Abstract
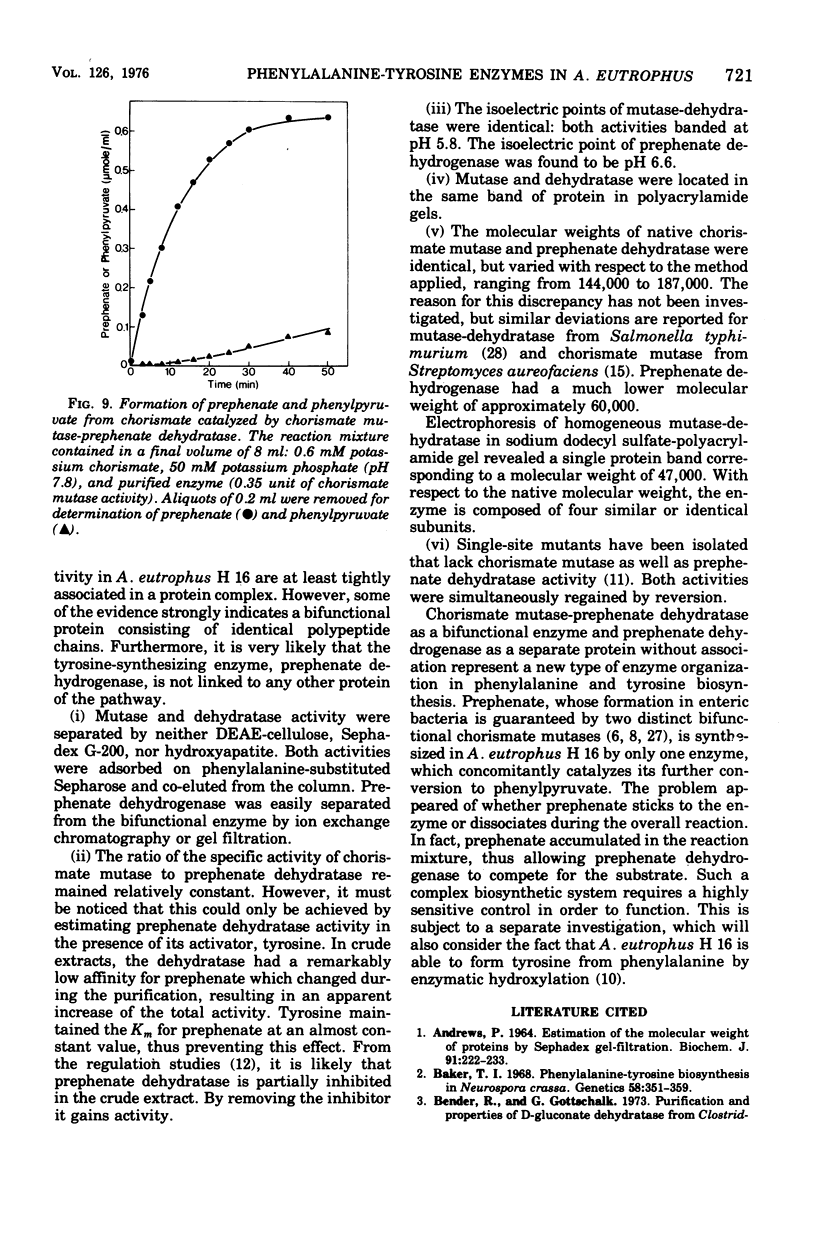
Chorismate mutase and prephenate dehydratase from Alcaligenes autophus H16 were purified 470-fold with a yield of 24%. During the course of purification, including chromatography on diethylaminoethyl (DEAE)-cellulose, phenylalanine-substituted Sepharose, Sephadex G-200 and hydrogyapatite, both enzymes appeared in association. The ratio of their specific activities remained almost constant. The molecular weight of chorismate mutase-prephenast dehydratase varied from 144,000 to 187,000 due to the three different determination methods used. Treatment of electrophoretically homogeneous mutase-dehydratase with sodium dodecyl sulfate dissociated the enzyme into a single component of molecular weight 47,000, indicating a tetramer of identical subunits. The isoelectric point of the bifunctional enzyme was 5.8. Prephenate dehydrogenase was not associated with other enzyme activities; it was separated from mutasedehydratase by DEAE-cellulose chromatgraphy. Chromatography on DEAE Sephadex, Sephadex G-200, and hydroxyapatite resulted in a 740-fold purification with a yield of 10%. The molecular weight of the enzyme was 55,000 as determined by sucrose gradient centrifugation and 65,000 as determined by gel filtration or electrophoresis. Its isoelectric point was pH 6.6. In the overall conversion of chorismate to phenylpyruvate, free prephenate was formed which accumulated in the reaction mixture. The dissociation of prephenate allowed prephenate dehydrogenase to compete with prephenate dehydratase for the substrate.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. I. Phenylalanine-Tyrosine Biosynthesis in NEUROSPORA CRASSA. Genetics. 1968 Mar;58(3):351–359. doi: 10.1093/genetics/58.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COTTON R. G., GIBSON F. THE BIOSYNTHESIS OF PHENYLALANINE AND TYROSINE; ENZYMES CONVERTING CHORISMIC ACID INTO PREPHENIC ACID AND THEIR RELATIONSHIPS TO PREPHENATE DEHYDRATASE AND PREPHENATE DEHYDROGENASE. Biochim Biophys Acta. 1965 Apr 12;100:76–88. doi: 10.1016/0304-4165(65)90429-0. [DOI] [PubMed] [Google Scholar]
- Calhoun D. H., Pierson D. L., Jensen R. A. Channel-shuttle mechanism for the regulation of phenylalanine and tyrosine synthesis at a metabolic branch point in Pseudomonas aeruginosa. J Bacteriol. 1973 Jan;113(1):241–251. doi: 10.1128/jb.113.1.241-251.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champney W. S., Jensen R. A. The enzymology of prephenate dehydrogenase in Bacillus subtilis. J Biol Chem. 1970 Aug 10;245(15):3763–3770. [PubMed] [Google Scholar]
