Abstract
Recent investigations have shown that the maintenance of genomic imprinting of the murine insulin-like growth factor 2 (Igf2) gene involves at least two factors: the DNA (cytosine-5-)-methyltransferase activity, which is required to preserve the paternal specific expression of Igf2, and the H19 gene (lying 90 kb downstream of Igf2 gene), which upon inactivation leads to relaxation of the Igf2 imprint. It is not yet clear how these two factors are related to each other in the process of maintenance of Igf2 imprinting and, in particular, whether the latter is acting through cis elements or whether the H19 RNA itself is involved. By using Southern blots and the bisulfite genomic-sequencing technique, we have investigated the allelic methylation patterns (epigenotypes) of the Igf2 gene in two strains of mouse with distinct deletions of the H19 gene. The results show that maternal transmission of H19 gene deletions leads the maternal allele of Igf2 to adopt the epigenotype of the paternal allele and indicate that this phenomenon is influenced directly or indirectly by the H19 gene expression. More importantly, the bisulfite genomic-sequencing allowed us to show that the methylation pattern of the paternal allele of the Igf2 gene is affected in trans by deletions of the active maternal allele of the H19 gene. Selection during development for the appropriate expression of Igf2, dosage-dependent factors that bind to the Igf2 gene, or methylation transfer between the parental alleles could be involved in this trans effect.
In mammals, the two parental haploid genomes are functionally nonequivalent (1, 2). The basis for this nonequivalence primarily originates from gamete-specific epigenetic modifications that define a parental imprint for some of the genes. The biological significance of this phenomenon appears obvious as defects in imprinting lead to developmental failure and are involved in a growing number of pathological states in humans (3). However, the selective advantages responsible for the appearance and maintenance of the parental imprinting in mammals are not understood and remain fervently debated (4, 5).
The best documented epigenetic marking of the chromosomes, thought to be involved in parental imprinting, is the methylation of CpG residues (6). Allele-specific DNA methylation has been described in the vicinity of most of the 16 endogenous imprinted genes described so far in the mouse and in the human (7). Targeted deletions of the maintenance methyltransferase have demonstrated that DNA methylation is required at least for the maintenance of the differential expression of the imprinted genes in the mouse (8). The methylated state is usually associated with the inactive allele, suggesting a transcriptional silencing of the genes. However, specific methylation of the active allele has been described for some imprinted genes such as Igf2r (9) and Igf2 (10, 11) in the mouse.
The Igf2 gene was the first endogenously imprinted gene to be identified in mammals (12). It is predominantly expressed from the paternal allele in the postimplantation stage embryo and encodes a major fetal growth factor. Several differentially methylated regions (DMRs) have been identified upstream and within the body of the gene (11). Because allele-specific methylation of these regions is associated with the paternally expressed copy of the Igf2 gene, we have suggested that the DMRs may represent constitutive silencers that could be inactivated by methylation (13).
The Igf2 gene lies in a cluster of at least five other imprinted genes on chromosome 7 in the mouse. Within this cluster, the H19 gene is located 90-kb downstream of the Igf2 and the two genes share a common set of enhancers. The competition between these genes for the use of the enhancers is now a well documented model that explains their opposite allele-specific expression patterns (14, 15). It is driven on the paternal allele by the allele-specific methylation (16) and silencing of the H19 gene (8). In recent experiments, deletions of the active maternal allele of the H19 gene have been shown to result in biallelic expression of the Igf2 gene, thus, demonstrating that the H19 gene expression is indeed required for the maintenance of the Igf2 imprinting (17, 18).
In this work, we attempt to shed light on the respective roles of the allele-specific methylation patterns of the DMRs and the H19 gene expression on the maintenance of Igf2 imprinting. To this end, we exploit two strains of mouse with distinct deletions within the H19 locus to investigate the precise allele-specific methylation patterns of the Igf2 gene in these animals.
MATERIALS AND METHODS
Mouse Crosses.
Cross I. Maternal inheritance of H19 gene region deletion (H19Δ13) (17). Heterozygous females were mated to SD7 males (SD7 is a C57BL/6 × CBA Mus musculus domesticus strain containing the distal portion of Mus spretus chromosome 7).
Cross II.
Paternal inheritance of H19 gene region deletion (H19Δ13) (17). SD7 females were mated to heterozygous males. In both cross I and II, tissue samples were collected from neonates 2 days after birth.
