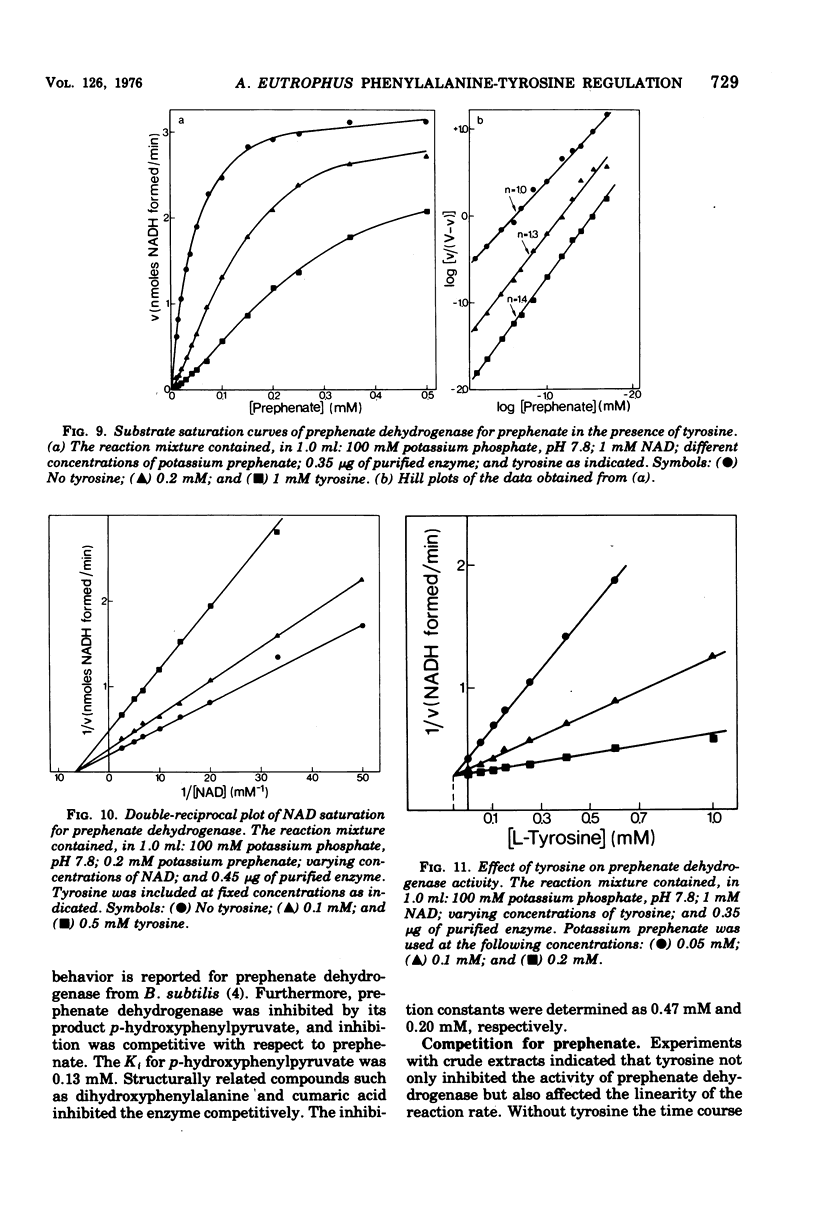
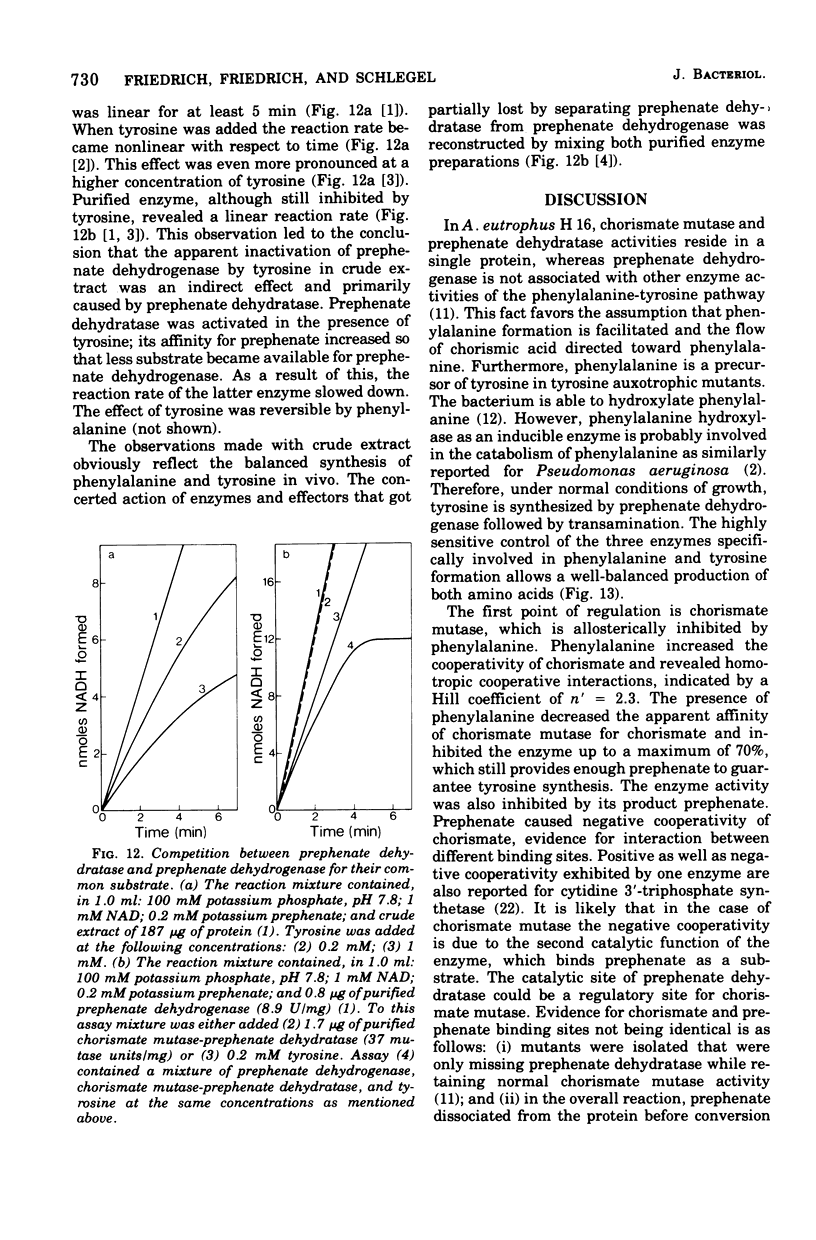
Abstract
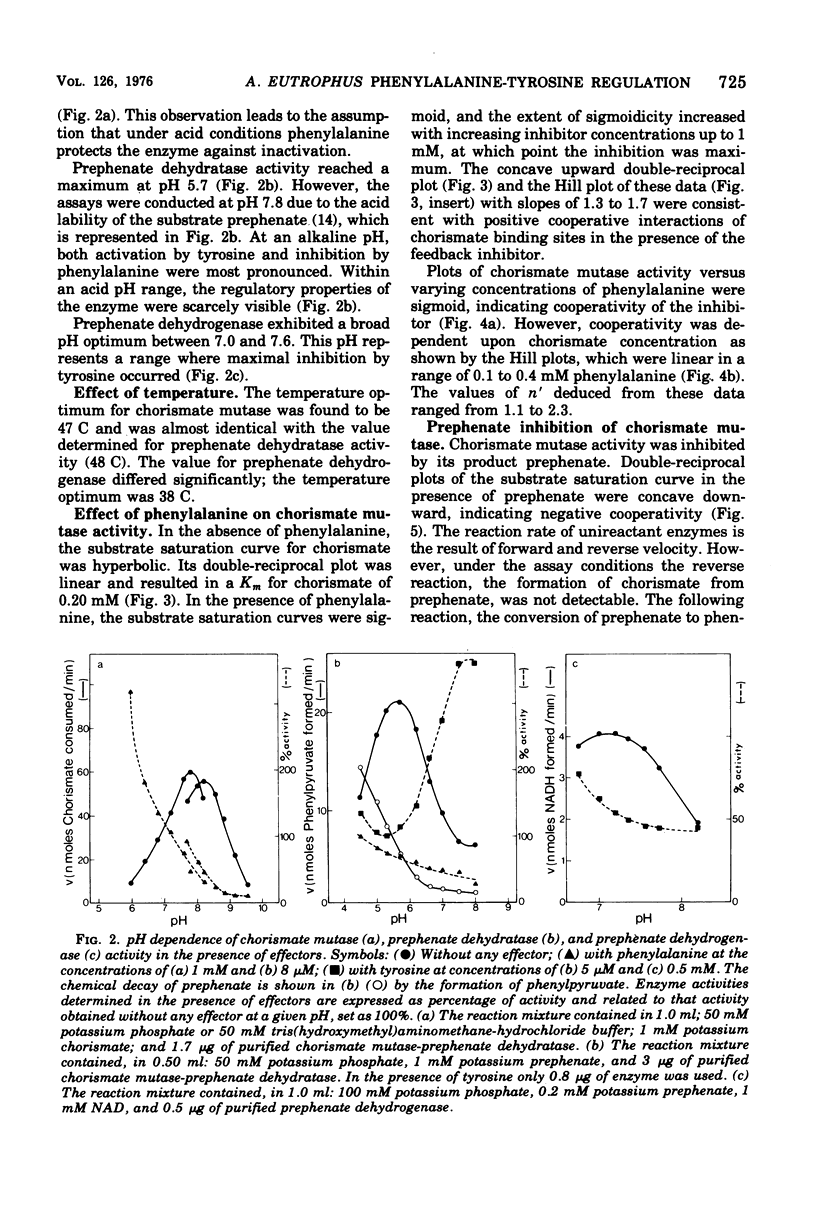
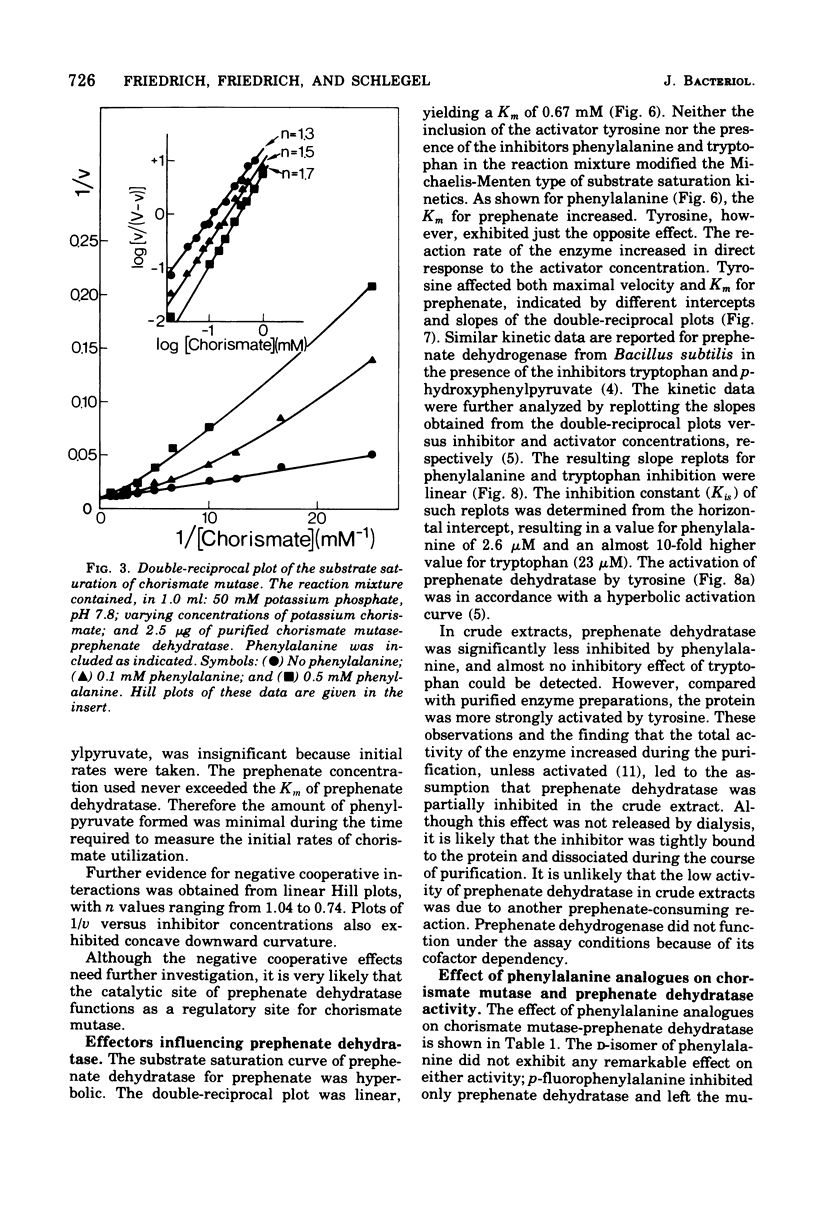
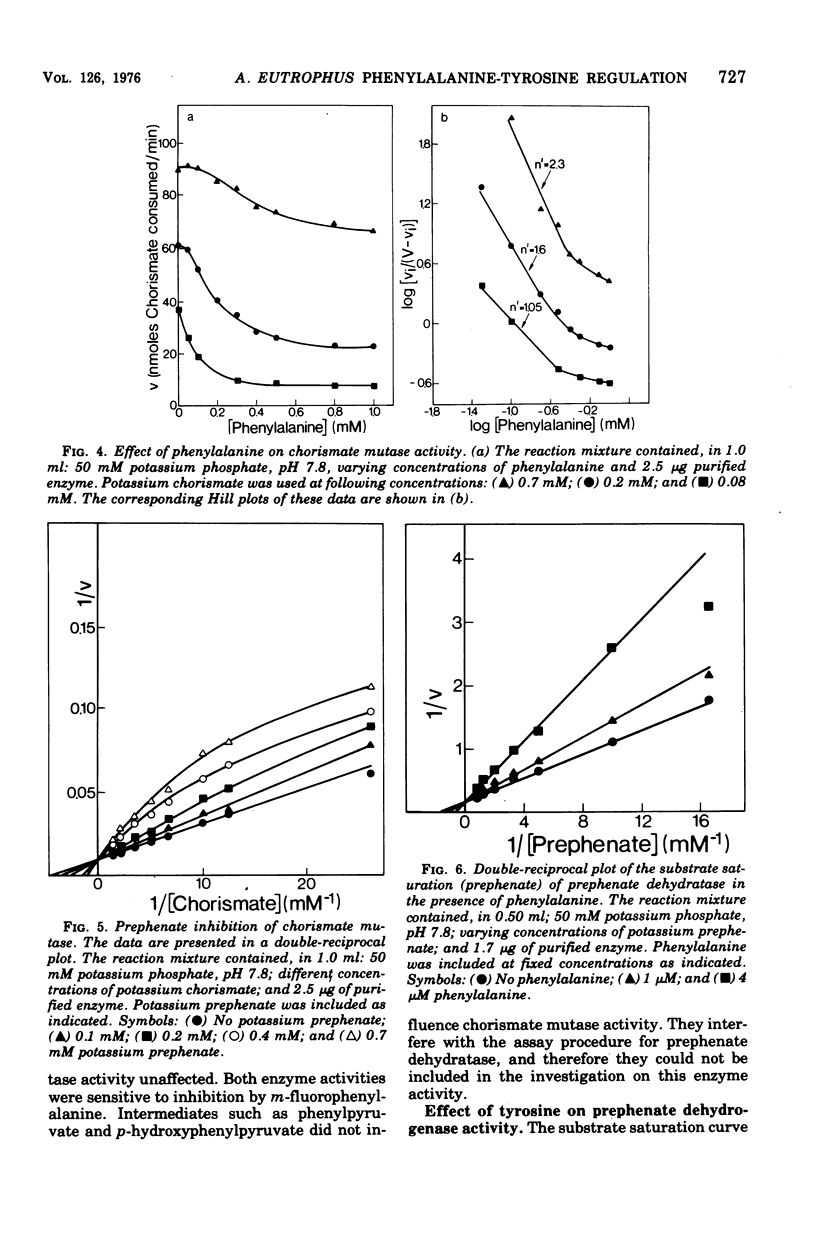
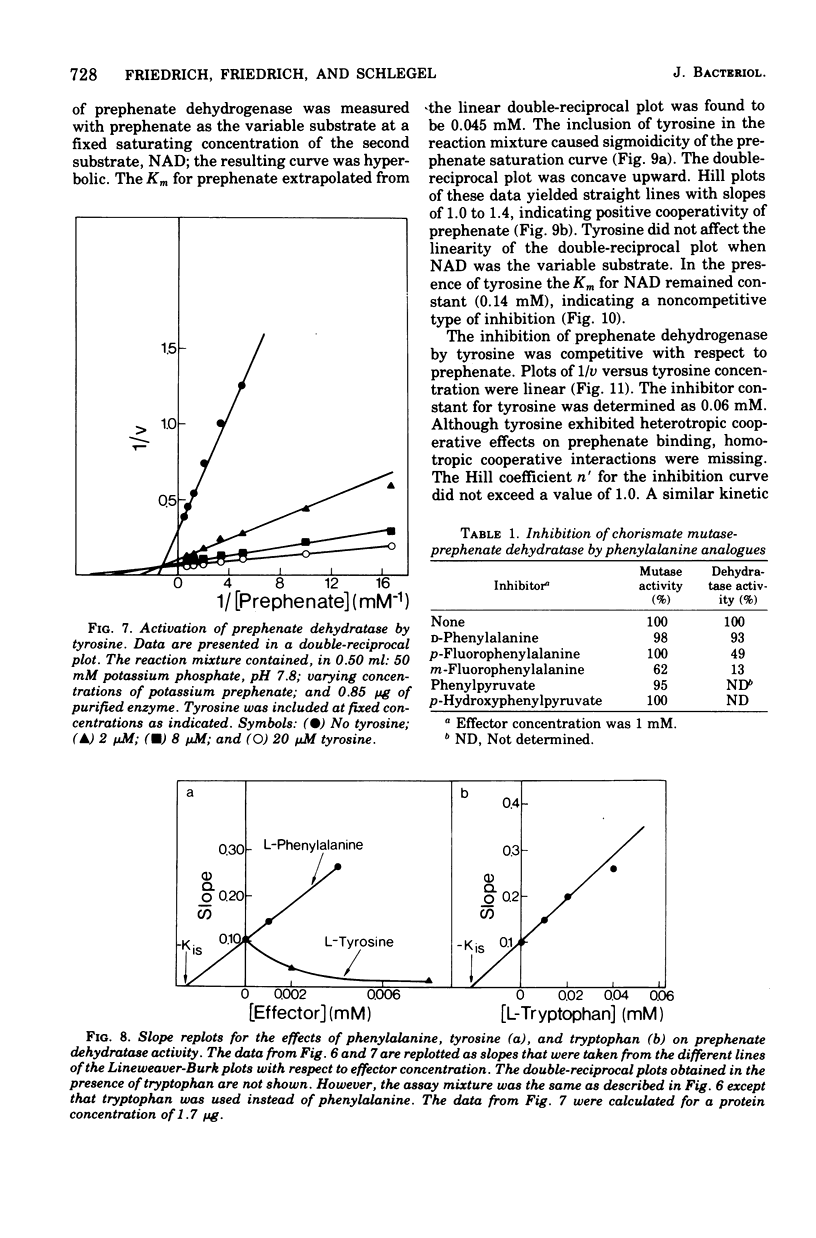
Highly purified enzymes from Alcaligenes eutrophus H 16 were used for kinetic studies. Chorismate mutase was feedback inhibited by phenylalanine. In the absence of the inhibitor, the double-reciprocal plot was linear, yielding a Km for chorismate of 0.2 mM. When phenylalanine was present, a pronounced deviation from the Michaelis-Menten hyperbola occurred. The Hill coefficient (n) was 1.7, and Hill plots of velocity versus inhibitor concentrations resulted in a value of n' = 2.3, indicating positive cooperativity. Chorismate mutase was also inhibited by prephenate, which caused downward double-reciprocal plots and a Hill coefficient of n = 0.7, evidence for negative cooperativity. The pH optimum of chorismate mutase ranged from 7.8 to 8.2; its temperature optimum was 47 C. Prephenate dehydratase was competitively