Abstract
The level and fate of hMSH3 (human MutS homolog 3) were examined in the promyelocytic leukemia cell line HL-60 and its methotrexate-resistant derivative HL-60R, which is drug resistant by virtue of an amplification event that spans the dihydrofolate reductase (DHFR) and MSH3 genes. Nuclear extracts from HL-60 and HL-60R cells were subjected to an identical, rapid purification protocol that efficiently captures heterodimeric hMutSα (hMSH2⋅hMSH6) and hMutSβ (hMSH2⋅hMSH3). In HL-60 extracts the hMutSα to hMutSβ ratio is roughly 6:1, whereas in methotrexate-resistant HL-60R cells the ratio is less than 1:100, due to overproduction of hMSH3 and heterodimer formation of this protein with virtually all the nuclear hMSH2. This shift is associated with marked reduction in the efficiency of base–base mismatch and hypermutability at the hypoxanthine phosphoribosyltransferase (HPRT) locus. Purified hMutSα and hMutSβ display partial overlap in mismatch repair specificity: both participate in repair of a dinucleotide insertion–deletion heterology, but only hMutSα restores base–base mismatch repair to extracts of HL-60R cells or hMSH2-deficient LoVo colorectal tumor cells.
Keywords: drug resistance, genetic instability
Methotrexate (Mtx) is widely used for the treatment of human malignancies (see refs. 1 and 2 for recent perspectives). The primary target for this compound is dihydrofolate reductase (DHFR) (3), an enzyme that catalyzes the reduction of dihydrofolate to tetrahydrofolate in a reaction essential to one-carbon metabolism (4). However, human tumor cells can acquire resistance to Mtx, a phenomenon that has been demonstrated both in vitro and in vivo (5–8). One well documented mechanism of resistance involves the amplification of a region of the human or rodent genome containing the DHFR gene (5–10), an event that leads to elevated expression of DHFR, which effectively circumvents the metabolic block produced by this agent.
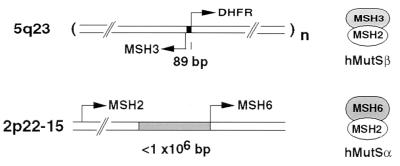
An unusual feature of the chromosomal organization of the human and murine DHFR gene is a shared promoter region with a second ORF (11, 12) that is transcribed in the opposite direction (Fig. 1). The divergently transcribed human gene, originally called DUG (divergent upstream gene) or MRP-1 (mismatch repair protein 1) displays homology to the bacterial MutS polypeptide, and now is called hMSH3 (human MutS homolog 3). Although it has not been demonstrated that hMSH3 is produced in human cells, a role for the homologous protein in Saccharomyces cerevisiae has been described (13). Yeast MSH3 shares overlapping function with yMSH6, each of which forms a molecular complex with yMSH2 and contributes to the maintenance of microsatellite stability (14–16). More recently, yMSH3 has been coexpressed with yMSH2 to produce a heterodimer that recognizes insertion mismatches in a mobility shift assay (17).
Figure 1.
Organization of human MutS homolog genes. The MSH3 gene is divergently transcribed from a promoter region shared with the DHFR gene (11, 41). The DHFR copy number (n) is about 200 in Mtxr HL-60R cells (11). Genes encoding the two polypeptides that comprise hMutSα, MSH2 and MSH6, reside in close proximity on chromosome 2 (20, 33). The relative orientation of transcription shown for MSH2 and MSH6 is arbitrary.
In vitro mismatch recognition activities of the corresponding human MSH2⋅MSH6 (hMutSα) and MSH2⋅MSH3 (hMutSβ) heterodimers have been more extensively examined. hMutSα binds both base–base and nucleotide insertion mismatches and restores correction of both types of mispairs to extracts of repair-defective cell lines (18). Both heterodimers have been generated from cDNA constructs by baculovirus expression (19) and in vitro transcription and translation (20). In vitro analysis has indicated that whereas both hMutSα and hMutSβ bind insertion–deletion mismatches, only hMutSα recognizes base–base mismatches (18–20). hMutSα and hMutSβ may have complementary functions in recognition of insertion–deletion mismatches, differentially recognizing mispairs of this class depending on heterology size and sequence context (19, 21).