Crosses III.
Maternal inheritance of H19 gene deletion (H19Δ3) (18). Heterozygous females from a mixed background (129/MFI/C57BL6) were mated to SD7 males. Tissue samples were collected from neonates 5 days after birth. For each cross, the F1 neonates were genotyped for the H19 gene deletion by screening for the presence of the neomycin-resistance transgene that was introduced by the targeting of the H19 gene.
Southern Blots.
DNA from each sample was digested with HpaII, DraI, and EcoRI (DMR 1) or HpaII and BamHI (DMR 2) (Fig. 1). The fragments were separated into a 1% agarose gel, transferred, and hybridized with the Igf2 specific probes as described in ref. 11.
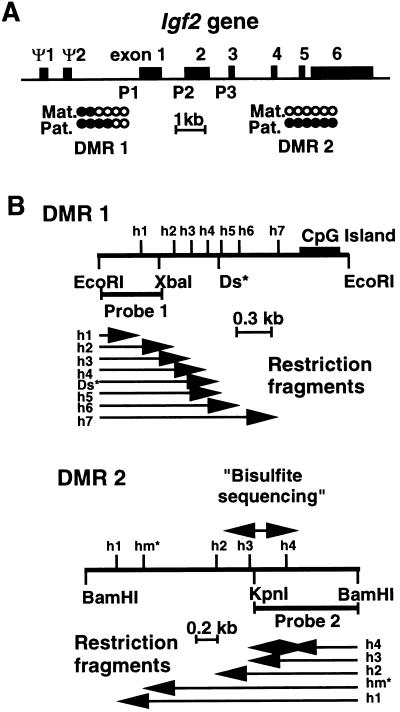
Figure 1.
Two DMRs of the Igf2 gene. (A) Map of the murine Igf2 gene. Solid bars depict the six exons and the two pseudoexons (Ψ1 and Ψ2) of the Igf2 gene. Exons 1–3 are the leader exons from which the expression is driven by promoters 1, 2, and 3 (P1–P3), respectively. Exons 4–6 are the coding exons. The two DMRs are located on the map: DMR 1 at ≈3 kb upstream of exon 1 and DMR 2 in the 3′ coding part of the gene. The relative levels of methylation on the maternal (Mat.) and paternal (Pat.) alleles in wild-type embryos are indicated: •, methylation; ○, absence of methylation (11). (B) Restriction maps of the DMR 1 and DMR 2 domains of the Igf2 gene. For DMR 1, the map shows the EcoRI, XbaI, and HpaII (h) restriction sites. Ds* is a polymorphic DraI site specific for Mus spretus. The location of the DMR 1-specific probe (probe 1) is shown; this probe is a 717-bp EcoRI–XbaI fragment. Below are shown the restriction fragments h1–h7 detected by probe 1 in EcoRI/HpaII-digested genomic DNA. For DMR 2, the map shows BamHI, KpnI, and HpaII (h) restriction sites. The location of the DMR 2-specific probe (probe 2) is shown; this probe is a 904-bp BamHI–KpnI fragment. Below are shown the restriction fragments h1–h4 detected by probe 2 in BamHI/HpaII-digested genomic DNA. The 703-bp sequence studied by the bisulfite genomic-sequencing technique (Fig. 3) is indicated on the map.
Bisulfite Genomic-Sequencing Technique.
The bisulfite genomic-sequencing technique was performed according to Olek et al. (19). Briefly, genomic DNA was digested with EcoRI, alkaline-denatured, and treated with bisulfite as described (19). This chemical modification mutates all unmethylated cytidine residues to thymidine residues whereas the methylated cytidines are not modified. PCR amplifications were carried out at an annealing temperature of 54.5°C and for 35 cycles (for primers, see ref. 19). The PCR products were cloned (TA cloning kit, Invitrogen) and sequenced by using the Applied Biosystems sequencing system.
RESULTS
Deletions of the Active H19 Gene Induce Methylation in Cis on the Maternal Allele of the Igf2 Gene.
