Abstract
The mouse Snrpn gene encodes the Smn protein, which is involved in RNA splicing. The gene maps to a region in the central part of chromosome 7 that is syntenic to the Prader–Willi/Angelman syndromes (PWS-AS) region on human chromosome 15q11-q13. The mouse gene, like its human counterpart, is imprinted and paternally expressed, primarily in brain and heart. We provide here a detailed description of the structural features and differential methylation pattern of the gene. We have identified a maternally methylated region at the 5′ end (DMR1), which correlates inversely with the Snrpn paternal expression. We also describe a region at the 3′ end of the gene (DMR2) that is preferentially methylated on the paternal allele. Analysis of Snrpn mRNA levels in a methylase-deficient mouse embryo revealed that maternal methylation of DMR1 may play a role in silencing the maternal allele. Yet both regions, DMR1 and DMR2, inherit the parental-specific methylation profile from the gametes. This methylation pattern is erased in 12.5-days postcoitum (dpc) primordial germ cells and reestablished during gametogenesis. DMR1 is remethylated during oogenesis, whereas DMR2 is remethylated during spermatogenesis. Once established, these methylation patterns are transmitted to the embryo and maintained, protected from methylation changes during embryogenesis and cell differentiation. Transfections of DMR1 and DMR2 into embryonic stem cells and injection into pronuclei of fertilized eggs reveal that embryonic cells lack the capacity to establish anew the differential methylation pattern of Snrpn. That all PWS patients lack DMR1, together with the overall high resemblance of the mouse gene to the human SNRPN, offers an excellent experimental tool to study the regional control of this imprinted chromosomal domain.
Keywords: gametogenesis, genomic imprinting, Snrpn methylation and expression
Genomic imprinting refers to a process that marks parental alleles and leads to monoallelic expression of specific genes. Aberrant imprinting of these genes or an imbalance in contribution of parental chromosomes in the embryo is implicated in abnormal embryonic development, genetic disorders. and tumor development (1–3).
Prader–Willi and Angelman syndromes (PWS and AS, respectively) provide a unique model system for the study of human imprinting mechanisms, because they result from opposite imprinting patterns on chromosome 15 q11-q13. PWS is caused by a deficiency of paternal contributions either by a paternal deletion, a maternal disomy, or an imprinting mutation in this region. In contrast, the AS genotype is characterized by a maternal deletion, a paternal disomy, an imprinting mutation, or a mutation in the AS gene located on the same chromosomal regions (2).
It has recently been suggested that a 100-kb domain in the PWS-AS region that includes the imprinted SNRPN gene and upstream sequences comprises an imprinting center (IC), which affects in cis DNA methylation, chromatin structure, and expression of imprinted genes throughout a 2-Mb chromosomal domain within the 15q11-q13 region (4). IC transcripts, which are alternative transcripts of the SNRPN gene and include newly discovered exons located tens of kilobases upstream to the SNRPN gene, now have been identified. These transcripts are paternally expressed, although at very low levels, and lack protein-coding potential (5). It was proposed that these transcripts are involved in the imprinting switch during gametogenesis, because mutation or deletions in the newly discovered exons impaired the paternal-to-maternal imprint switch, whereas deletions around the SNRPN exon 1 affect the maternal-to-paternal switch.
This recently discovered central role of the human imprinted SNRPN gene in the imprinting control of the PWS-AS region prompted us to study thoroughly the mouse Snrpn gene.
The mouse Snrpn gene is located in the central region of chromosome 7 proximal to the loci of pink-eyed dilution and the γ-aminobutyric acid receptor b3 (Gabrb3) (6). This region is homologous to the PWS-AS region on human chromosome 15q11-q13 and shares in common many structural features. As in the case for human SNRPN, the mouse gene is paternally expressed (6, 7) primarily in brain and heart (8), coding for a protein (Smn) that is thought to be involved in splicing (9).
In the present communication a detailed description of the structural features of the entire mouse Snrpn gene is provided. Special emphasis is put on the allele-specific methylation patterns of the gene, because localized areas of allele-specific methylation have been observed in all imprinted genes examined so far (10) and some of these allele-specific modifications are inherited from the gametes and may therefore mark the parental alleles and determine which allele will be expressed.
The significance of DNA methylation in monoallelic expression has been demonstrated experimentally in DNA methyltransferase-deficient mice (Dnmtase). Genes such as H19 and Xist that normally express only the unmethylated allele are unable to maintain monoallelic expression in Dnmtase-deficient mouse embryos. Genes that express the methylated allele, such as Igf2 and Igf2r, show no expression in Dnmtase-deficient mice (11). Here we describe two differentially methylated regions in the Snrpn gene, one in the 5′ end of the gene, which is methylated specifically on the silent maternal allele, and a second in the 3′ end of the gene, which is methylated on the active paternal allele. These two differentially methylated domains are separated by a region that is methylated biallelically. The establishment of the parent-specific methylation patterns during gametogenesis and their stable inheritance throughout development of the preimplantation embryo are also demonstrated. In addition, our results imply that the monoallelic methylation patterns of Snrpn must be established prior to gamete maturation.