The contribution of MSH3 to genetic stability in human cells is uncertain. Loss of MSH3 expression, but not associated hypermutability, has been reported in marrow cells from patients with hematological malignancies (22). More recently, the HHUA endometrial tumor cell line has been shown to contain mutations in both MSH3 and MSH6 (21). Introduction of chromosome 5 with a functional MSH3 gene into HHUA cells restored microsatellite stability at dinucleotide and tetranucleotide repeat sequences but not at mononucleotide or trinucleotide repeats. Extracts of chromosome 5-complemented HHUA cells were found to be proficient in repair of selected mononucleotide and tetranucleotide insertion–deletion mispairs, but to exhibit only limited activity on base–base mismatches.
We show here that extensive overproduction of hMSH3 in Mtxr HL-60R promyelocytic leukemia cells sequesters virtually all of the nuclear hMSH2 into the hMutSβ heterodimer. This phenomenon is associated with a defect in base–base mismatch repair and hypermutability at the HPRT locus.
MATERIALS AND METHODS
Cell Lines and Mismatch Repair Assays.
Cell lines HL-60 and HL-60R were obtained from T. Shimada (Nippon Medical School, Tokyo) and cultured according to published procedures (11). HeLa S3 and LoVo cells were grown as described previously (18, 23). Mismatch repair assays contained 100 μg of nuclear extract protein and 24 fmol of heteroduplex DNA (23, 24). Extract complementation used 200 ng hMutSα or 100 ng hMutSβ.
Gels and Western Analysis.
Electrophoresis of protein samples was performed on 6% polyacrylamide in the presence of sodium dodecylsulfate. Protein bands were visualized with Coomassie stain or transferred to a poly(vinylidene difluoride) membrane (Immobilon P, Millipore), typically for 100 V/hr at 4°C in Tris/glycine buffer (25 mM Tris base plus 192 mM glycine). Blots were incubated in 100 mM Tris, pH 7.5/0.9% NaCl/0.1% Tween 20 containing 5% nonfat dry milk and probed with the specified antibody according to the enzyme chemiluminescence protocol supplied by Amersham. Antibodies used in this study include a mouse monoclonal anti-hMSH2 (Calbiochem, Ab-1) and a goat polyclonal anti-hMSH6 (anti-GTBP, Santa Cruz Biotechnology, N-20). A polyclonal antibody was raised in rabbits to the peptide sequence TEIDRRKKRPLENDGPVKKK (residues 81–100), derived from the deduced peptide sequence for human hMSH3 (11).
Resolution of hMutSα and hMutSβ.
All steps were performed at 4°C. Samples of nuclear extract prepared from HL-60 or HL-60R (0.42 g/ml ammonium sulfate fractions; ref. 23) containing 18 mg of total protein (600 μl of a 30 mg/ml solution) were diluted 1:10 into buffer A (25 mM Hepes⋅KOH, at pH 7.5/200 mM KCl/0.1 mM EDTA/2 mM DTT/1 μg/ml leupeptin/0.1% phenylmethysulfonyl fluoride (PMSF) (a 1:1,000 dilution of a saturated PMSF stock solution in isopropanol at room temperature that was added immediately before use). Samples were centrifuged (12,000 × g) for 10 min and applied to a column of single-stranded DNA cellulose (Sigma, 3.5 mg of DNA per gram of resin), equilibrated overnight in buffer A without DTT, leupeptin, or PMSF, and poured into a 0.5-cm2 × 4-cm disposable column (Bio-Rad). The flow-through was passed over the column a second time, and the resin then washed with 15 ml of buffer A and 15 ml of buffer A plus 2.5 mM MgCl2. Bound protein then was eluted in 300-μl fractions with buffer B (25 mM Hepes⋅KOH/650 mM KCl/0.1 mM EDTA/2.5 mM MgCl2/1 mM ATP) containing DTT, leupeptin, and PMSF as above. Protein-containing fractions were combined and diluted with 25 mM Hepes⋅KOH, pH 7.5, to a conductivity equivalent to that of the Hepes buffer containing 100 mM KCl. This fraction was loaded at 0.5 ml/min onto a 1-ml MonoQ anion exchange FPLC column (Pharmacia) equilibrated with 25 mM Hepes⋅KOH, pH 7.5 containing 100 mM KCl and 0.1 mM EDTA. Proteins were eluted with a linear salt gradient of 100–660 mM KCl in the same buffer over 40 min. Fractions of 0.5 ml were collected and analyzed as described in the text and figure legends.
Purified Proteins.