We found previously that the methylation patterns of two domains, DMR 1 and 2, of the Igf2 gene in the mouse are monoallelic and are associated with the paternal expression of this gene (Fig. 1A) (11). To verify if, in mice carrying a deletion of the H19 gene, the loss of imprinting (LOI) of the Igf2 gene is correlated with the loss of allelic methylation (LOAM) at this locus, we used the HpaII-methylation-sensitive enzyme in combination with polymorphic restriction sites (Fig. 1B) to study by Southern blot analysis the changes of the levels of methylation in both DMR 1 and 2.
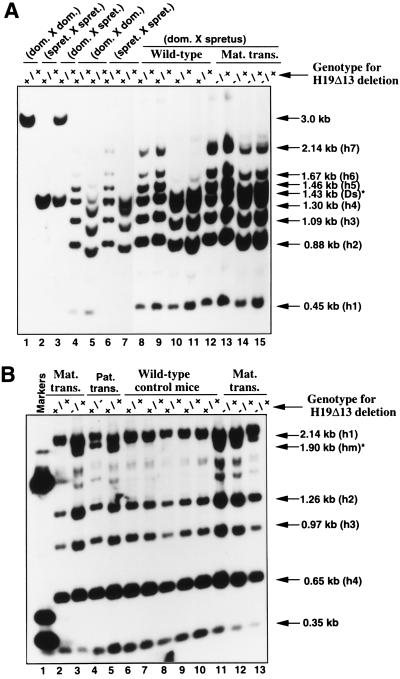
The first H19 deletion that we studied is a targeted disruption of 13 kb encompassing not only H19 but also 10 kb of the 5′ upstream region of the gene (17). We refer to this mutation as the H19Δ13 deletion. DNAs from neonates with maternal transmission of this deletion (cross I) were analyzed. By using a M. spretus-specific DraI site (10), we were able to study the methylation status of the maternal alleles from HpaII fragments 6 and 7 (h6 and h7) in DMR 1 (see Fig. 1B). In liver (Fig. 2A) and kidney (data not shown) of wild-type neonates, the residual methylation at the level of h6 and h7 was very low on the maternal allele (Fig. 2A, compare lanes 8 and 9 with lanes 10 and 11). By contrast, in neonates with maternal transmission of the deletion (H19Δ13), the same sites were highly methylated on the maternal allele in the liver (Fig. 2A, compare lanes 12 and 13 with lanes 14 and 15) and in the kidney (data not shown). No effect of the deletion was observed in the brain (data not shown).
Figure 2.
LOAM of the Igf2 gene in mice heterozygous for an H19 gene deletion (H19Δ13). (A) Methylation analyses of the DMR 1 domain. Heterozygous F1 females were mated to SD7 males (cross I) and the neonates were screened for the H19 deletion. DNA was then extracted from liver of two wild-type and two mutant mice and incubated with EcoRI and HpaII in the absence (lanes 8, 9, 12, and 13) or the presence (lanes 10, 11, 14, and 15) of the DraI restriction enzyme. The DNA was then separated in a 1% agarose gel, transferred to a nitrocellulose membrane, and hybridized with a probe specific to DMR 1 (probe 1). Lanes 1–7 show control liver DNA digestions from 2-day-old wild-type neonates from the following crosses: M. musculus domesticus × M. musculus domesticus (lanes 1, 4, and 5), M. spretus × M. spretus (lanes 2, 6, and 7), and M. musculus domesticus × M. spretus (lanes 3). DNA was digested with EcoRI and DraI (lanes 1, 2, and 3) and with EcoRI and HpaII in the absence (lanes 4 and 6) or in the presence (lanes 5 and 7) of the DraI enzyme. (Ds* designates the polymorphic restriction site only present in the M. spretus alleles.) (B) Methylation analyses of the DMR 2 domain. DNA from cross I (maternal transmission of the deletion) was digested with BamHI and HpaII restriction enzymes, separated in a 1% agarose gel, transferred, and hybridized with a DMR 2-specific probe (probe 2). Lanes 2 and 6–10 show the digests from wild-type animals (+/+) and lanes 3 and 11–13) show digests from mutant mice (−/+). Lanes 4 and 5 show the analysis of DNA samples obtained from crossing heterozygous males with SD7 females (paternal transmission of the deletion, cross II). The digests were obtained from 2-day-old neonate liver DNAs of wild-type (+/+) and mutant mice (+/−), respectively. hm* is a polymorphic HpaII fragment specific to M. musculus domesticus. Lane 1 shows marker DNA fragments (the sizes are 390, 510, and 1,630 bp).