inhibited by phenylalanine and activated by tyrosine. Tyrosine stimulated its activity up to 10-fold and decreased the Km for prephenate, which was 0.67 mM without effectors. Tryptophan inhibited the enzyme competitively. Its inhibition constant (Ki = 23 muM) was almost 10-fold higher than that determined for phenylalanine (Ki = 2.6 muM). The pH optimum of prephenate dehydratase was pH 5.7; the temperature optimum was 48 C. Prephenate dehydrogenase was feedback inhibited by tyrosine. Inhibition was competitive with prephenate (Ki = 0.06 mM) and noncompetitive with nicotinamide adenine dinucleotide. The enzyme was further subject to product inhibition by p-hydroxyphenylpyruvate (Ki = 0.13 mM). Its Km for prephenate was 0.045 mM, and that for nicotinamide adenine dinucleotide was 0.14 mM. The pH optimum ranged between 7.0 and 7.6; the temperature optimum was 38 C. It is shown how the sensitive regulation of the entire enzyme system leads to a well-balanced amino acid production.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker T. I. Phenylalanine-Tyrosine Biosynthesis in NEUROSPORA CRASSA. Genetics. 1968 Mar;58(3):351–359. doi: 10.1093/genetics/58.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COTTON R. G., GIBSON F. THE BIOSYNTHESIS OF PHENYLALANINE AND TYROSINE; ENZYMES CONVERTING CHORISMIC ACID INTO PREPHENIC ACID AND THEIR RELATIONSHIPS TO PREPHENATE DEHYDRATASE AND PREPHENATE DEHYDROGENASE. Biochim Biophys Acta. 1965 Apr 12;100:76–88. doi: 10.1016/0304-4165(65)90429-0. [DOI] [PubMed] [Google Scholar]
- Calhoun D. H., Pierson D. L., Jensen R. A. Channel-shuttle mechanism for the regulation of phenylalanine and tyrosine synthesis at a metabolic branch point in Pseudomonas aeruginosa. J Bacteriol. 1973 Jan;113(1):241–251. doi: 10.1128/jb.113.1.241-251.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catcheside D. E. Prephenate dehydrogenase from Neurospora: feedback activation by phenylalanine. Biochem Biophys Res Commun. 1969 Aug 15;36(4):651–656. doi: 10.1016/0006-291x(69)90355-6. [DOI] [PubMed] [Google Scholar]
- Champney W. S., Jensen R. A. The enzymology of prephenate dehydrogenase in Bacillus subtilis. J Biol Chem. 1970 Aug 10;245(15):3763–3770. [PubMed] [Google Scholar]