Human MutSα was purified from HeLa cells as previously described (18), except that a 0.19–0.32 g/ml ammonium sulfate fraction was used instead of the 0.215–0.42 g/ml fraction described previously (18). Human MutSβ was purified using a variation on this method, and full details will be provided elsewhere (J.G., S. Littman, and P.M., unpublished work).
Determination of HPRT Mutation Rates.
Determination of HPRT mutation rates was performed by minor modifications of the procedure of Eshleman et al. (25). Mutants of HeLa, LoVo, HL-60, and HL-60R cells were identified after 1 month of growth in medium containing 5 μg/ml 6-thioguanine, where clonal survival was visualized by staining viable cells with methylene blue dye (HeLa and LoVo) or nitro-blue tetrazolium dye (HL-60 and HL-60R). HeLa and LoVo were propagated in Eagle’s minimal essential medium with 10% fetal bovine serum, and HL-60 and HL-60R were grown in RPMI 1640 medium plus 15% fetal bovine serum. Cultures of HL-60R cells typically were supplemented with 1 μM Mtx to ensure maintenance of the high copy number of the DHFR/MSH3 amplicon, but mutation rates were determined in both the presence and the absence of the drug. In the latter case, cultures to be used for fluctuation analysis were established by inoculation of drug-free medium with about 100 cells from culture samples that had been centrifuged and washed to remove Mtx.
Mutation rates were calculated using equation 8 (r = aNtln[NtCa]) of Luria and Delbrück (26) and the numerical tables of Capizzi and Jameson (27), where r is the average number of mutants per culture; a is the intrinsic mutation rate per locus per generation; Nt is the average number of mutants per culture; and C is the number of replicate cultures.
RESULTS
MSH3 Amplification Is Associated with a Reduction in Levels of hMSH6.
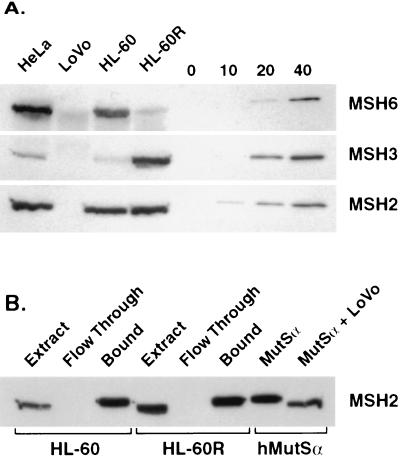
Using near-homogeneous hMutSα and hMutSβ as standards, the four cell lines used in this study were subjected to Western analysis to obtain rough estimates of the relative expression levels of hMSH2, hMSH3, and hMSH6. As shown in Fig. 2A, hMSH3 is present at much lower levels than hMSH2 or hMSH6 in repair-proficient HeLa and HL-60 cell lines. The most striking differences were observed for HL-60 and HL-60R cells. As compared with HeLa or HL-60 cells, amplification of the MSH3/DHFR locus in the HL-60R cell line is associated with a large increase in hMSH3 and a corresponding decrease in hMSH6, an effect presumably due either to reduced hMSH6 expression or instability of the hMSH6 polypeptide in the absence of available hMSH2 for complex formation. Evidence for coregulation or protein instability effects also was obtained with LoVo cells, which harbor an MSH2 deletion (28). Western analysis of LoVo nuclear extracts indicated absence of hMSH2 polypeptide, with hMSH3 levels below the detection limit and hMSH6 present only in trace amounts. Levels of the three MutS homologs are therefore interdependent, with the amount of hMSH2 comparable to the sum of the hMSH3 and hMSH6 proteins.
Figure 2.
Western analysis of MutS homologs. (A) In each of the first four lanes, 75 μg of the indicated nuclear extract was loaded and probed independently for hMSH2, hMSH3, or hMSH6. In the latter four lanes, the indicated amount (ng) of purified hMutSα or hMutSβ was loaded to demonstrate the lower detection limits of the experiment. (B) The efficiency of retention of hMSH2 on single-stranded DNA cellulose is shown for extracts derived from HL-60 and HL-60R cells. The volume loaded was normalized in each case to that of the extract sample. In the two right lanes, purified hMutSα (100 ng) was subjected to electrophoresis in the presence or absence of LoVo extract (50 μg) to demonstrate the effect of high protein concentration on electrophoretic mobility and blotting efficiency.
