Abstract
Telomerase, a ribonucleoprotein complex, adds hexameric repeats called “telomeres” to the growing ends of chromosomal DNA. Characterization of mammalian telomerase has been elusive because of its low level of expression. We describe a bioinformatics approach to enrich and characterize the human telomerase complex. Using local sequence homology search methods, we detected similarity of the Tetrahymena p80 subunit of telomerase with the autoantigen Ro60. Antibodies to Ro60 immunoprecipitated the telomerase activity. Ro60 and p80 proteins were cross-recognizable by antibodies to either protein. Telomerase activity and the RNA component of telomerase complex were localized to a doublet in a native gel from the Ro60 antibody-precipitated material. The enriched material showed specific binding to a TTA GGG probe in vitro in an RNA template-dependent manner. Polyclonal antibodies to the doublet also immunoprecipitated the telomerase activity. These results suggest an evolutionary conservation of the telomerase proteins.
Keywords: bioinformatics, Ro60, RNP, autoantigen, telomeres
The eukaryotic chromosomes are made up of an array of tandem repeats of the hexanucleotide 5′-TTA GGG-3′, called “telomeres,” which protect the ends of chromosomes against many challenges (for a review see refs. 1–4). Telomerase, a ribonucleoprotein enzyme, uses its own RNA as a template to add telomeres to the ends of chromosomes (2). Telomerase activity has been detected in a vast majority of human tumors but in few normal somatic cells (5–8). Attempts to characterize telomerase complex have been largely based on ciliates because of their high levels of telomerase activity. Two subunits of Tetrahymena telomerase complex have been cloned (9). The p80 subunit has been shown to bind to the telomerase RNA component, and the p95 subunit has been shown to bind to DNA. Recently characterization of telomerase from Euplotes aeoiculatus has been reported (10).
The RNA components of diverse ciliate and eukaryotic telomerases have been cloned, and the ciliate RNA shows an evolutionary conservation at the secondary structure level (11, 12). We reasoned that such conservation might extend to the proteins. Using a sequence homology search strategy specific for Tetrahymena proteins, we detected a human protein, Ro60, with apparent sequence similarity with p80. The Ro60 protein is an RNA-associated protein, a subunit of a ribonucleo particle (RNP), and an autoantigen that has been implicated in systemic autoimmune diseases (13, 14). The gene for the Ro60 protein encodes RNP-consensus motifs found in several RNA-binding proteins (15). Intrigued by the coincidence of homology between Tetrahymena p80 and Ro60 (both RNA-binding proteins), we tested the ability of anti-Ro60 antibodies to recognize the human telomerase complex.
We report that antibodies to Ro60 and Tetrahymena p80 cross-recognize either protein; Ro60 antibodies immunoprecipitate functional telomerase activity in human cells. We used this approach to enrich mammalian telomerase activity, and we describe here the characterization of telomerase complex.
MATERIALS AND METHODS
Cell Lines and Reagents.
SW-480, human colon carcinoma cells, and 293, immortal human kidney cells, were maintained as described (16, 17). Anti-Ro60 rabbit polyclonal antibody (18), Ro60 protein [bovine (19, 20) and recombinant (21)], and anti-Ro60 mAb 2G10 (22) have been described. Tetrahymena p80 polyclonal antibody, 1589 (C terminus) was generated in rabbits (K.C., unpublished work). Tetrahymena S-100 lysates were made as described (9).
Freeze-Thaw Lysate and Immunoprecipitation.
Cell pellets were washed with PBS (pH 7.4) and pelleted at 6000 × g for 7 min. The supernatant was removed, and the cell pellet was resuspended in 1 vol of immunoprecipitation buffer containing PBS (pH 7.4), 1 mM MgCl2, 1 mM EDTA, 0.05% Nonidet P-40, 1% glycerol, 10 units of RNase inhibitor, 1 μM leupeptin, 10 μM pepstatin, 100 units of aprotinin, and 4.3 mM β-mercaptoethanol. The suspension was freeze-thawed (10 min, 3 cycles), and lysates were prepared (17).