- Cuatrecasas P., Wilchek M., Anfinsen C. B. Selective enzyme purification by affinity chromatography. Proc Natl Acad Sci U S A. 1968 Oct;61(2):636–643. doi: 10.1073/pnas.61.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Davidson B. E., Blackburn E. H., Dopheide T. A. Chorismate mutase-prephenate dehydratase from Escherichia coli K-12. I. Purification, molecular weight, and amino acid composition. J Biol Chem. 1972 Jul 25;247(14):4441–4446. [PubMed] [Google Scholar]
- Friedrich B., Schlegel H. G. Aromatic amino acid biosynthesis in Alcaligenes eutrophus H16. II. The isolation and characterization of mutants auxotrophic for phenylalanine and tyrosine. Arch Microbiol. 1975 Apr 7;103(2):141–149. doi: 10.1007/BF00436341. [DOI] [PubMed] [Google Scholar]
- Friedrich B., Schlegel H. G. Die Hydroxylierung von Phenylalanin durch Hydrogenomanas eutropha H16. Arch Mikrobiol. 1972;83(1):17–31. [PubMed] [Google Scholar]
- Friedrich C. G., Friedrich B., Schlegel H. G. Regulation of Chorismate mutase-prephenate dehydratase and prephenate dehydrogenase from alcaligenes eutrophus. J Bacteriol. 1976 May;126(2):723–732. doi: 10.1128/jb.126.2.723-732.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg A., Stadtman E. R. Multienzyme systems. Annu Rev Biochem. 1970;39:429–472. doi: 10.1146/annurev.bi.39.070170.002241. [DOI] [PubMed] [Google Scholar]
- Görisch H., Lingens F. Chorismate mutase from Streptomyces. Purification, properties, and subunit structure of the enzyme from Streptomyces aureofaciens Tü 24. Biochemistry. 1974 Aug 27;13(18):3790–3794. doi: 10.1021/bi00715a027. [DOI] [PubMed] [Google Scholar]
- Hedrick J. L., Smith A. J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968 Jul;126(1):155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- Huang L., Montoya A. L., Nester E. W. Characterization of the functional activities of the subunits of 3-deoxy-D-arabinoheptulosonate 7-phosphate synthetase-chorismate mutase from Bacillus subtilis 168. J Biol Chem. 1974 Jul 25;249(14):4473–4470. [PubMed] [Google Scholar]
- Huang L., Nakatsukasa M., Nester E. Regulation of aromatic amino acid biosynthesis in Bacillus subtilis 168. Purification, characterization, and subunit structure of the bifunctional enzyme 3-deoxy-D-arabinoheptulosonate 7-phosphate synthetase-chorismate mutase. J Biol Chem. 1974 Jul 25;249(14):4467–4472. [PubMed] [Google Scholar]