Overproduction of hMSH3 Drives hMSH2 into the hMutSβ Complex.
To examine the fate of the hMSH2 polypeptide in HL-60 and HL-60R, a rapid fractionation protocol was performed concurrently on nuclear extract prepared from each cell line. The hMSH2 was effectively recovered (≥90%) from extracts of both cell lines by passage over single-stranded DNA cellulose (Fig. 2B), a procedure that captures both hMutSα and hMutSβ. These then were eluted efficiently into a small volume (see Materials and Methods). The hMutSα and hMutSβ heterodimers were cleanly resolved by subsequent chromatography on MonoQ, providing a measure of the relative amounts and activities of these heterodimers (Fig. 3).
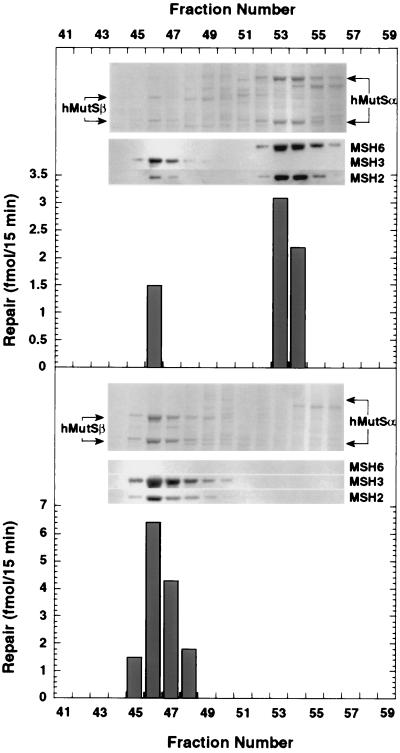
Figure 3.
Resolution of hMutSα and hMutSβ on MonoQ. MonoQ anion exchange FPLC chromatography was performed on DNA cellulose eluates (see Materials and Methods) prepared from HL-60 extract (Upper) or HL-60R extract (Lower). In each case, the upper section shows results of a Coomassie-stained gel, and the middle section shows the corresponding Western blot for the hMSH2, hMSH3, and hMSH6 polypeptides. The positions of subunit polypeptides of hMutSα and hMutSβ in stained gels are indicated by arrows. Each fraction (2.5 μl of 500 μL) was tested for its ability to complement repair-deficient LoVo extract on a 3′ /CA/ insertion–deletion heteroduplex.
This analysis indicated distinct fates of hMSH2 in HL-60 and HL-60R cell lines. In HL-60 cells, hMutSα predominates and hMutSβ is a minor component, accounting for approximately 15% of the hMSH2 in the cell extract (Fig. 3A). Similar results were obtained upon fractionation of HeLa nuclear extract, where hMutSβ also was found to comprise a small fraction of the MutS heterodimers (<10% of MSH2, not shown). Conversely, overexpression of hMSH3 in HL-60R cells resulted in a dramatic shift in the hMutSα/hMutSβ ratio (Fig. 3B). Virtually all of the hMSH2 recovered from HL-60R extracts eluted with hMutSβ, with the hMutSα level below the limit of detection (≈1% of the heterodimers). The small amount of hMSH6 protein detectable in HL-60R extracts (Fig. 2A) was apparently free of the hMSH2 polypeptide and did not bind to DNA cellulose.
The identity and repair activities of hMutSα and hMutSβ were confirmed by Western analysis and complementation assay (Fig. 3). Western analysis of MonoQ fractions indicated that hMSH2 was partnered with either hMSH3 or hMSH6 and was not detected in other column fractions, including those not shown in Fig. 3. Mismatch repair activity, which was scored by complementation of LoVo extract to restore repair on a /CA/ insertion–deletion heteroduplex, was associated only with hMutSα and hMutSβ, with levels of repair consistent with the amount of protein present in each fraction. As discussed above, LoVo cells are free of hMSH2, essentially free of hMSH3, and contain only trace levels of hMSH6, rendering them useful for preparation of receptor extracts to assess the activity of either hMutSα or hMutSβ.
Results like those summarized in Figs. 2 and 3 were reproducible, with extracts prepared from three independent cultures of HL-60 and HL-60R cells yielding similar quantitative results with respect to mismatch repair activities and distribution of hMSH2 between hMutSα and hMutSβ heterodimers.
A Mismatch Repair Defect in HL-60R Cells and Repair Specificity of hMutSα and hMutSβ.