All of the immunoprecipitation steps were performed at 4°C. Five μl of the Ro60 antiserum was incubated with 50 μg of freeze-thaw lysate. After 4 h, 25 μl of protein-A Sepharose beads (50% wt/vol in PBS; Sigma) was added to bring the final volume to 50 μl. After 18–19 h of incubation, the mixture was spun at 1000 × g for 30 s, the supernatant was stored, and the pellet was washed (5×) using 10 volumes of the immunoprecipitation buffer. The last wash and the pellet resuspended to the original volume of 50 μl were collected for the telomeric repeat amplification protocol (TRAP) assay. Elution was performed with 1% Triton X-100 in PBS for 30 min at 4°C. Fifty micrograms of cell lysate diluted to 50 μl served as the original extract in the TRAP assay.
Telomerase Assay.
Telomerase activity was measured essentially as described (5, 16, 17) by the PCR-based TRAP assay. Quantification of telomerase activity was done by picogreen assay (17). The gel slices were incubated in TRAP buffer for 30 min in ice before being analyzed for telomerase activity.
Reverse Transcriptase-PCR (RT-PCR).
RNA was extracted from gel slices and analyzed by RT-PCR for expression of hTR, human Ro60-associated RNA (hY3), and glyceraldehyde phosphate dehydrogenase (GAPDH). The PCR primers used follow: (i) hTR, sense, 5′-CAA CGA ACG GCC AGC AGC TGA CAT T-3′; antisense, 5′-GTC TAA CCC TAA CTG AGA AGG GCG TAG; amplicon, 158 bp; (ii) hY3, sense, 5′-GGC TGG TCC GAG TGC AG; antisense, 5′-AAA GGC TAG TCA AGT GAA GC-3′; amplicon, 101 bp; (iii) GAPDH, sense, 5′-CCA TGG AGA AGG CTG GGG-3′; antisense, 5′-CAA AGT TGT CAT GGA TGA CC-3′; amplicon, 208 bp.
Mobility-Shift Assays.
Material eluted from 500 μg of lysate was used to ascertain binding of the 32P-labeled (TTA GGG)12 probe by an electrophoretic mobility-shift assay (EMSA). A probe mixture consisting of 1.0 μl of the probe (1 ng), 3.0 μl of mobility-shift buffer (33 mM Tris acetate, pH 8.0/3.3 mM MgCl2/33% vol/vol glycerol), 1.5 μl of 100 ng/ml d(T)18, and 4.5 μl of sterile water was added to 10 μl of the eluate. For competition reactions, 1 μl of unlabeled competitor was added to the mixture. The samples were incubated at 4°C for 30 min, followed by UV irradiation at 254 nm for 20 min at 4°C. The reaction products were electrophoresed and resolved on 8% native PAGE, and the dried gel was autoradiographed. Control probes were pBR, 5′-AGC CAC TAT CGA CTA CGC GAT CAT-3′ and M13, 5′-TTT CAC CAG GAA ACA GCT ATG ACC AT-3′.
Antibody Generation.
Doublet was isolated on native gel from anti-Ro60 immunoprecipitation from 300 mg of total protein from SW-480 cells, and the gel slices were used as immunogen to generate polyclonal antiserum in rabbits (Lampire Biologicals, Pipersville, PA). Rabbit IgG was purified with an Immunopure A/G0 IgG purification kit (Pierce) using the manufacturer’s protocol.
RESULTS
Primary Structure Homology of Tetrahymena p80 with Ro60.
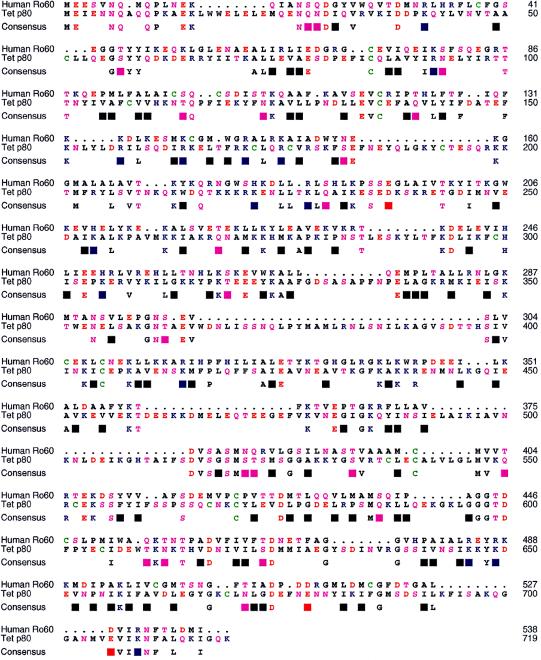
Searches of nucleotide and protein databases for sequence similarity using the published telomerase protein subunits from Tetrahymena p80 and p95 as query sequences were undertaken. The search parameters were tuned to account for the relatively AT-rich Tetrahymena genome (p80 = 65% AT; p95 = 73% AT) and the unusual codon usage of the ciliates, in addition to biases in Tetrahymena amino acid preferences. Use of the Smith–Waterman algorithm with optimized gap opening and extension penalties detected a human ribonucleoprotein, Ro60/SSA, with apparent homology to the RNA-binding subunit of Tetrahymena telomerase. A global alignment (BLOSUM 62 scoring matrix) revealed a 21% amino acid identity between the two proteins with five conserved cysteine residues (Fig. 1). The precise three-dimensional structures of the Ro60 and telomerase p80 proteins have not been determined; however, the arrangement of predicted secondary structural elements was also similar between these two proteins (data not shown). Hence, we next tested whether antibodies to Ro60 would recognize telomerase activity.