To investigate the allele-specific methylation of the DMR 2 in the same cross, we digested the same DNA samples with HpaII and used a DMR 2-specific probe (probe 2). In liver (Fig. 2B) and kidney (data not shown) of wild-type neonates, the maternal allele was not methylated (absence of hm* band in Fig. 2B, lanes 2 and 6–10). In contrast, mice carrying the H19 deletion were methylated on the maternal allele since a fragment was now detected at the level of hm* (Fig. 2B, lanes 3 and 11–13). Again, the deletion did not affect methylation levels in brain.
As a control, the effect on DMR 1 and DMR 2 methylation of the deletion of the inactive (paternal) copy of H19 was determined. This deletion did not have any effect on Igf2 methylation (data not shown, but see Fig. 3B). Therefore, deletion of an active (maternal) copy of H19 is needed to effect changes of allelic methylation in Igf2. However, because this deletion is rather large, the observed changes could be due to factors other than the H19 gene itself. We therefore examined a second deletion of H19 (H19Δ3) that is restricted to the 3 kb encompassing the H19 transcription unit itself (18). The effects of maternal transmission of this deletion on DMR 1 and 2 methylation were qualitatively identical to that of the H19Δ13 deletion, but the extent of methylation of the maternal Igf2 allele was lower (data not shown, but see Table 1). The quantitative difference may be due to the different reporter construct used for H19 deletion or to elements situated upstream of H19 that are deleted in the H19Δ13 but not in the H19Δ3 deletion (20, 21).
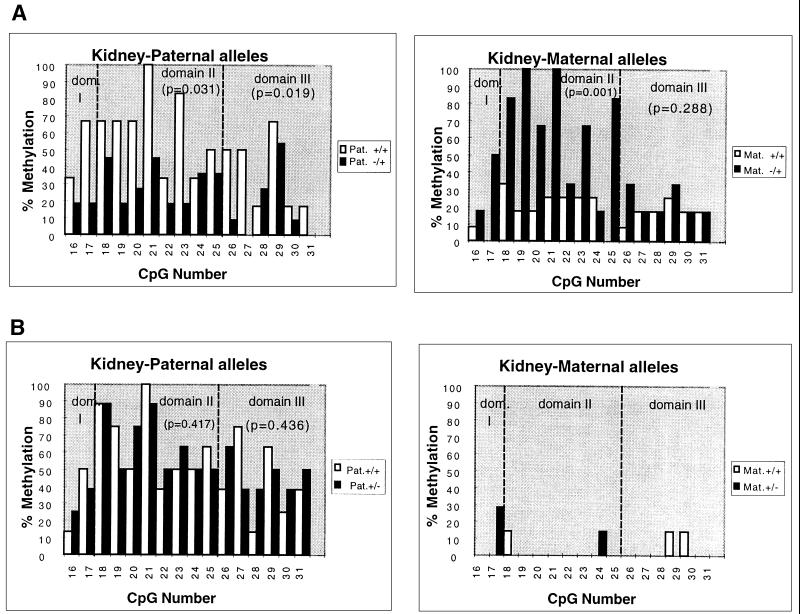
Figure 3.
Methylation analyses on individual maternal and paternal chromosomes in the DMR 2 of the Igf2 gene by the bisulfite genomic-sequencing technique. (A) H19Δ13 deletion, maternal transmission. DNA samples from the kidney of neonates issued from cross I were modified by sodium bisulfite (NaHSO3), PCR amplifications were performed, and the fragments were cloned and sequenced. The level of methylation was then calculated (in percent) for each of the 16 CpGs analyzed. (Left) Comparison of the methylation patterns of the paternal alleles from wild-type mice (+/+) with those from mice with a maternal transmission of the H19Δ13 deletion (−/+). (Right) Comparison of the methylation patterns of the maternal alleles from the same samples. (B) H19Δ13 deletion, paternal transmission. The same analysis as above was performed with DNA samples from the kidney of neonates issued from cross II (paternal transmission, +/−). In both cases, the wild-type and mutant mice were from the same litter. The hatched bars delineate the “domain II” identified within the DMR 2 (see text). The P values were obtained after a statistical analysis of the data using the Mann–Whitney U test.
Table 1.