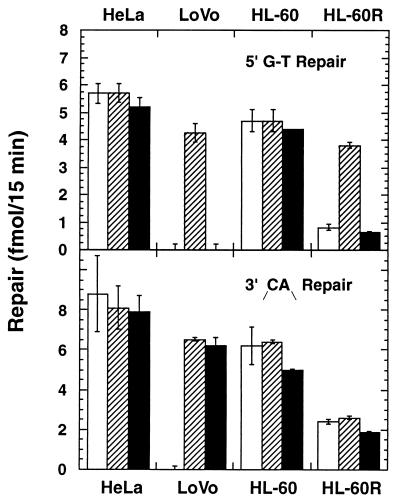
Supplementation of extracts derived from repair-proficient HeLa and HL-60 cells with purified hMutSα or hMutSβ neither stimulated nor inhibited repair of a G-T base–base mismatch or a /CA/ dinucleotide insertion–deletion heteroduplex, indicating that these activities are not limiting for the extract reaction (Fig. 4). However, a functional difference emerged when LoVo extracts were supplemented with amounts of hMutSα or hMutSβ similar to those found in repair-proficient extracts. Only hMutSα restored G-T repair to LoVo extracts, but both hMutSα and hMutSβ restored similar levels of repair on the heteroduplex with an unpaired /CA/dinucleotide. The hMutSα-dependent repair of the latter substrate cannot be due to exchange of hMSH2 between the hMSH2⋅hMSH6 heterodimer and hMSH3 in the extract because LoVo extracts are virtually free of the latter protein (Fig. 2). These results are consistent with the mismatch binding specificity of hMutSβ, which recognizes /CA/ or /GT/ insertion heteroduplexes, but fails to bind to base–base mismatches (19, 20).
Figure 4.
Complementation activity of hMutSα and hMutSβ. Circular heteroduplexes containing a G-T base–base mismatch (Upper) or a /CA/ insertion–deletion mispair (Lower) were used to determine extract repair activities in the absence of exogenous proteins (empty bars), in the presence of added hMutSα (hatched bars), or hMutSβ (filled bars). Error bars are ± 1 SD. The G-T substrate had a strand break at the Sau96I site located 181 nucleotides 5′ to the mismatch (shorter path in the circular substrate), whereas the /CA/ heteroduplex had a strand break 125 nucleotides 3′ to mispair (23, 42).
These observations and the finding that virtually all of the hMSH2 in HL-60R extracts is associated with hMSH3 suggests that HL-60R cells might be defective in base–base mismatch repair. This was confirmed by in vitro assay on a G-T heteroduplex, and repair was restored to normal levels by addition of hMutSα but not by hMutSβ (Fig. 4). Despite the apparent deficiency of hMutSα in HL-60R cells, extracts nevertheless retain a low level of repair activity on the G-T heteroduplex (compare with hMSH2-deficient LoVo cells in Fig. 4). It is possible that the extremely high levels of hMutSβ in HL-60R extracts contribute to the residual base–base repair, despite the failure of more modest levels of hMutSβ to complement LoVo extracts on the G-T substrate. Alternatively, residual base–base mismatch repair may reflect trace amounts of hMutSα that we are unable to detect by the separation method used in Fig. 3. Repair activity of HL-60R extracts on the /CA/ insertion–deletion heteroduplex was consistently found to be about 40% of that observed with HL-60 extracts, but in contrast to the defect in base–base mismatch repair, extract activity was not affected by addition of hMutSα or hMutSβ (Fig. 4).
HL-60R Cells Are Hypermutable.
The shift of hMSH2 from the hMutSα to the hMutSβ complex that occurs in HL-60R cells and the accompanying defect in base–base mismatch repair suggests that this cell line might be hypermutable. We therefore determined mutation rates at the HPRT locus (29) for each of the four cell lines used in this work (Table 1). Mutation rates for HeLa and HL-60 cells were determined to be 1–5 × 10−8, values similar to those previously determined by others (29, 30). By contrast, the mutation rate of HL-60R cells was found to be 3–5 × 10−5, similar to that of LoVo cells, which lack both hMutSα and hMutSβ due to deletion of the MSH2 gene (18, 28). A large increase in HPRT mutability has been previously observed by Caligo et al. (31) for a Mtxr Chinese hamster ovary cell line with a 500- to 1,000-fold amplification of the DHFR region. In view of the hMutSα defect of HL-60R cells reported here, it is interesting to note that the mutation spectrum of the Mtxr Chinese hamster ovary cell line is comprised almost entirely of transitions and transversions.