- Cotton R. G., Gibson F. The biosynthesis of tyrosine in Aerobacter aerogenes: partial purification of the T protein. Biochim Biophys Acta. 1967 Oct 23;147(2):222–237. doi: 10.1016/0005-2795(67)90401-1. [DOI] [PubMed] [Google Scholar]
- Davidson B. E., Blackburn E. H., Dopheide T. A. Chorismate mutase-prephenate dehydratase from Escherichia coli K-12. I. Purification, molecular weight, and amino acid composition. J Biol Chem. 1972 Jul 25;247(14):4441–4446. [PubMed] [Google Scholar]
- Dopheide T. A., Crewther P., Davidson B. E. Chorismate mutase-prephenate dehydratase from Escherichia coli K-12. II. Kinetic properties. J Biol Chem. 1972 Jul 25;247(14):4447–4452. [PubMed] [Google Scholar]
- Friedrich B., Schlegel H. G. Aromatic amino acid biosynthesis in Alcaligenes eutrophus H16. II. The isolation and characterization of mutants auxotrophic for phenylalanine and tyrosine. Arch Microbiol. 1975 Apr 7;103(2):141–149. doi: 10.1007/BF00436341. [DOI] [PubMed] [Google Scholar]
- Friedrich B., Schlegel H. G. Die Hydroxylierung von Phenylalanin durch Hydrogenomanas eutropha H16. Arch Mikrobiol. 1972;83(1):17–31. [PubMed] [Google Scholar]
- Gibson F. Chorismic acid: purification and some chemical and physical studies. Biochem J. 1964 Feb;90(2):256–261. doi: 10.1042/bj0900256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson F., Pittard J. Pathways of biosynthesis of aromatic amino acids and vitamins and their control in microorganisms. Bacteriol Rev. 1968 Dec;32(4 Pt 2):465–492. [PMC free article] [PubMed] [Google Scholar]
- Görisch H., Lingens F. Chorismate mutase from Streptomyces. Purification, properties, and subunit structure of the enzyme from Streptomyces aureofaciens Tü 24. Biochemistry. 1974 Aug 27;13(18):3790–3794. doi: 10.1021/bi00715a027. [DOI] [PubMed] [Google Scholar]
- Jensen R. A. Metabolic interlock. Regulatory interactions exerted between biochemical pathways. J Biol Chem. 1969 Jun 10;244(11):2816–2823. [PubMed] [Google Scholar]