Relative amounts of methylation of Igf2 gene in mice with H19 gene deletions
| Section | Genotype | Tissue | Allele | DMR2 domain | Mutant/wild-type DNA methylation |
|---|---|---|---|---|---|
| A | H19Δ13/+ | Liver | I + II + III | 1.29 | |
| Kidney | I + II + III | 1.04 | |||
| H19Δ3/+ | Liver | I + II + III | 1.18 | ||
| Kidney | I + II + III | 0.60 | |||
| Gut | I + II + III | 1.18 | |||
| B | H19Δ13/+ | Liver | Paternal | I + III | 0.73 (P = 0.326; n = 6) |
| II | 0.80 (P = 0.301; n = 6) | ||||
| Maternal | I + III | 3.99 (P = 0.035; n = 6) | |||
| II | 11.00 (P = 0.027; n = 6) | ||||
| Kidney | Paternal | I + III | 0.43 (P = 0.019; n = 11) | ||
| II | 0.49 (P = 0.031; n = 11) | ||||
| Maternal | I + III | 1.85 (P = 0.288; n = 6) | |||
| II | 3.31 (P = 0.001; n = 6) | ||||
| H19Δ3/+ | Liver | Paternal | I + III | 0.80 (P = 0.212; n = 10) | |
| II | 0.87 (P = 0.330; n = 10) | ||||
| Maternal | I + III | 4.39 (P = 0.026; n = 6) | |||
| II | 3.74 (P = 0.031; n = 6) | ||||
| Kidney | Paternal | I + III | 0.29 (P = 0.023; n = 8) | ||
| II | 0.53 (P = 0.051; n = 8) | ||||
| Maternal | I + III | 1.00 (P = 0.440; n = 10) | |||
| II | 2.42 (P = 0.021; n = 10) | ||||
| Gut | Paternal | I + III | 0.73 (P = 0.028; n = 10) | ||
| II | 0.94 (P = 0.298; n = 10) | ||||
| Maternal | I + III | 1.85 (P = 0.145; n = 5) | |||
| II | 3.44 (P = 0.004; n = 5) |
For each sample, the sequences obtained from the bisulfite-treated DNAs were used to calculate a value for the average of methylation levels throughout DMR2 (Section A) or in domains I or domains I + III of DMR2 separately for each parental allele (Section B). A ratio between the mutant and wild-type values was calculated for each tissue analyzed. Statistical analysis was performed with the Mann–Whitney U test.
To gain more detailed and quantitative insights into these methylation changes, bisulfite genomic sequencing (19) was performed on DMR 2. DMR 2 was chosen because allelic methylation is consistently associated with Igf2 imprinting (ref. 11 and unpublished results) and because allelic methylation in this region is conserved in humans (22).
We amplified a 703-bp fragment containing 31 CpG dinucleotides (numbered 1 to 31), and the allelic methylation in a 280-bp region including 16 CpGs numbered 16 to 31 was analyzed because allelic methylation differences are most pronounced in this region (unpublished results). The H19 heterozygous mutant and wild-type control animals that were compared were littermates. The results confirm that, in all tissues analyzed, the maternal allele of the Igf2 gene became methylated in mice carrying a deletion of the active allele of H19 (Fig. 3A Right, kidney) and remained unmethylated in mice carrying a deletion of the inactive paternal H19 allele (Fig. 3B Right, kidney) (liver, not shown). Interestingly, mathematical analysis of these data showed that the highest increase of methylation of the Igf2 maternal allele in mice with a deletion of the H19 gene were found between CpGs 18 and 25 (domain II) in all tissues analyzed and that the increase was less pronounced for the other CpGs (domains I and III, Fig. 3). Indeed, statistical analysis (Mann–Whitney U test) showed that, in some tissues such as the gut and the kidney, the increase in methylation on maternal chromosomes was not significant in domains I and III but was highly significant for domain II in the same samples (Table 1, lower parts of section A and B). These experiments show unequivocally that the H19 locus controls in cis the allelic methylation patterns of the DMR 2 region in Igf2. Furthermore, the domain II, as defined in this study and which is only 150 bp long, appears to be a preferential target for this phenomenon.
Deletions of H19 Decrease Methylation in Trans on the Paternal Allele of Igf2.