MATERIALS AND METHODS
Isolation and Characterization of the Mouse Genomic Clone.
P1 genomic clones containing the complete mouse Snrpn gene were custom-made (Genome Systems, St. Louis). The P1 genomic clone was digested by XbaI, BamHI, or SacI. To construct a P1 subclone library the digested fragments were ligated to Puc19. The library was screened by the complete Snrpn cDNA for exon/intron junctions. Exon/intron boundaries were identified by sequencing PCR fragments using primers derived from the Snrpn cDNA.
Cell Cultures and Transfection Experiments.
Mouse embryonic stem (ES) cells were grown in a DMEM:Ham’s F-12 medium supplemented with 20 pg/ml LIF (GIBCO/BRL).
Transfection of ES cells was performed with a lipofectin transfection kit (Boehringer Mannheim) according to the manufacturer’s instructions. Neo-resistent clones were pooled, grown to mass culture, and used to extract DNA. DNA was prepared and Southern blotted, and methylation was analyzed by PCR as described previously (12).
DNA samples were methylated in vitro by incubation overnight at 37°C with 16 μm SAM and 5–10 units/1 μg DNA of M·HhaI, M·HpaII, or dam methylase. The DNA was then purified, and the extent of methylation was evaluated by digestion with HhaI, HpaII, or MboI, respectively.
Preparation of Biological Material.
Sperm was collected from the vas deferens, oocytes were collected from oviducts of superovulated CB6F1 females, and parthenogenetic, androgenetic, and normal preimplantation embryos were prepared as described before (12).
Injection into the Pronuclei of the Fertilized Egg.
Injection experiments were performed as described before (13). DNA samples were methylated in vitro by dam methylase at GATC sites located adjacent to the studied CpG site. This site is sensitive to digestion by DpnI only when methylated on both strands or by MboI when the site is unmethylated on both strands. Hemimethylated GATC sites are refractory to digestion by either DpnI or MboI. We took advantage of this fact, and DNA extracted from blastocysts derived from the injected zygotes was digested with DpnI to eliminate the fully methylated unintegrated DNA and with MboI to eliminate the unmethylated endogenous sequence and integrated molecules that underwent several rounds of replication. DNA molecules that represent the original integrated DNA (hemimethylated at GATC site) should survive digestion by both DpnI and MboI.
Zygotes were isolated from mated superovulated females 20 hr after injection of chorionic gonadotropin. Approximately 1 pl of DNA (10 ng/μl) was injected into one of the pronuclei. The injected zygotes were grown in culture in M-16 medium (Sigma) for several days to obtain blastocysts. The PCR primers used for the above analyses were as follows: M1 site, 5′-CCCTCTCCCACATAGTAAAAATCTGT and 3′-CGTCCCAGGCAATGGCTGC; H1 site, 5′-TACTGGTGGCAATGGGTTTCAGAG and 3′-CACGGGGAAGGGATATAAATAAAGGTTCG; H2 site, 5′-CTCAACGTGCTATGTAAGC and 3′- GCTAGCGACTATAAGTCCCT; H3 site, 5′- TTGGACTTCCCCCTGCTCGTG and 3′-GCAGTAAGAGGGGTCAAAAGC.
RESULTS
Genomic Structure and Complete cDNA of the Mouse Snrpn Gene.
The published cDNA sequence of the mouse Snrpn gene consists of 1.1 kb (8) but is inconsistent with the 1.3-kb-long reported size of the Snrpn mRNA (6). Using 5′ rapid amplification of cDNA ends and primer extension analysis, we have observed and sequenced an additional upstream sequence of 200 bp (Fig. 1). It should be noted that this 5′ region of the mouse cDNA shows good homology with the 5′ end of the human SNRPN cDNA.
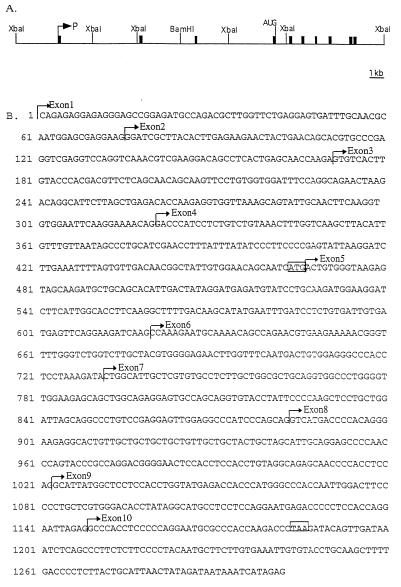
Figure 1.
