22 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126 2387–2396 [DOI] [PubMed] [Google Scholar]
- Broadhvest J, Baker SC, Gasser CS (2000) SHORT INTEGUMENTS 2 promotes growth during Arabidopsis reproductive development. Genetics 155 895–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209 [DOI] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA 76 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley RT, Rogers SG, Horsch RB, Eichholtz DA, Flick JS, Fink CL, Hoffmann NL, Sanders PR (1985) The SEV system: a new disarmed Ti plasmid vector for plant transformation. Biotechnology (N Y) 3 629–635 [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 1203–1207 [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR (1990) Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8 528–535 [PubMed] [Google Scholar]
- Kanaya E, Watanabe K, Nakajima N, Okada K, Shimura Y (2001) Zinc release from the CH2C6 zinc finger domain of FILAMENTOUS FLOWER protein from Arabidopsis thaliana induces self-assembly. J Biol Chem 276 7383–7390 [DOI] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236 1299–1302 [DOI] [PubMed] [Google Scholar]
- Kranz AR, Kirchheim B (1987) Handling of Arabidopsis. In AR Kranz, ed, Arabidopsis Information Service, v. 24: Genetic Resources in Arabidopsis. Arabidopsis Information Service, Frankfurt, pp 4.1.1–4.2.7
- Meister RJ, Kotow LM, Gasser CS (2002) SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129 4281–4289 [DOI] [PubMed] [Google Scholar]
- Meister RJ, Oldenhof H, Bowman JL, Gasser CS (2005) Multiple protein regions contribute to differential activities of YABBY proteins in reproductive development. Plant Physiol 137 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister RJ, Williams LA, Monfared MM, Gallagher TL, Kraft EA, Nelson CG, Gasser CS (2004) Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter region. Plant J 37 426–438 [DOI] [PubMed] [Google Scholar]
- Morikawa T, Terao A, Iwasaki M (1996) Power evaluation of various modified Bonferroni procedures by a Monte Carlo study. J Biopharm Stat 6 343–359 [DOI] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Kanaya E, Morita EH, Okada K (1999) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Petrascheck M, Barberis A, Schneitz K (2004) Organ polarity in Arabidopsis. NOZZLE physically interacts with members of the YABBY family. Plant Physiol 135 2172–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews DN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 128 4117–4128 [DOI] [PubMed] [Google Scholar]
- Sonnenberg A (1985) Bonferroni-Holm sequential test procedure. Z Gastroenterol 23 703–704 [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS (1999) INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev 13 3160–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SP (1992) Adjusted P-values for simultaneous inference. Biometrics 48 1005–1013 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.