While studying the cis effect of H19 on the maternal allele of Igf2, we had made the assumption that methylation on the paternal allele remained unaffected. Surprisingly, however, the paternal allele of Igf2 in mice carrying a deletion of the maternal H19 gene was found to be less methylated than the paternal allele in wild-type mice (Fig. 3A Left and Table 1). This difference was statistically significant (except in liver samples); its magnitude varied with the tissue and seemed to be less affected by the deletion of the 5′ part of the H19 gene than the cis difference (Table 1, compare sections A and B). Finally, as an important control, allelic methylation in Igf2 was affected neither in cis nor in trans by a paternally inherited deletion in H19 (Fig. 3B, kidney).
In conclusion, the deletion of the active H19 gene affects the allelic methylation patterns of the Igf2 gene in two opposite ways: it has a positive effect in cis, on the maternal allele, and a negative effect in trans, on the paternal allele. Both effects contribute to the LOAM at this locus. In contrast to the cis effect, which is more pronounced in domain II, the trans effect affects the whole region (Table 1). Furthermore, the magnitudes of the trans and the cis effect vary depending on the tissue analyzed and seem to be influenced in an opposite manner: in tissues where the cis effect is large, the trans effect is small and vice versa. This is exemplified by the results obtained from the liver samples for which the cis effect is the largest while the trans effect is small and not statistically significant (Tables 1, lower parts of sections A and B). These results suggest that the two consequences of the H19 gene deletions on the methylation patterns (cis and trans effects) arise by two different but interdependent mechanisms that are both influenced by H19 gene expression.
DISCUSSION
In this study, we used mice carrying deletions in the H19 locus as a model to investigate the process of LOAM in the mouse. We confirm that LOAM in these animals correlates with LOI of the Igf2 gene (15). We show that deletions of the active maternal allele of the H19 gene have both cis and trans effects on the methylation patterns of the Igf2 gene but deletions of the nonexpressed paternal H19 allele have no effects. These results led us to conclude the expression of the H19 gene, and/or cis-acting sequences within the gene, is directly or indirectly required for the maintenance of the monoallelic patterns of methylation in the DMR 1 and 2 of the Igf2 gene. Overall, our work provides insight into the molecular process of LOAM in the mouse, a process of fundamental importance for the understanding of the somatic maintenance of genomic imprinting in mammals and that is known to be associated with some pathological defects with LOI in humans (Wilms tumor and Beckwith–Wiedemann Syndrome) (23–25).
Competition for Common Regulatory Factors and Enhancers.
Our results emphasize the role of the H19 gene expression in the maintenance of the allelic methylation patterns of the Igf2 gene. The cis-acting effect is consistent with the proposed model of competition for a common set of enhancers (14, 15). The interaction of the enhancers with regulatory elements of Igf2 would lead to the activation of transcription and methylation of the Igf2 gene in cis through the interaction with the de novo methylation/demethylation system (26, 27). Alternatively, or in addition, the H19 RNA could itself be involved in regulating in cis (20) the accessibility to methylase/demethylase activities. Upon H19 inactivation, the methylation/demethylation activities would now be acting equally on both parental alleles leading to an equalization of the levels of methylation (LOAM). Even though the model of competition for the enhancers, as described above, is able to explain in full the cis effect on the methylation patterns, it is not sufficient by itself to explain the trans effect.
The trans effect may reflect selection at the cellular level for a reduction in the overall Igf2 expression in the H19 deleted mice. In that case, cells in which the paternal Igf2 allele is less methylated will be selected for in the H19 deleted animals. Alternatively the two alleles of the Igf2 gene may compete for a limiting pool of regulatory factors affecting either transcription or methylation (Fig. 4A). When the H19 gene is deleted, activation of the second (maternal) Igf2 allele by interaction with the H19 enhancers would partly deplete the paternal allele of the regulatory factors (Fig. 4B). One of these regulatory factors could be a repressor complex that binds to the unmethylated DMRs on the maternal chromosome (13). Repressor competition is further supported from the observation that when Igf2 transgenic constructs, without the H19 enhancers, are introduced into mice, the transgene is inactivated and the endogenous Igf2 gene is hyperactivated (unpublished observations). Introduction of ectopic copies of other imprinted genes such as Xist and U2af in transgenic mice also results in deregulation of the endogenous copy of the gene (28, 29). In addition, the dependence on chromosome counting of some allelically methylated sites in imprinted genes (30) also seems to support the “competition for a limiting factor” model.