Figure 1.
Primary structure homology of p80 and Ro60. Amino acid homology of Tetrahymena telomerase p80 subunit (Tet p80) to human Ro60 is 21%. The telomerase subunit (accession no. U25641) was aligned globally to the human Ro60 (accession no. M25077) by the sequence-structure alignment tool XSAE 9 (v.1.4) and the BLOSUM 62 scoring matrix, then adjusted manually. Identical amino acids are displayed beneath the alignment; similar amino acids are denoted by blocks. The color scheme is AGILMPV, black; C, green; DE, red; FWY, dark grey; HKR, blue; and NQST, magenta.
Telomerase Activity Is Recognizable by Ro60 Antibody.
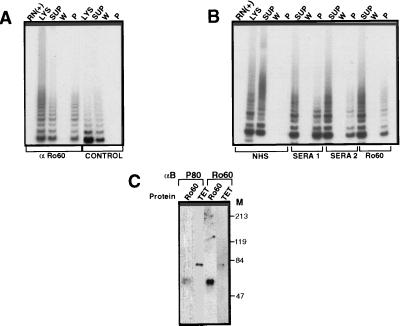
Total cell lysates from SW-480 cells were immunoprecipitated by anti-Ro60 antibody (polyclonal) or with no antibody, and equal volumes of supernatant, wash, and protein A beads were tested for telomerase activity by the TRAP assay (Fig. 2A). Telomerase activity was depleted in the cell lysates after immunoprecipitation (compare lysates vs. supernatant) and was readily detectable in the bead-bound Ro60 immunoconjugate. In contrast, with no antibody controls, telomerase activity was negligible in the pellet. Quantification of telomerase activity by picogreen assay showed a recovery of 20–25% after a single immunoprecipitation with Ro60 antibody. The residual activity in the supernatant was reimmunodepletable with an additional two rounds of immunoprecipitation with the recovery of intact telomerase activity in the beads (data not shown).
Figure 2.
The homology of p80 and Ro60 is functionally relevant. (A) Anti-Ro60 immunoprecipitates telomerase activity. Fifty micrograms of freeze-thawed lysates from SW-480 cells were immunoprecipitated with or without Ro60 antisera (rabbit, polyclonal) and the lysates (Lys), supernatant (Sup), wash (W), and pellet (P) were analyzed for telomerase activity by TRAP. RN (+) indicates pretreatment of lysate with RNase A (10 mg/ml). (B) Recognition of telomerase complex by Ro60-positive human sera. Fifty micrograms of freeze-thawed lysates from SW-480 cells were immunoprecipitated with either normal human serum (NHS), anti-Ro60-positive human sera (sera 1 or sera 2), or Ro60 polyclonal antisera, and the lysates (Lys), supernatant (Sup), and pellet (P) were analyzed for telomerase activity by TRAP. RNase A control as in A. (C) Antibodies to p80 and Ro60 cross-recognize respective proteins. Purified Ro60 (bovine, 0.25 μg) or Tetrahymena S-100 extract (10 μg) was analyzed by Western blot using Ro60 (polyclonal) or Tetrahymena p80 (C terminus) antibody. M, molecular weight markers.