(A) The genomic structure of the mouse Snrpn gene. Solid boxes are exons 1–10. P, transcription start site. (B) The complete sequence of the mouse Snrpn cDNA is presented. The first 309 nucleotides are data from our laboratory. Nucleotides 310-1302 are previously published data (8). The start and stop codons for the Smn protein are marked in boxes ATG and TAA, respectively. The start of exons 1–10, designated by horizontal arrows, were obtained by PCR sequencing of the exon/intron junctions, as described in Materials and Methods. Exons 1–3 were identified by long-range PCR using subclones of our P1 genomic clone containing the Snrpn gene.
To determine the genomic structure of the mouse Snrpn gene, PCRs were performed using primers derived from the cDNA sequence to identify the exact location of the intron/exon boundaries. The results of this analysis were further confirmed using cDNA probes to screen a subclone library of a P1 genomic clone that covers the entire mouse Snrpn gene. When the library was screened using exon 1 as a probe we isolated a genomic clone that includes an upstream sequence, rich in CpG and alternating GTs. This sequence shows promoter activity in a transfection experiment with a CAT reporter gene (to be published elsewhere). As can be seen in the scheme presented in Fig. 1 the gene contains 10 exons and extends over a region of 22 kb of genomic DNA, similar to the human SNRPN gene (7).
Maternal Methylation of the Snrpn 5′ Region and Intron 1(DMR1).
One important feature that is likely to be involved in the regulation of the gene is the methylation status of the 5′ region, not only with regard to its effect on Snrpn expression but also for the possible involvement of methylation in imprinting regulation of the entire region. We have therefore analyzed the methylation pattern of this region in several somatic tissues and cell lines. Genomic DNA was digested with a combination of the restriction enzymes PstI and HpaII, and the restriction fragments were electrophoresed, Southern blotted, and probed with a 1.6-kb genomic fragment spanning exon 1 of the Snrpn gene (Fig. 2A).
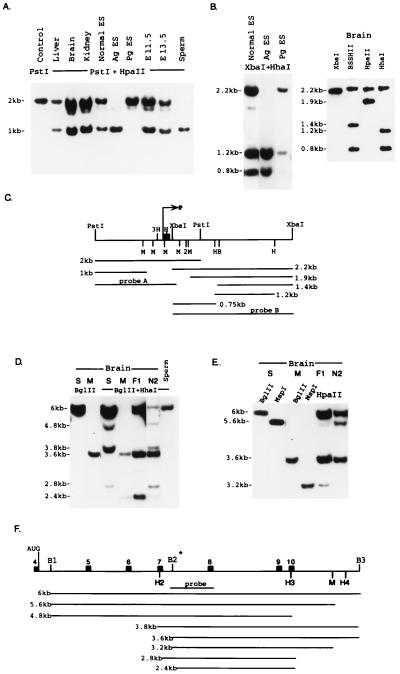
Figure 2.
Differential methylation in Snrpn. (A–C) Differential methylation of the 5′ part of Snrpn (DMR1). (A) DNA was extracted from adult tissues, embryonic stem cells, and 11.5- and 13.5-dpc embryos. Extracted DNA was digested with PstI (lane 1) or PstI plus HpaII (lane 2–9), Southern blotted, and probed with the 1.6-kb fragment (probe A). E11.5 and E13.5 are 11.5- and 13.5-dpc embryos, respectively. (B) DNA was extracted from embryonic stem cells (ES), androgenetic ES cells (Ag ES), parthenogenetic ES cells (Pg ES), and brain. DNA samples were digested with XbaI, XbaI plus HhaI (HhaI), XbaI plus BSSHII (BssHII), or XbaI plus HpaII (HpaII). The digested DNA was blotted and probed with a 2.2-kb XbaI fragment (probe B). The lower part of the figure displays the size of the expected restriction fragments. Exon 1 is indicated by the solid box. M, HpaII/MspI sites; B, BSSHII site; and H, HhaI sites. (D–F) Differential methyalion of the 3′ part of Snrpn (DMR2). Brain DNA samples were prepared from Mus spretus (S) and Mus musculus (M), from offspring (F1) of a cross between female Mus musculus × male Mus (maternally derived Mus musculus Snrpn allele), or from offspring of a back-cross between a F1 female and Mus musculus male (N2) (maternally derived Mus spretus allele). All DNA samples were digested with either BglII plus HhaI (BglII) (D) or BglII plus MspI (MspI) (E) and BglII plus HpaII (HpaII) and probed with an intron 7 fragment. F shows the probe and size of the expected restriction fragments. B1, B2, and B3 are BglII sites. B2* is BglII polymorphic site absent in Mus spretus and present in Mus musculus. H2, H3, and H4 are HhaI sites; M is an HpaII site. Solid boxes are exons 4–10.