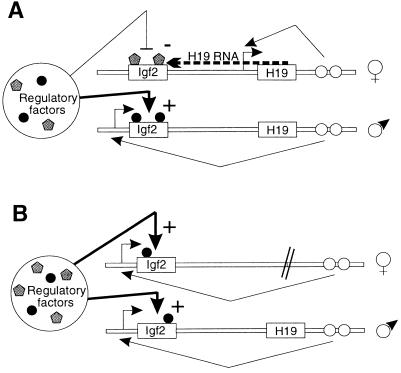
Figure 4.
Double competition model to explain the communication between the two parental chromosomes at the Igf2/H19 locus. (A) The two Igf2 alleles would be competing for both the H19 enhancers in cis and a common limited stock of regulatory factors (transcription/methylation) in trans. These latter are able to act on both Igf2 alleles but with different efficiencies. On the paternal allele interaction of the Igf2 gene with cis regulatory sequences (enhancers, ○) blocks the access to demethylating agents and/or favors the methylating factors (•). Conversely, on the maternal allele, because of the competition of the H19 gene for the enhancers and/or of the presence of the H19 RNA (acting in cis) (20), the methylating factors would be blocked and/or demethylating agents favored (shaded pentagons). (B) Upon deletion of the maternal H19 allele, the competition for the enhancers is lost on the maternal chromosome, and in consequence, both Igf2 alleles are now able to interact with the regulatory factors with similar efficiencies. Because these factors are in a limiting amount, a part would be depleted from one Igf2 allele to interact with the other. This would lead to lower levels of methylation on the paternal chromosome and higher on the maternal.
LOAM by Methylation Transfer.
A third model to explain the trans effects of the H19 deletions on Igf2 methylation is suggested from recent observations in the fungus Ascobolus immersus, where methylation can be transferred from methylated to unmethylated alleles through physical interactions between the homologous alleles (31). This is mechanistically linked to meiotic recombination and gene conversion (32) and leads to both cis and trans effects on the methylation patterns of the genes involved. A model for LOAM by methylation transfer has been proposed for mammals (31, 33, 34) that could account for LOI at the Igf2/H19 locus. Although meiotic and mitotic gene conversion have been described in the mouse (35, 36), it is difficult to believe that these events occur at such a frequency that they become detectable in our system. Indeed the frequency of interchromosomal mitotic gene conversion is known to be very low compared with meiotic or intrachromosomal gene conversion events in mammals (36, 37). In fact no gene conversion events have been detected in our experiments, although this fact alone does not exclude methylation transfer, as it has been shown in Ascobolus that this process may also occur independently of gene conversion (31).
It is thought that the low level of mitotic recombination in mammals is due to the segregation of mammalian chromosomes into different subnuclear sites during interphase, thus restricting their opportunity for contact (37). Interestingly, LaSalle and Lalande (38) recently discovered an association of imprinted domains (trans sensing) during the late S phase of the cell cycle in humans (38) that would be compatible with the formation of heteroduplex DNA and methylation transfer between oppositely imprinted domains during the cell cycle in somatic cells. However, it is important to point out that the methylation transfer we have observed at the Igf2 gene in the H19 deletion mice is not observed in wild-type mice, where both DMR I and DMR 2 maintain their allele-specific methylation patterns (31). Clearly, further experiments are required to investigate this intriguing phenomenon.
Acknowledgments
We thank Robert Feil, Gavin Kelsey, Ben Pickard, Alexander Olek, Vincent Colot, Guy Cathala, and Claude Brunel for helpful comments on the manuscript and David Brown for help with the statistical analysis. T.F. is a recipient of European Molecular Biology Organization and European Union fellowships. W.R. acknowledges support from the Biotechnology and Biological Sciences Research Council, Medical Research Council, European Union, Ministry of Agriculture, Fisheries, and Food, and Action Research.