Encouraged by these findings, we next tested whether anti-Ro60-positive human sera would recognize telomerase activity (Fig. 2B). Two different anti-Ro60-positive reference sera but not normal human serum immunoprecipitated telomerase activity. In addition, a mAb to Ro60 (2G10) immunoprecipitated the telomerase activity, albeit weakly. The anti-Ro60 antibody (polyclonal) immunoprecipitated telomerase activity in diverse human and murine tumor cell lines. In contrast, an irrelevant autoimmune serum containing antibodies to an amino acyl t RNA synthetase, a polyclonal antiserum to an RNA-binding protein, and to poly(A)DP-ribose polymerase and several varied normal rabbit and normal human sera did not immunoprecipitate telomerase activity (not shown).
We next reasoned that if the homology of Tetrahymena p80 and Ro60 is relevant, then antibodies to either should cross-recognize the respective proteins. Western blot analysis of purified Ro60 and Tetrahymena S-100 lysates probed with anti-Ro60 or p80 antibodies revealed that purified Ro60 protein is readily recognized by p80 antibody, and a very weak recognition of p80 protein from the Tetrahymena extract was seen by the Ro60 antibodies (Fig. 2C). These results suggested that the homology of p80 to Ro60 detected through a bioinformatics approach was more than coincidental and might provide a means to characterize the telomerase complex.
Telomerase Activity Is Localized to a Doublet.
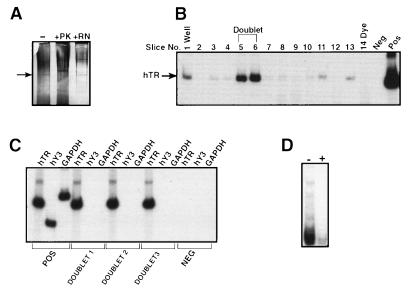
The bead-bound telomerase activity was eluted with Triton X-100, and the eluate was analyzed by native PAGE (Fig. 3A). A silver stainable doublet region, which migrated half way through the gel, was readily seen in the eluate. The protein nature of the doublet was established by sensitivity to proteinase K but lack of sensitivity to RNase A treatment (Fig. 3A). Purification of Tetrahymena telomerase also revealed a doublet when analyzed under native gel conditions using a mobility-shift assay (23). Hence we reasoned that the doublet seen in Fig. 3A might encompass hTR. The triton eluate was electrophoresed on native PAGE and cut into 0.5-mM slices, and the RNA was isolated from the slices and analyzed for the RNA component of human telomerase (hTR) by RT-PCR (Fig. 3B). The hTR expression peaked at the region on the gel that corresponded to the doublet. The mobility of free hTR (in vitro transcribed) was closer to the bottom of the gel (data not shown). Although the doublet region was positive for hTR, it was negative for hY3 RNA, a Ro60-associated RNA, and for control GAPDH (Fig. 3C). The doublet was also negative for other Ro60-associated RNAs (hY1, hY4, and hY5; data not shown). The unstained doublet region, when analyzed for telomerase activity by the TRAP assay, showed an RNase A-sensitive telomerase activity (Fig. 3D).
Figure 3.
Localization of telomerase activity to a doublet. (A) Analysis of Triton X-100 eluate of anti-Ro60 immunoprecipitates. Analysis was performed on an 8% native PAGE gel. Immunoprecipitates from 1 mg of lysates from SW-480 cells were either untreated (−), RNase A-treated (+ RN) or proteinase K-treated (+ PK) and detected by silver staining. Arrow indicates position of doublet. (B) Doublet peaks for hTR expression. RNA was extracted from the gel slices (0.5 cm) from the native gel and analyzed by RT-PCR for hTR expression. POS, positive control; NEG, template minus control. (C) Lack of Ro60-Associated hY3 RNA in the Doublet. RNAs from doublet excised from three different gel slices were analyzed by RT-PCR for hTR, hY3, and GAPDH expression. POS, positive control; NEG, negative control. (D) Telomerase activity comigrates with the doublet. A gel slice containing the doublet was analyzed for telomerase activity in the absence (−) or presence (+) of RNase A pretreatment.
Specific Binding of Anti-Ro60-Enriched Telomerase to TTA GGG Probe in Vitro.