Table 1.
HPRT mutation rates
| Cell line | Affected MMR locus* | Biochemical deficiency | Mutation frequency† | HPRT mutation rate mutations/generation‡ |
|---|---|---|---|---|
| HL-60 | — | — | 9.6 ± 3.5 × 10−8 | 4.7 ± 1.4 × 10−8 |
| HL-60R§ | MSH3 amplified | hMutSα | 2.2 ± 0.9 × 10−4 (+ Mtx) | 2.6 ± 1.2 × 10−5 (+ Mtx) |
| 4.4 × 10−4 (no Mtx) | 5.0 × 10−5 (no Mtx) | |||
| HeLa | — | — | 1.4 × 10−8 | 1.0 × 10−8 |
| LoVo | MSH2 deletion | hMutSα, hMutSβ | 3.7 × 10−5 | 4.9 × 10−6 |
Mutation rates were determined as described in Materials and Methods.
Mismatch repair (MMR) defect.
Mutation frequency is expressed as HPRT mutants per colony forming unit (CFU) ± 1 SD. Total CFUs from which 6-thioguanine mutants were selected ranged from 2-10 × 107 cells.
± One standard deviation.
HL-60R cells were cultured in the presence or absence of 1 μM Mtx as indicated.
DISCUSSION
What Controls the Levels of hMSH3 and hMSH6?
Our results indicate that the stability and ultimate fate of hMSH2, hMSH3, and hMSH6 are interdependent. Both hMSH3 and hMSH6 were reduced to the limits of detection in extracts of LoVo cells, which lack hMSH2 due to a partial deletion of the structural gene. In the repair-proficient cell lines HeLa and HL-60, both hMSH3 and hMSH6 were present in Western blots, and in each case they were associated with hMSH2 in a 1:1 complex. In HL-60R, where hMSH3 is greatly overexpressed, virtually all of the protein was found in the hMutSβ complex, without any substantial change in the level of hMSH2. One interpretation that accounts for these data posits that hMSH2 is stably expressed at a given level and is captured by either hMSH6 or hMSH3 to yield hMutSα or hMutSβ, respectively. In this model, uncomplexed hMSH3 or hMSH6 are subject to degradation. Instability of hMSH6 has been noted previously (18), and baculovirus expression of human MutS homologs has indicated that although hMSH2 can be stably expressed alone, expression of hMSH3 or hMSH6 required hMSH2 for stability (19).
Contributions of hMutSα and hMutSβ to Mismatch Repair.
In both yeast (14–16) and human cells (21), genetic studies on MSH3 function suggest that this protein functions in stabilization of repetitive sequence elements, which are prone to slippage events during replication (32). The activity of hMutSβ in vitro reported here is consistent with these findings. The protein was active in correction of an unpaired dinucleotide within a short microsatellite run, with an efficiency similar to that of hMutSα. A comprehensive description of the specificity of hMutSβ in mismatch repair will be presented elsewhere (J.G., S. Littman, and P.M., unpublished work).
Given that the majority (≥ 85%) of hMSH2 in HeLa and HL-60 cells is present in hMutSα, which is active in the repair of base–base and insertion–deletion mispairs, this heterodimer is arguably responsible for the recognition and repair of most DNA biosynthetic errors in these cell lines under culture conditions used in this work. This assertion also is supported by the finding that a MSH3-deficient Chinese hamster ovary cell line is not detectably hypermutable at the HPRT locus (G. Crouse, personal communication) and the observation that HPRT mutability in HL-60R is greatly elevated despite the presence of abnormally high levels of hMutSβ. This possibility is not necessarily inconsistent with the suggestion of complementary action of hMutSα and hMutSβ on insertion–deletion mispairs, with the latter protein recognizing heterologies that may not be well recognized by hMutSα (19, 21). In addition, our observations do not exclude mutation avoidance functions for hMutSβ that are not readily evident with the cell lines that we have examined.