- Kawahara K., Tanford C. The number of polypeptide chains in rabbit muscle aldolase. Biochemistry. 1966 May;5(5):1578–1584. doi: 10.1021/bi00869a018. [DOI] [PubMed] [Google Scholar]
- Koch G. L., Shaw D. C., Gibson F. Studies on the subunit structure of chorismate mutase-prephenate dehydrogenase from Aerobacter aerogenes. Biochim Biophys Acta. 1970 Sep 16;212(3):387–395. doi: 10.1016/0005-2744(70)90244-5. [DOI] [PubMed] [Google Scholar]
- Koch G. L., Shaw D. C., Gibson F. The purification and characterisation of chorismate mutase-prephenate dehydrogenase from Escherichia coli K12. Biochim Biophys Acta. 1971 Mar 23;229(3):795–804. doi: 10.1016/0005-2795(71)90298-4. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lorence J. H., Nester E. W. Multiple molecular forms of chorismate mutase in Bacillus subtillis. Biochemistry. 1967 May;6(5):1541–1553. doi: 10.1021/bi00857a041. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- SCHLEGEL H. G., KALTWASSER H., GOTTSCHALK G. [A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies]. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- Schmit J. C., Artz S. W., Zalkin H. Chorismate mutase-prephenate dehydratase. Evidence for distinct catalytic and regulatory sites. J Biol Chem. 1970 Aug 25;245(16):4019–4027. [PubMed] [Google Scholar]
- Schmit J. C., Zalkin H. Chorismate mutase-prephenate dehydratase. Partial purification and properties of the enzyme from Salmonella typhimurium. Biochemistry. 1969 Jan;8(1):174–181. doi: 10.1021/bi00829a025. [DOI] [PubMed] [Google Scholar]
- Schmit J. C., Zalkin H. Chorismate mutase-prephenate dehydratase. Phenylalanine-induced dimerization and its relationship to feedback inhibition. J Biol Chem. 1971 Oct 10;246(19):6002–6010. [PubMed] [Google Scholar]
- Sprössler B., Lenssen U., Lingens F. Eigenschaften der Chorismat-Mutase aus Saccharomyces cerevisiae S 288 C. Hoppe Seylers Z Physiol Chem. 1970 Oct;351(10):1178–1190. [PubMed] [Google Scholar]
- Vesterberg O., Wadström T., Vesterberg K., Svensson H., Malmgren B. Studies on extracellular PROTEINS FROM Staphylococcus aureus. I. Separation and characterization of enzymes and toxins by isoelectric focusing. Biochim Biophys Acta. 1967 Apr 11;133(3):435–445. doi: 10.1016/0005-2795(67)90547-8. [DOI] [PubMed] [Google Scholar]
- Weber H. L., Böck A. Chorismate mutase from Euglena gracilis. Purification and regulatory properties. Eur J Biochem. 1970 Oct;16(2):244–251. doi: 10.1111/j.1432-1033.1970.tb01078.x. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]