Two bands representing 2- and 1-kb fragments were observed with restriction enzyme digests of DNA from adult liver, brain, kidney, and total DNA from 11.5-dpc embryos and head DNA from 13.5-dpc embryos. The observation of the intact 2-kb fragment implies that all five HpaII (M) sites in this region are methylated, whereas the 1-kb product represents a complete HpaII digest, meaning that the entire region is hypomethylated. Because both bands appear in similar intensities we assume that the HpaII sites in this region are monoallelically methylated. That this is indeed the case is corroborated by the following observations: Although both bands (1 and 2 kb) are observed with normal ES cell DNA, only the 1-kb product is seen when androgenetic ES cell DNA or sperm DNA is analyzed. These results are consistent with the observation that only the 2-kb product is observed in parthenogenetic ES cell DNA. Identical results were obtained for the four HhaI (H) sites at the 5′ region of the gene. Taken together these results indicate that the paternal allele is unmethylated throughout the 5′ region, whereas the maternal allele is completely methylated.
This differentially methylated region, which will be designated DMR1, is not limited to the 5′ end of the gene but rather extends 6-kb downstream, constituting a region that includes the entire Snrpn intron 1 (Fig. 2B and Fig. 3). The normal ES cells and brain DNA digested with XbaI and HhaI and probed with probe B revealed both the methylated maternal fragment (2.2 kb) and the unmethylated paternal fragments (1.2 and 0.8 kb). In contrast, the same treatment of parthenogenetic ES cell DNA produced an intact 2.2-kb band consistent with methylation of the two HhaI sites, whereas digestion of androgenetic ES cell DNA and sperm DNA produced 1.2- and 0.8-kb bands, consistent with an unmethylated state of the HhaI sites. The same methylation profile was observed for the HpaII site (M) and the BSSHII site (B), indicating that the entire 6-kb region (DMR1) is differentially methylated on the maternal allele (Fig. 3). Restriction enzyme analyses of sites in the central part of the gene (exons 2–4) showed biallelic methylation (data not shown; see Fig. 3). This region, which contains two HpaII and two HhaI sites, was completely resistant to digestion by HpaII or HhaI and thus is a biallelic methylated region (BMR).
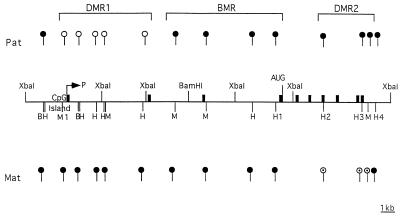
Figure 3.
Summary of the DNA methylation pattern of the mouse Snrpn gene. The methylation analysis of the Snrpn gene revealed methylation differences between the paternal (Pat) and the maternal (Mat) allele. Open circles indicate unmethylated sites. Solid circles indicate methylated sites. Partial methylation is indicated by dotted circles. Exons 1–10 are indicated by solid boxes. B, BSSHII; H, HhaI; M, HpaII. The restriction sites indicated by numbers (e.g., H1) are those further studied during gametogenesis and following injection into zygote pronuclei and transfection experiments.
Paternal Methylation of Snrpn 3′ Region (DMR2).
In contrast to the maternal methylation described above for DMR1, a 3.5-kb region spanning exons 7–10 (DMR2) turned out to be preferentially methylated on the paternal allele (Figs. 2 D–F). In this region, two HhaI sites (H2, H3) were found to be partially methylated in brain (Fig. 2D) and other adult tissues (data not shown). To determine whether this partial methylation is allele-specific, we analyzed brain DNA from the offspring (F1) of a cross between Mus musculus female and Mus spretus male and brain DNA from a mouse with a maternally derived spretus allele (N2) that was obtained by a F1 female back-crossed with a Mus musculus male. A unique BglII polymorphic restriction site (B2*) located in intron 7 of the Mus musculus DNA allowed us to distinguish between the parental alleles. BglII digestion of the Mus musculus DNA (M) generates a 3.6-kb fragment, and BglII digestion of the Mus spretus DNA (S) generates a 6-kb fragment (Fig. 2D). Digestion of Mus spretus DNA with a combination of BglII + HhaI resulted in three additional bands of 4.8, 3.8, and 2.8 kb, indicating that H2 and H3 are partially methylated in Mus spretus brain. BglII + HhaI digestion of Mus musculus brain DNA (M) results in two bands of 3.6 kb and 2.4 kb, indicating partial methylation of H3. We were not in a position to analyze the methylation status of H2 in Mus musculus because the polymorphic BglII restriction site lies 3′ to the H2 site. BglII + HhaI digestion of brain DNA from F1 mice (Mus musculus maternal allele) revealed, in addition to the 6-kb band, only the 3.6- and 2.4-kb bands. The absence of the 4.8- and 3.8-kb bands (Mus spretus) indicates that H2 and H3 are methylated on the paternal allele, and the 2.4-kb band indicates that H3 is unmethylated on the maternal allele. BglII + HhaI digestion of brain DNA from N2 mice (Mus spretus maternal allele) revealed 4.8-, 3.8-, and 2.8-kb bands in addition to the maternally derived 6-kb band and the paternally derived 3.6-kb band, indicating that H2 and H3 sites are undermethylated on the maternal allele. The faint 2.4-kb band should represent the B1-H2 fragment. If that is indeed the case and the 2.4-kb fragment (B2-H3) is absent, this again will indicate that H3 is methylated on the paternal allele.