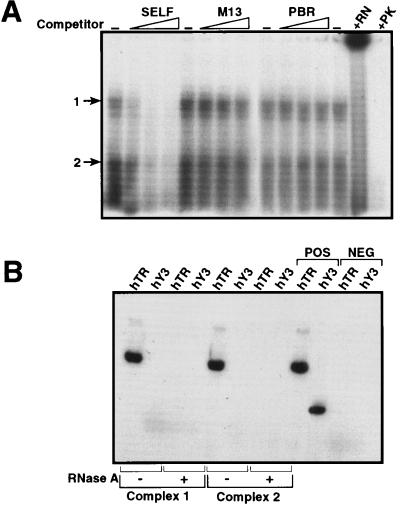
We next investigated whether the isolated human telomerase complex binds to the TTA GGG probe using gel shift-EMSA and UV cross-linking (Fig. 4A). The triton eluate of anti-Ro60 immunoprecipitate was incubated with radiolabeled (TTA GGG)12 probe, UV cross-linked, and analyzed by EMSA. Two distinct complexes were detected whose binding to the TTA GGG probe was competed by self-competitor and was not competed by two different irrelevant competitors (pBR 322 and M13; Fig. 4A).
Figure 4.
EMSA-UV cross linking of telomerase complex to TTA GGG probe. (A) Specificity of binding. The Triton X-100 eluate from anti-Ro60 immunoprecipitates was treated with 32P-labeled (TTA GGG)12 probe in the absence of competitor DNA (−) or presence of 10-, 100-, and 200-fold excess of unlabeled DNA (self) or irrelevant competitors (M13 and PBR), UV cross-linked, and analyzed by EMSA on 8% native gel. Two distinct complexes are denoted by 1 and 2. Eluate pretreatment with RNase A (+ RN) or proteinase K (+ PK) before cross-linking and EMSA. (B) Cross-linked complexes are hTR-positive. RNA was extracted from gel slices containing the cross-linked complexes (1 and 2) corresponding to the ±-RNase A lanes in A and was analyzed by RT-PCR for hTR and hY3 expression. POS, positive control (RNA from SW-480 cells); NEG, negative control (template minus).
The specific binding of the TTA GGG probe to the enriched telomerase complex was abolished by pretreatment with both proteinase K and RNase A. Both of these mobility complexes were positive for hTR expression but not for the hY3 (Ro60-associated) RNA as analyzed by RT-PCR (Fig. 4B); the hTR expression was abolished by pretreatment of the telomerase-enriched eluate with RNase A.
Antibody to the Doublet Immunoprecipitates Telomerase Activity.
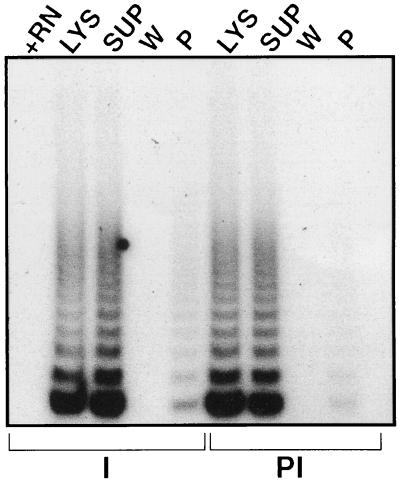
We next generated a rabbit polyclonal antibody to the doublet region by using the gel slices as an immunogen. The doublet antiserum immunoprecipitated telomerase activity in 293 cells; in comparison, the preimmune serum showed only background levels (Fig. 5). Rabbit polyclonal antibodies against the total eluate (from which the doublet was generated) also immunoprecipitated the telomerase activity (data not shown). The bead-bound activity was elutable by Triton X-100, showed the doublet when analyzed by native gel, and was hTR-positive and -negative for hY3 RNA expression.
Figure 5.
Antibody to doublet immunoprecipitates telomerase activity. Affinity-purified rabbit polyclonal antiserum raised against the doublet was used to immunoprecipitate telomerase activity as in Fig. 2A, and the lysate, supernatant, wash, and pellet were analyzed for telomerase activity by TRAP. PI, preimmune serum; I, immune serum; +RN, RNase A-inactivated lysate.
DISCUSSION
The widespread detection of telomerase activity in the vast majority of human tumors but in few normal tissues has raised the possibility that telomerase may be a novel cancer target (5). However, validation of the role of telomerase in cancer is limited by the lack of molecular reagents. Although the RNA component of the telomerase RNP has been cloned from several organisms including humans (24), characterization of telomerase protein subunits has been difficult because of its relatively low abundance. In this report, we describe a method to enrich the telomerase activity through a molecular lead predicted by bioinformatics efforts.