- Jensen R. A., Nasser D. S., Nester E. W. Comparative control of a branch-point enzyme in microorganisms. J Bacteriol. 1967 Nov;94(5):1582–1593. doi: 10.1128/jb.94.5.1582-1593.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch G. L., Shaw D. C., Gibson F. Studies on the subunit structure of chorismate mutase-prephenate dehydrogenase from Aerobacter aerogenes. Biochim Biophys Acta. 1970 Sep 16;212(3):387–395. doi: 10.1016/0005-2744(70)90244-5. [DOI] [PubMed] [Google Scholar]
- Koch G. L., Shaw D. C., Gibson F. The purification and characterisation of chorismate mutase-prephenate dehydrogenase from Escherichia coli K12. Biochim Biophys Acta. 1971 Mar 23;229(3):795–804. doi: 10.1016/0005-2795(71)90298-4. [DOI] [PubMed] [Google Scholar]
- Lingens F., Goebel W., Uesseler H. Regulation der Biosynthese der aromatischen Aminosäuren in Saccharomyces cerevisiae. 2. Repression, Induktion und Aktivierung. Eur J Biochem. 1967 May;1(3):363–374. doi: 10.1111/j.1432-1033.1967.tb00083.x. [DOI] [PubMed] [Google Scholar]
- MONOD J., WYMAN J., CHANGEUX J. P. ON THE NATURE OF ALLOSTERIC TRANSITIONS: A PLAUSIBLE MODEL. J Mol Biol. 1965 May;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Rebello J. L., Jensen R. A. Metabolic interlock. The multi-metabolite control of prephenate dehydratase activity in Bacillus subtilis. J Biol Chem. 1970 Aug 10;245(15):3738–3744. [PubMed] [Google Scholar]
- Schmit J. C., Artz S. W., Zalkin H. Chorismate mutase-prephenate dehydratase. Evidence for distinct catalytic and regulatory sites. J Biol Chem. 1970 Aug 25;245(16):4019–4027. [PubMed] [Google Scholar]
- Schmit J. C., Zalkin H. Chorismate mutase-prephenate dehydratase. Partial purification and properties of the enzyme from Salmonella typhimurium. Biochemistry. 1969 Jan;8(1):174–181. doi: 10.1021/bi00829a025. [DOI] [PubMed] [Google Scholar]
- Sprössler B., Lenssen U., Lingens F. Eigenschaften der Chorismat-Mutase aus Saccharomyces cerevisiae S 288 C. Hoppe Seylers Z Physiol Chem. 1970 Oct;351(10):1178–1190. [PubMed] [Google Scholar]
- Weber H. L., Böck A. Chorismate mutase from Euglena gracilis. Purification and regulatory properties. Eur J Biochem. 1970 Oct;16(2):244–251. doi: 10.1111/j.1432-1033.1970.tb01078.x. [DOI] [PubMed] [Google Scholar]