The HpaII site (M) located 0.8 kb 3′ to exon 10 is also partially methylated on the maternal allele. The 3.2-kb band revealed with F1 DNA digested with BglII + HpaII and the 5.6-kb band obtained with N2 DNA digested with BglII + HpaII indicate that the M site is completely methylated on the paternal allele and partially methylated on the maternal allele (Fig. 2E). Taken together, all the results that are presented in Fig. 2 (D–E) reveal a region in the Snrpn gene that is preferentially methylated on the paternal allele (DMR2).
In conclusion, the Snrpn gene consists of two differentially methylated regions, the 5′ end methylated on the maternal allele (DMR1) and the 3′ end methylated on the paternal allele (DMR2). These two regions are separated by a third region between exons 2 and 4 that is methylated on both alleles (BMR) (Fig. 3). A similar methylation pattern was observed in human SNRPN gene (7).
Involvement of DMR1 and DMR2 in Silencing the Maternal Snrpn Allele.
To examine whether the methylation of Snrpn DMR1 and DMR2 are involved in silencing the maternal allele of the Snrpn gene, we have compared the level of mRNA extracted from E9.5 normal embryos (+/+), S mutants of Dnmtase (−/−), and heterozygous embryos (+/−). As seen in Fig. 4, Snrpn mRNA levels are not reduced but rather seem to be elevated in the Dnmtase mutant as compared with normal or heterozygous embryos. This result may imply that DMR1 methylation in itself is responsible for silencing the maternal allele. Similar results are shown here and have been observed before for the H19 gene (11). Our observation cannot support a simple repressor hypothesis for DMR2, as it was suggested previously for Igf2 (14) and Igf2r (15), which do not show any expression in Dnmtase embryos (Fig. 4 and ref. 11).
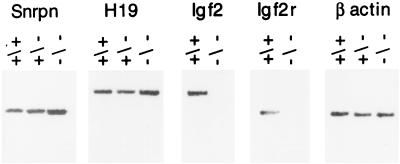
Figure 4.
Expression of Snrpn in DNA methyltransferase-deficient mice. RNA samples of normal (+/+), Dnmtase-deficient (−/−), and heterozygous (−/+) E9.5 embryos were obtained from R. Jaenisch (Massachusetts Institute of Technology, Cambridge, MA) and reverse transcribed by reverse transcriptase–PCR (RT-PCR) using oligo(dT) as primer. The RT-PCR products were further amplified by PCR using primers described by Szabo and Mann (19), and primers for β actin were 5′-CAGCTTCTTTGCAGCTCCTT and 3′-TCACCCACATAGGAGTCCTT.
The Snrpn Differential Methylation Patterns Are Established During Gametogenesis, Inherited from the Gametes, and Maintained in the Preimplantation Embryo.
Having shown that the Snrpn gene is differentially methylated in upstream (DMR1) and downstream (DMR2) sequences prompted us to investigate the origin of this allele-specific methylation pattern and determine the precise time of its establishment. If differential modifications DMR1 and DMR2 serve as an imprinting signal, they should be inherited from the gametes or established in the zygote prior to syngamy.
Four sites were chosen for methylation analysis in sperm, oocytes, and in the preimplantation embryo: M1, H1 and H2, and H3, representing DMR1, BMR, and DMR2, respectively. In the oocytes, in contrast to sperm, the M1 site (DMR1) was methylated, whereas M1 is partially methylated in eight cell embryos, morulae, and blastocysts. To examine which of the parental alleles is methylated in the early embryo, we used parthenogenetic and androgenetic blastocysts. The M1 site was completely methylated in parthenogenetic blastocysts but completely unmethylated in androgenetic blastocysts (Fig. 5A). This observation clearly shows that the partial methylation is exclusively on the maternal allele, indicating that the methylation observed in the gametes is preserved throughout preimplantation development. The sites in DMR2 (H2 and H3) were methylated in sperm, unmethylated in oocytes, and partially methylated in the blastocyst (Fig. 5B). We expected that this partial methylation resides on the paternal allele. We were able to show that this is indeed the case by the analysis of methylation of site H2 in F1 blastocysts derived from a cross between Mus spretus males (S) and Mus musculus females (M). Following HhaI digestion and PCR amplification of a fragment containing H2, the PCR product was found to be resistant to digestion with BglII (BglII is a polymorphic site on Mus spretus). The H1 site, located in BMR, was used as a control and found to be unmethylated in the blastocyst (Fig. 5B). In conclusion, the methylation patterns of DMR1 and DMR2 are inherited from the gametes and preserved throughout preimplantation development. To examine whether these parentally inherited patterns are reestablished during gametogenesis, we have analyzed by PCR the methylation status of M1, H1, and H2 at various stages of spermatogenesis and oogenesis.
Figure 5.