ABBREVIATIONS
- LOI
loss of imprinting
- LOAM
loss of allelic methylation
- DMR
differentially methylated region
References
- 1.McGrath J, Solter D. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 2.Surani M A, Barton S C, Norris M L. Nature (London) 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 3.Lalande M. Annu Rev Genet. 1997;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- 4.Moore T, Haig D. Trends Genet. 1991;7:47–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 5.Hurst L D. In: Frontiers in Molecular Biology. Reik W, Surani M A, editors. London: Oxford Univ. Press; 1997. , in press. [Google Scholar]
- 6.Neumann B, Barlow D P. Curr Opin Genet Dev. 1996;6:159–163. doi: 10.1016/s0959-437x(96)80045-1. [DOI] [PubMed] [Google Scholar]
- 7.Barlow D P. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Beard C, Jaenisch R. Nature (London) 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 9.Stöger R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow D P. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 10.Brandeis M, Kafri T, Ariel M, Chaillet J R, McCarrey J, Razin A, Cedar H. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil R, Walter J, Allen N, Reik W. Development (Cambridge, UK) 1994;120:2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- 12.DeChiara T M, Robertson E J, Efstratiadis A. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 13.Walter J, Allen N, Kruger T, Engemann S, Kelsey G, Feil R, Forné T, Reik W. In: Epigenetic Mechanisms of Gene Regulation. Russo V E A, Martienssen R A, Riggs A D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 195–213. [Google Scholar]
- 14.Bartolomei M S, Webber A L, Brunkow M E, Tilghman S M. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 15.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Nature (London) 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 16.Tremblay K D, Saam J R, Ingram R S, Tilghman S M, Bartolomei M S. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 17.Leighton P A, Saam J R, Ingram R S, Stewart C L, Tilghman S. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 18.Ripoche M A, Kress C, Poirier F, Dandolo L. Genes Dev. 1997;11:1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 19.Olek A, Oswald J, Walter J. Nucleic Acids Res. 1996;24:5064–5066. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeifer K, Leighton P A, Tilghman S M. Proc Natl Acad Sci USA. 1996;93:13876–13883. doi: 10.1073/pnas.93.24.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elson D A, Bartolomei M S. Mol Cell Biol. 1997;17:309–317. doi: 10.1128/mcb.17.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, Hensle T, Weiss L, McMorrow L, Loew T, Kraus W, Gerald W, Tycko B. Nat Genet. 1994;7:440–447. doi: 10.1038/ng0794-440. [DOI] [PubMed] [Google Scholar]
- 23.Steenman M J C, Rainier S, Dobry C, Grundy P, Horon I L, Feinberg A P. Nat Genet. 1994;7:433–439. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- 24.Reik W, Brown K W, Schneid H, Le Bouc Y, Bickmore W, Maher E R. Hum Mol Genet. 1995;12:2379–2385. doi: 10.1093/hmg/4.12.2379. [DOI] [PubMed] [Google Scholar]
- 25.Schneid H, Seurin D, Vazquez M-P, Gourmelen M, Cabrol S, LeBouc Y. J Med Genet. 1993;30:353–362. doi: 10.1136/jmg.30.5.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei H, Oh S P, Okano M, Jüttermann R, Goss K A, Jaenisch R, Li E. Development (Cambridge, UK) 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 27.Weiss A, Keshet I, Razin A, Cedar H. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 28.Herzing L B, Romer J T, Horn J M, Ashworth A. Nature (London) 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 29.Hatada, I., Nabetani, A., Arai, Y., Ohishi, S., Suzuki, M., Miyabara, S., Nishimune, Y. & Mukai, T. (1997) J. Biol. Chem., in press. [DOI] [PubMed]
- 30.Shemer R, Birger Y, Dean W L, Reik W, Riggs A, Razin A. Proc Natl Acad Sci USA. 1996;93:6371–6376. doi: 10.1073/pnas.93.13.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colot V, Maloisel L, Rossignol J L. Cell. 1996;86:855–864. doi: 10.1016/s0092-8674(00)80161-0. [DOI] [PubMed] [Google Scholar]
- 32.Szostak J W, Orr-Weaver T L, Rothstein R, Stahl F. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 33.Holliday R. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 34.Bestor T, Tycko B. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 35.Liskay M, Stachelek J L. Cell. 1983;35:157–165. doi: 10.1016/0092-8674(83)90218-0. [DOI] [PubMed] [Google Scholar]
- 36.Murti R, Bumbulis M, Schimenti J C. Mol Cell Biol. 1992;12:2545–2552. doi: 10.1128/mcb.12.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shulman M J, Collins C, Connor A, Read L R, Baker M D. EMBO J. 1995;14:4102–4107. doi: 10.1002/j.1460-2075.1995.tb00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaSalle J, Lalande M. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]