Sequence homology comparisons of the Tetrahymena telomerase p80 subunit against the protein and nucleotide databases revealed a match with a protein called Ro60. The Ro/SSA protein belongs to a family of autoantigens targeted by antibodies in sera from patients with systemic lupus erythematosus and Sjögren syndrome (14, 25). Expressed ubiquitously, the precise role of Ro60 is not known, and it has been suggested that it may form a part of a larger protein complex (26). Intrigued by the findings that Ro60 is an RNP similar to telomerase and shows homology to the Tetrahymena telomerase p80 subunit, we reasoned that antibodies against Ro60 might recognize telomerase.
Telomerase activity was immunoprecipitable with anti-Ro60 antibodies (polyclonal, human sera, and monoclonal). The inability of normal human sera and other control sera to immunoprecipitate functional telomerase activity adds to the specificity of Ro60-based recognition of telomerase. Purified Ro60 protein was readily recognized by p80 antibody whereas the recognition of the Tetrahymena p80 protein by Ro60 antibody was very weak. This probably reflects differences in the nature of the epitopes these two antibodies recognize. Nevertheless, these observations of cross-recognition support the starting premise of sequence homology between these two proteins.
We have localized telomerase activity from the anti-Ro60 immunoprecipitates to a doublet in native gel. The absence of Ro60-associated RNA (hY3) in the doublet argues against Ro60 being a requirement for telomerase activity; further support is the failure of purified Ro60 protein to bind to labeled hTR (data not shown). Thus, it is unlikely that Ro60 is an integral component of telomerase complex. Instead, these two proteins share sequence homology that can be exploited for cloning.
The functioning of telomerase requires binding to telomeric DNA in a template-dependent manner (RNA) to synthesize new repeats (2). Harrington et al. (23) have used gel-shift and UV cross-linking assays to monitor the purification of Tetrahymena telomerase. These authors have demonstrated specific single-strand DNA binding to the enriched telomerase; moreover, the binding was RNase-sensitive. These results suggested that the DNA-binding complex contains an RNA component necessary for complex stability. In contrast, Collins et al. (9) observed that primer binding was independent of RNase treatment.
Using a similar strategy, we observed that TTA GGG binding to the anti-Ro60-enriched telomerase was specific and sensitive to RNase A treatment. Two distinct complexes were seen in these gel shift–UV cross-linking experiments, and both of these complexes were abolished by pretreatment with RNase A. The detection of hTR (but not hY3 RNA) in both of these complexes could be due to degradation of proteins after cross-linking. Alternatively, there may be more than one form of telomerase complex in mammalian cells. Neither 5S RNA nor oligomers complementary to the immediate 5′ or 3′ end of the RNA motif (TTA GAC and TCT CAG, both 36 mers) competitively inhibited the complex formation (data not shown). Furthermore, the proteinase K sensitivity of the complex suggested that the TTA GGG binding involves the complementary motif present in the hTR of the telomerase-RNP.
The immunoprecipitation of telomerase activity by the polyclonal antibody raised against the doublet lends support to the possibility that telomerase components are located in the doublet region. Because of the high sensitivity of the TRAP assay, low levels of telomerase activity were seen occasionally with the control sera, similar to levels reported by Collins et al. (9).
Recently, a human homolog of a telomerase-associated protein (TP1) was described (27). TP1 shows significant amino acid homology to the Tetrahymena p80 protein, and antiserum to TP1 immunoprecipitates telomerase activity. Interesting to note, one region of TP1 (amino acids 484–554) shows 18% homology to the Ro60 protein. These authors also have inferred that the corresponding sequences of Tetrahymena p80 show 29% identity to Ro60. These results, taken in the context of our own, support the notion that telomerase proteins are conserved from ciliates to humans. Our results further indicate that Ro60 is neither the telomerase gene nor a part of the telomerase complex. Instead, there is simply a vestigial sequence conservation between these two proteins that, although less exciting in its implications, can still be harnessed as a valuable tool in telomerase research. Our ability to use the computer-aided homology prediction (p80 vs. Ro60) to characterize the telomerase complex in human cells opens new avenues for molecular cloning of telomerase genes.
Acknowledgments
We thank Dr. T. Gordon for recombinant Ro60, R. Hsiao and K. Higgins-Sochaski for technical assistance, and J. Narayanan for editorial help. The manuscript was produced using IBM Voice Type Dictation in os/2 warp 4. This work was supported in part by a grant from the Arthritis Foundation to A.D.F.