Methylation changes during gametogenesis and preimplantation embryo development. (A) Methylation of site M1 was analyzed by PCR (see Materials and Methods) in sperm, oocyte, eight cell embryos, morulae, blastocyst, parthenogenetic blastocyst (Pg), and androgenetic blastocyst (Ag). DNA used for PCR analysis was predigested with BamHI (−) or BamHI plus HpaII (+). (B) Sites H2 and H3 in DMR2 were analyzed in sperm, oocytes, and blastocysts. Methylation analysis of site H1 located at the exon 2–4 region was included as a control. This site, in contrast to M1, H2, and H3, is not methylated in the blastocyst stage and becomes methylated on both alleles in the postimplantation embryo (Fig. 3). Site H2 was assayed in a similar manner using blastocysts DNA from Mus spretus (S), Mus musculus (M), and from a cross between a Mus spretus male and Mus musculus female, (F1). The amplification products were digested with BglII and electrophoresed on agarose gel. At the bottom of the figure, filled boxes represent exons 1–10. BglII* is a polymorphic site present in Mus musculus. (C) The methylation status of sites M1, H1, and H2 was analyzed during gametogenesis. DNA samples were prepared from male and female 12.5-, 15.5-, 18.5-, and 21.5-dpc primordial germ cells that were cut with PvuII (−) or PvuII plus HpaII (+) for site M1 and with PvuII/HhaI (+) for site H2. The digested DNA was subjected to PCR using primers listed in Materials and Methods.
Primordial germ cells emerge from the epiblast, are seen first at the root of the allantoic mesoderm at 7.5 dpc, and then eclipse until seen again in the genital ridges at 12.5 dpc. All genes studied to date were found to be unmethylated when they reach the genital ridges. This is true also for the entire Snrpn gene (Fig. 5C); all three regions (DMR1, DMR2, and BMR) are unmethylated in the primordial germ cells of 12.5-dpc embryos. However, site M1 (DMR1) becomes gradually methylated during oogenesis but stays unmethylated throughout spermatogenesis.
In contrast, H1 (BMR) undergoes rapid de novo methylation and stays methylated during both spermatogenesis and oogenesis. Similarly, site H2 (representing DMR2) undergoes rapid de novo methylation during spermatogenesis as well as oogenesis but undergoes demethylation toward maturation of the oocyte (Fig. 5 B and C).
Parent-Specific Methylation in DMR1 and DMR2 Cannot Be Achieved Postfertilization.
To examine whether DMR1 and DMR2 can acquire their methylated patterns even postfertilization, we have injected these sequences in their methylated or unmethylated state into the male or female pronucleus of a fertilized egg and determined the methylation status of individual sites in the blastocyst (for details see injection experiments in Materials and Methods). If that were the case, we would expect that de novo methylation will be observed when M1 (representing DMR1) is injected into the female pronucleus and demethylation will be observed when M1 is injected into the male pronucleus. In practice, no changes in methylation were observed with M1, regardless of the pronucleus to which it was injected (Fig. 6A). Similarly, a fragment containing H2 (representing DMR2), which is expected to undergo de novo methylation when injected to the male pronucleus, remained unmethylated regardless of which pronucleus was injected (Fig. 6A).
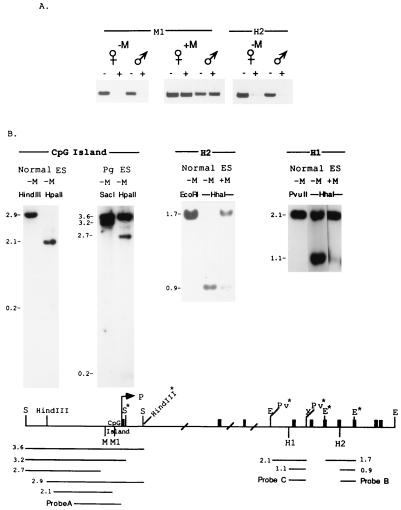
Figure 6.
Differntial methylation of DMR1 and DMR2 cannot be established postfertilization. (A) A SacI fragment (S-S) that includes M1 was methylated in vitro by HpaII methylase and injected methylated (+M) or unmethylated (−M) into the female pronucleus (♀) or into the male pronucleus (♂) of a fertilized egg. An EcoRI fragment (E*-E*) that includes H2 was also injected as above. The injected zygotes were grown in culture to obtain blastocysts. DNA was extracted from pooled blastocysts, digested with HpaII (+) or undigested (−), and subjected to PCR analysis. The strategy designed to distinguish between the endogenous and the injected fragment is described in Materials and Methods. (B) Plasmid constructs were prepared containing the SacI fragment (includes M1) (construct A), EcoRI fragment (E*-E*) containing site H2 (construct B), and EcoRI-XbaI fragment (E-X) containing site H1 (construct C). The plasmids were methylated in vitro with HpaII methylase (construct A) or with HhaI methylase (constructs B and C). The plasmids were transfected methylated (+M) or unmethylated (−M) into embryonic stem cells (ES) or into parthenogenetic ES cells (Pg). DNA samples from pooled neomycin-resistant colonies were digested with HindIII or HindIII/HpaII (HpaII), with SacI or SacI/HpaII (HpaII), with EcoRI or EcoRI/HhaI(HhaI), and with PvuII or PvuII/HhaI (HhaI). Hybridizations were with probes A, B, or C, respectively. S, SacI; E, EcoRI; Pv, PvuII. Stars designate restriction sites located only in the exogenous constructs and used for distinguishing between the endogenous and the exogenous DNA fragments.