The elevated level of hMSH3 in HL-60R cells sequesters hMSH2 into the hMutSβ complex, resulting in deficiency of hMutSα. This biochemical deficiency suggests that HL-60R cells may be similar to HCT-15 and MT1 human cell lines that lack hMutSα due to genetic inactivation of MSH6 (18, 33). Both HCT-15 and MT1 cells display an elevated mutation rate at the HPRT locus (34, 35). However, one caveat with respect to attribution of function to hMSH3 or hMSH6 based on mutant studies must be considered. In cells harboring MSH3 or MSH6 mutations that block heterodimer formation with hMSH2, available hMSH2 is apparently free to associate with and stabilize the remaining homolog. For example, the consequence of an MSH6 mutation might not be restricted to inactivation of hMutSα, but could result in a substantial increase in hMutSβ level as well, due to increased availability of hMSH2. Thus, care must be taken in assigning function to these hMSH3 and hMSH6 polypeptides based on phenotype associated with inactivation of the corresponding genes.
Ramifications of the Genetic Organization of DHFR and MSH3 Genes.
Human and mouse DHFR and MSH3 genes are controlled by a divergent promoter element (11, 12). Although the genes are subject to similar growth-dependent regulation, recent evidence indicates differential response to E2F control (36, 37). The potential for some degree of coregulation is noteworthy given that DHFR is essential for biosynthesis of thymidylate and purine precursors and hence for maintenance balanced pools of the precursors of DNA synthesis (4). Indeed, folate deficiency has been shown to confer genetic instability (38).
Due to its proximity to DHFR, the MSH3 locus can be amplified in Mtxr cells, resulting in a defect in base–base mismatch repair and genetic destabilization. Mismatch repair deficiency also is associated with resistance to the cytotoxic effects of several other chemotherapeutic agents, including DNA methylators, cisplatin, and adriamycin, but in these cases resistance has been attributed to genetic inactivation of the hMSH2 or hMSH6 subunits of hMutSα, or the hMLH1 subunit of hMutLα (reviewed in ref. 39). The depletion of hMutSα activity resulting from MSH3 amplification described here thus raises the possibility that some Mtxr cells may be resistant to other chemotherapeutic agents as well. Although hMutSα has been shown to recognize the cytotoxic lesions produced by cisplatin and DNA methylating agents (40), the activity of MutSβ on these lesions has not been addressed.
Acknowledgments
We thank Dr. James Eshleman for his advice on performance of mutation rate assays. This work was supported in part by Grant GM45190 from the National Institute of General Medical Sciences.
ABBREVIATIONS
- DHFR
dihydrofolate reductase
- hMSH
human MutS homolog
- HPRT
hypoxanthine phosphoribosyltransferase
- Mtx
methotrexate
References
- 1.Huennekens F M. Adv Enzyme Regul. 1994;34:397–419. doi: 10.1016/0065-2571(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 2.Bertino J R. J Clin Oncol. 1993;11:5–15. doi: 10.1200/JCO.1993.11.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Osborn M J, Freeman M, Huennekens F M. Proc Soc Exp Biol Med. 1958;97:429–431. doi: 10.3181/00379727-97-23764. [DOI] [PubMed] [Google Scholar]
- 4.Blakley R L. In: Folates and Pterins. Blakley R L, Benkovic S J, editors. Vol. 1. New York: Wiley; 1984. pp. 191–253. [Google Scholar]
- 5.Carman M D, Schomagel J H, Rivest R S, Srimatkandada S, Portlock C S, Duffy T, Bertino J R. J Clin Oncol. 1984;2:16–20. doi: 10.1200/JCO.1984.2.1.16. [DOI] [PubMed] [Google Scholar]
- 6.Curt G A, Carney D N, Cowan R H, Jolivet J, Bailey B D, Drake J C, Kao-Shan C S, Minna J D, Chabner B A. New Eng J Med. 1983;308:199–202. doi: 10.1056/NEJM198301273080406. [DOI] [PubMed] [Google Scholar]
- 7.Horns R C, Dower W J, Schimke R T. J Clin Oncol. 1984;2:2–7. doi: 10.1200/JCO.1984.2.1.2. [DOI] [PubMed] [Google Scholar]
- 8.Trent J M, Buick R N, Olson S, Horns R C, Schimke R T. J Clin Oncol. 1984;2:8–15. doi: 10.1200/JCO.1984.2.1.8. [DOI] [PubMed] [Google Scholar]