ABBREVIATIONS
- RNP
ribonucleo particle
- EMSA
electrophoretic mobility-shift assay
- GAPDH
glyceraldehyde phosphate dehydrogenase
- hY3
human Ro60-associated RNA
- PAGE
polyacrylamide gel electrophoresis
- RT-PCR
reverse transcriptase PCR
- TRAP
telomeric repeat amplification protocol
References
- 1.Harley C B. Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E H. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. Proc Natl Acad Sci USA. 1994;91:2882–2885. doi: 10.1073/pnas.91.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma H W, Maltese J Y, Zhu X, Kaiser H E, Narayanan R. Anticancer Res. 1996;16:511–516. [PubMed] [Google Scholar]
- 5.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 6.Broccoli D, Young J W, de Lange T. Proc Natl Acad Sci USA. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Counter C M, Gupta J, Harley C B, Leber B, Bacchetti S. Blood. 1995;85:2315–2320. [PubMed] [Google Scholar]
- 8.Hsiao R, Sharma H W, Ramakrishnan S, Keith E, Narayanan R. Anticancer Res. 1997;17:827–832. [PubMed] [Google Scholar]
- 9.Collins K, Kobayashi R, Greider C W. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 10.Lingner J, Cech T R. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero D P, Blackburn E H. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 12.Lingner J, Hendrick L L, Cech T R. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Chetrit E, Gandy B J, Tan E M, Sullivan K F. J Clin Invest. 1989;83:1284–1292. doi: 10.1172/JCI114013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reichlin M. J Clin Immunol. 1986;6:339–348. doi: 10.1007/BF00915372. [DOI] [PubMed] [Google Scholar]
- 15.Deutscher S L, Harley J B, Keene J D. Proc Natl Acad Sci USA. 1988;85:9479–9483. doi: 10.1073/pnas.85.24.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma H W, Sokoloski J A, Perez J R, Maltese J Y, Sartorelli A C, Stein C A, Nichols C G, Khaled Z, Telang N T, Narayanan R. Proc Natl Acad Sci USA. 1995;92:12343–12346. doi: 10.1073/pnas.92.26.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X, Kumar R, Mandal M, Sharma N, Sharma H W, Dhingra U, Sokoloski J A, Hsiao R, Narayanan R. Proc Natl Acad Sci USA. 1996;93:6091–6095. doi: 10.1073/pnas.93.12.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mamula M J, O’Brien C A, Harley J B, Hardin J A. Clin Immunol Immunopathol. 1989;52:435–446. doi: 10.1016/0090-1229(89)90158-x. [DOI] [PubMed] [Google Scholar]
- 19.Mamula M J, Fox O F, Yamagata H, Harley J B. J Exp Med. 1986;164:1889–1901. doi: 10.1084/jem.164.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickey W O, van Egmond J E, Hardgrave K L, Harley J B, Scofield R H. J Invest Dermatol. 1993;100:412–416. doi: 10.1111/1523-1747.ep12472055. [DOI] [PubMed] [Google Scholar]
- 21.Beer R G, Rischmueller M, Coates T, Purcell A W, Keech C L, McCluskey J, Gordon T P. Clin Immunol Immunopathol. 1996;79:314–318. doi: 10.1006/clin.1996.0084. [DOI] [PubMed] [Google Scholar]
- 22.Veldhoven C H, Pruijn G J, Meilof J F, Thijssen J P, Van der Kemp A W, Van Venrooij W J, Smeenk R J. Clin Exp Immunol. 1995;101:45–54. doi: 10.1111/j.1365-2249.1995.tb02275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington L, Hull C, Crittenden J, Greider C. J Biol Chem. 1995;270:8893–8901. doi: 10.1074/jbc.270.15.8893. [DOI] [PubMed] [Google Scholar]
- 24.Feng J, Funk W D, Wang S S, Weinrich S L, Avilion A A, Chiu C P, Adams R R, Chang E, Allsopp R C, Yu J, Le S, West M D, Harley C B, Andrews W H, Greider G W, Villeponteau B. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 25.Harley J B, Alexander E L, Bias W B, Fox O F, Provost T T, Reichlin M, Yamagata H, Arnett F C. Arthritis Rheum. 1986;29:196–206. doi: 10.1002/art.1780290207. [DOI] [PubMed] [Google Scholar]
- 26.Boire G, Craft J. J Clin Invest. 1990;85:1182–1190. doi: 10.1172/JCI114551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]