These results indicate that the differential methylation patterns of DMR1 and DMR2 cannot be acquired postfertilization. This was supported by results of transfection experiments in ES cells, which were carried out with unmethylated or in vitro methylated (by M·HpaII or M·HhaI) fragments. Transfection with the DMR1 fragment in its unmethylated state resulted in a 2.9-kb HindIII band being replaced by 2.1- and 0.2-kb bands in the HindIII/HpaII digest, indicating that the HpaII sites in the 2.9-kb fragment did not undergo de novo methylation (Fig. 6B). Although DMR1 is fully methylated in parthenogenetic ES cells (Fig. 2A), the introduction of a SacI fragment into parthenogenetic ES cells did not result in de novo methylation of the HpaII sites as judged by the digestion of the 3.2-kb band to 2.7- and 0.2-kb bands. (The 3.6-kb band represents the fully methylated endogenous Snrpn 5′ region.)
To examine the behavior of DMR2 upon transfection to ES cells we introduced a 1.7-kb EcoRI fragment flanking the H2 site in its methylated and unmethylated state into ES cells. As shown above for DMR1, the results presented in Fig. 6B demonstrate that ES cells are incapable of methylating de novo or demethylating the H2 site (DMR2). In contrast, the H1 site, which represents the BMR region, remained fully modified when introduced methylated, as indicated by the 2.1-kb band (Fig. 6B) and underwent de novo methylation when introduced unmethylated, as seen by the appearance of a strong 2.1-kb band in addition to the 1.1-kb band. That the H1 site as well as sites in nonimprinted genes undergo de novo methylation in ES cells testifies for the existence of the methylation machinery in ES cells; however, both monoallelic methylated regions, DMR1 and DMR2, are recognized as sequences that must be protected from this machinery.
DISCUSSION
It is generally believed that DNA methylation is a major epigenetic determinant in the establishment and maintenance of the imprinted state (10). We have observed two differentially methylated regions in Snrpn: a maternally methylated domain in the 5′ region (DMR1), which inversely correlates with the paternal activity of the gene, and a paternally methylated region (DMR2) covering the 3′ part of the gene (Figs. 2 and 3). This allele-specific methylation originates from the gametes. Therefore, for this methylation to play a role in the imprinting process it must be erased and reestablished during gametogenesis. Our observations presented in Fig. 5 demonstrate that this is indeed the case.
We next asked whether DMR1 and DMR2 methylation patterns can be reestablished postfertilization in the case of an aberrant methylation process during gametogenesis. Transfection experiments into ES cells and injections of DMR1 or DMR2 into the male or female pronuclei of the fertilized egg revealed that embryonic cells are not capable of reestablishing the appropriate differential methylation pattern of either of these regions (Fig. 6). These results are consistent with previously published data obtained with Dnmtase mice (16). Nevertheless, embryonic cells seem to recognize each of these sequences and protect them from de novo methylation, which is characteristic of these cells (17). In fact, these results are consistent with the striking stability of the methylation patterns of DMR1 and DMR2. These patterns prevail in all embryonic cells starting at the onset of zygotic expression (Fig. 5) and are maintained in all somatic cells (Fig. 2). Thus, DMR1 and DMR2 escape the genome-wide demethylation at the precavitation stage, the global de novo methylation at the postimplantation stage, and cell-specific demethylations during cell differentiation (18).
Interestingly, the embryonic cells seem to recognize the sequences independent from each other because they were injected and transfected individually. This implies that the methylation of each region is regulated locally and is not dependent on sequences outside the DMR. However, this observation does not rule out the possibility that the establishment of the parent-specific methylation patterns requires additional signals outside the DMR sequences during gametogenesis, where DMR1 becomes methylated in the female gonads and DMR2 becomes methylated in the male gonads (Fig. 5).
In addition to the feasible role of DMR1 and DMR2 in the imprinting process, it is possible that the differential methylation of these regions is associated with the monoallelic expression of the Snrpn gene. Our observation of elevated levels of Snrpn mRNA in the methyl-deficient mice (Fig. 4) strongly suggests that methylation of the maternal allele at DMR1 plays a role in silencing this allele. In contrast, our results do not allow us to decide whether the preferential methylation of the paternal allele in the DMR2 sequence participates in this silencing.