- 9.Schimke R T, Kaufman R J, Alt F W, Kellems R F. Science. 1978;102:1051–1055. doi: 10.1126/science.715457. [DOI] [PubMed] [Google Scholar]
- 10.Schimke R T. Cancer. 1986;57:1912–1917. doi: 10.1002/1097-0142(19860515)57:10<1912::aid-cncr2820571004>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 11.Fujii H, Shimada T. J Biol Chem. 1989;264:10057–10064. [PubMed] [Google Scholar]
- 12.Linton J P, Yen J-Y J, Selby E, Chen Z, Chinsky J M, Liu K, Kellems R E, Crouse G F. Mol Cell Biol. 1989;9:3058–3072. doi: 10.1128/mcb.9.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.New L, Liu K, Crouse G F. Mol Gen Genet. 1993;239:97–108. doi: 10.1007/BF00281607. [DOI] [PubMed] [Google Scholar]
- 14.Johnson R E, Kovvali G K, Prakash L, Prakash S. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 15.Marsischky G T, Filosi N, Kane M F, Kolodner R. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 16.Strand M, Earley M C, Crouse G F, Petes T D. Proc Natl Acad Sci USA. 1995;92:10418–10421. doi: 10.1073/pnas.92.22.10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habraken Y, Sung P, Prakash L, Prakash S. Curr Biol. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 18.Drummond J T, Li G-M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 19.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 20.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risinger J I, Umar A, Boyd J, Berchuck A, Kunkel T A, Barrett J C. Nat Genet. 1996;14:102–105. doi: 10.1038/ng0996-102. [DOI] [PubMed] [Google Scholar]
- 22.Koiti I, Ikejima M, Watanabe A, Nakajima E, Orimo H, Nomura T, Shimada T. Biochem Biophys Res Comm. 1995;214:171–179. doi: 10.1006/bbrc.1995.2271. [DOI] [PubMed] [Google Scholar]
- 23.Holmes J, Clark S, Modrich P. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsons R, Li G M, Longley M J, Fang W H, Papadopoulos N, Jen J, de la Chapelle A, Kinzler K W, Vogelstein B, Modrich P. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 25.Eshleman J R, Lang E Z, Bowerfind G K, Parsons R, Vogelstein B, Willson J K, Veigl M L, Sedwick W D, Markowitz S D. Oncogene. 1995;10:33–37. [PubMed] [Google Scholar]
- 26.Luria S E, Delbrück M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capizzi R L, Jameson J W. Mutat Res. 1973;17:147–148. doi: 10.1016/0027-5107(73)90265-0. [DOI] [PubMed] [Google Scholar]
- 28.Umar A, Boyer J C, Thomas D C, Nguyen D C, Risinger J I, Boyd J, Ionov Y, Perucho M, Kunkel T A. J Biol Chem. 1994;269:14367–14370. [PubMed] [Google Scholar]
- 29.Monnat R J. Cancer Res. 1989;49:81–87. [PubMed] [Google Scholar]
- 30.Aquilina G, Hess P, Fiumicino S, Ceccotti S, Bignami M. Cancer Res. 1995;55:2569–2575. [PubMed] [Google Scholar]
- 31.Caligo M A, Armstrong W, Rossiter B J, Meuth M. Mol Cell Biol. 1990;10:6805–6808. doi: 10.1128/mcb.10.12.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 33.Papadopoulos N, Nicolaides N C, Liu B, Parsons R, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson J K V, Kinzler K, Jiricny J, Vogelstein B. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharyya N P, Skandalis A, Ganesh A, Groden J, Meuth M. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kat A, Thilly W G, Fang W H, Longley M J, Li G M, Modrich P. Proc Natl Acad Sci USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farnham P J, Schimke R T. Mol Cell Biol. 1986;6:365–371. doi: 10.1128/mcb.6.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schilling L J, Farnham P J. Cell Growth Differ. 1995;6:541–548. [PubMed] [Google Scholar]
- 38.Blount B C, Mack M M, Wehr C M, MacGregor J T, Hiatt R A, Wang G, Wickramasinghe S N, Everson R B, Ames B N. Proc Natl Acad Sci USA. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modrich, P. (1997) J. Biol. Chem. 272, in press. [DOI] [PubMed]
- 40.Duckett D R, Drummond J T, Murchie A I H, Reardon J T, Sancar A, Lilley D M J, Modrich P. Proc Natl Acad Sci USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinya E, Shimada T. Nucleic Acids Res. 1994;22:2143–2149. doi: 10.1093/nar/22.11.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang W-h, Modrich P. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]