Although methylation of DMR1 correlates perfectly well with the paternal expression of Snrpn in the embryo and adult tissues, it should be noted that Snrpn has been reported to be expressed biallelically during oogenesis (19), while, as we show here, DMR1 becomes gradually methylated (Fig. 5). However, the biallelic expression during oogenesis may reflect accumulation of RNA produced at 12.5 dpc at a stage when both alleles are unmethylated (Fig. 5).
The evidence presented here (Figs. 5 and 6), that the differential methylation patterns must be erased and reestablished during gametogenesis, strengthens the hypothesis that the mechanism of the imprinting switch involves changes in methylation during gametogenesis. For the maternal-to-paternal imprinting switch to occur, DMR2 has to undergo de novo methylation, whereas the paternal-to-maternal switch should involve de novo methylation of DMR1. In fact, in PWS patients carrying microdeletions including SNRPN exon1, the entire region is characterized by an abnormal methylation pattern (4). DMR1 includes exon 1 and, therefore, may participate in the regulation of two processes: affecting the activity of the Snrpn promoter, thereby regulating production of the Smn protein (9), and playing a role in the imprinting switch mechanism of the entire region (5).
In light of the fact that the imprinting switch is a process occurring during gametogenesis, it is clear that further study of the mechanism of the imprinting switch with regard to the role played by Snrpn in this process and the relevance of methylation to the switch mechanism must be carried out in a mouse model. Our studies have set the groundwork for such a model, considering the striking similarity of the mouse Snrpn to the human SNRPN gene.
Acknowledgments
We are grateful to Drs. Mira Ariel and John McCarrey for providing us with cells from various stages of gametogenesis, Dr. Rudolf Jaenisch for providing RNA from Dnmtase-deficient mice (S mutant), and to Drs. Wolf Reik and Wendy Dean for providing us with androgenetic embryos. This work was supported by National Institutes of Health Grant GM 20483 and CTR Grant 3022R1 to A.R.
ABBREVIATIONS
- ES
embryonic stem cell
- dpc
days postcoitum
- BMR
biallelic methylated region
- PWS
Prader–Willi syndrome
- AS
Angelman syndrome
References
- 1.Beechey C V, Cattanach B M. Mouse Genome. 1996;94:96–99. [Google Scholar]
- 2.Nicholls R D. Am J Hum Genet. 1994;54:733–740. [PMC free article] [PubMed] [Google Scholar]
- 3.Feinberg A P. Nat Genet. 1993;4:110–113. doi: 10.1038/ng0693-110. [DOI] [PubMed] [Google Scholar]
- 4.Buiting K, Saitoh S, Gross S, Schwartz S, Nicholls R D, Horsthemke B. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- 5.Dittrich B, Buiting K, Korn B, Richard S, Buxton J, Saitoh S, Nicholls R D, Poustka A, Winterpacht A, Zabel B, Horsthemke B. Nat Genet. 1996;14:165–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- 6.Leff S E, Brannan C I, Reed M L, Ozcelik T, Francke U, Copeland N G, Jenkins N A. Nat Genet. 1992;2:259–264. doi: 10.1038/ng1292-259. [DOI] [PubMed] [Google Scholar]
- 7.Glenn C C, Saitoh S, Jong M T C, Filbrandt M M, Surti U, Driscoll D J, Nicholls R D. Am J Hum Genet. 1996;58:335–336. [PMC free article] [PubMed] [Google Scholar]
- 8.Gerrelli D, Sharpe N G, Latchman D S. Nucleic Acids Res. 1991;19:6642. doi: 10.1093/nar/19.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steitz J A, Black D L, Gerke V, Parker K A, Kramer A, Frendewey D, Keller W. In: Structure and Function of Major and Minor Small Nuclear Ribonuclear Ribonucleoprotein Particles. Birnstiel M L, editor. New York: Springer; 1988. pp. 115–154. [Google Scholar]
- 10.Razin A, Cedar H. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 11.Li E, Beard C, Jaenisch R. Nature (London) 1993;366:2282–2292. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 12.Shemer R, Birger Y, Dean W L, Reik W, Riggs A D, Razin A. Proc Natl Acad Sci USA. 1996;93:6371–6376. doi: 10.1073/pnas.93.13.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kafri T, Gao X, Razin A. Proc Natl Acad Sci (USA) 1993;90:10558–10562. doi: 10.1073/pnas.90.22.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki H, Jones P A, Chaillet J R, Ferguson-Smith A C, Barton S C, Reik W, Surani M A. Genes Dev. 1992;6:1843–1856. doi: 10.1101/gad.6.10.1843. [DOI] [PubMed] [Google Scholar]
- 15.Stoger R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow D P. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 16.Tucker K L, Beard C, Dausman J, Jackson-Grusby L, Laird P W, Lei H, Li E, Jaenisch R. Genes Dev. 1996;10:1008–1020. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- 17.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Nature (London) 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 18.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 19.Szabo P E, Mann J R. Genes Dev. 1995;9:1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]