Abstract
Markers on the non-recombining portion of the human Y chromosome continue to have applications in many fields including evolutionary biology, forensics, medical genetics, and genealogical reconstruction. In 2002, the Y Chromosome Consortium published a single parsimony tree showing the relationships among 153 haplogroups based on 243 binary markers and devised a standardized nomenclature system to name lineages nested within this tree. Here we present an extensively revised Y chromosome tree containing 311 distinct haplogroups, including two new major haplogroups (S and T), and incorporating approximately 600 binary markers. We describe major changes in the topology of the parsimony tree and provide names for new and rearranged lineages within the tree following the rules presented by the Y Chromosome Consortium in 2002. Several changes in the tree topology have important implications for studies of human ancestry. We also present demography-independent age estimates for 11 of the major clades in the new Y chromosome tree.
In 2002, the Y Chromosome Consortium published a single most parsimonious phylogeny of 153 binary haplogroups based on markers genotyped in a globally representative set of samples (Y Chromosome Consortium 2002). A simple set of rules was developed to unambiguously label the different clades nested within this tree. This hierarchical nomenclature system, which has been widely accepted by the research community, unified all past nomenclatures and allowed the inclusion of additional mutations and haplogroups yet to be discovered. Subsequently, Jobling and Tyler-Smith (2003) published a modified version of the Y Chromosome Consortium (YCC) tree; however, since 2003, there has been no unified effort to update the Y chromosome binary haplogroup tree with the hundreds of polymorphisms that have been discovered and surveyed in global human populations. Here, we attempt to bring the Y chromosome tree up to date and present a modified nomenclature based on the rules put forward by the Y Chromosome Consortium in 2002.
The original YCC tree was constructed on the basis of 243 unique polymorphisms, representing most of the binary markers known at the time. Since then, more than 400 binary polymorphisms have been discovered. While this has led to increased phylogeographic resolution for many Y chromosome studies, without occasional unified efforts to incorporate these markers into a fully resolved Y chromosome tree, there is once again the possibility that multifarious and erroneous naming systems will appear in the literature. As we show, many of the newly discovered polymorphisms require topological changes to the tree, as well as new nomenclature to define the lineages. As pointed out by the Y Chromosome Consortium (2002), newly discovered mutations may have the effect of splitting clades or joining previously separated clades, while new samples may contain intermediate haplogroups. The overall goals of this study are to produce an updated tree showing the evolutionary relationships among lineages marked by newly discovered and published Y chromosome markers, to name these lineages, to make mutation information available to the Y chromosome research community, and to date 11 of the major clades in this tree.
Methods
To bring the Y Chromosome Consortium (2002) tree up to date, we used three different approaches to map recently discovered mutations on the Y chromosomal binary haplogroup tree. Many of these mutations came from the laboratories of Michael Hammer and Peter Underhill (“P” and “M” mutations, respectively). See Supplemental Table 1 for a list of all markers tested. Markers numbered P45–P122 were discovered in the course of various resequencing projects in the Hammer lab (Hammer et al. 2003; Wilder et al. 2004a, b), while Underhill’s group published markers in the range of M226–M450 in the past 5 yr (Cruciani et al. 2002; Semino et al. 2002, 2004; Cinnioglu et al. 2004; Rootsi et al. 2004, 2007; Shi et al. 2005; Kayser et al. 2006; Regueiro et al. 2006; Sengupta et al. 2006; Hudjashov et al. 2007). Other mutations (e.g., most P mutations from 123–297) were mined from public databases in the following manner. In a recent study aimed at characterizing patterns of common DNA variation in three populations (Hinds et al. 2005), 334 Y-linked SNPs were typed in 33 males (13 European-Americans, 11 African-Americans, and nine Asian-Americans). The set of typed markers included SNPs ascertained during that study, as well as previously reported SNPs, some of which had been mapped onto the Y chromosome tree (Y Chromosome Consortium 2002). Using information from mapped SNPs, we provisionally assigned Y chromosome haplogroups for these 33 samples. For example, according to the state they had for M9 (rs3900:C → G), it was possible to assign 18 males as belonging and 15 as not belonging to the KT branch of the Y chromosome tree. To confirm and better resolve the position of the newly reported SNPs in the tree, we performed further genotyping (either by direct resequencing or PCR-RFLP). During the process of mapping, several new SNPs were discovered and also mapped. Mutations that define major haplogroups are referred to as “defining” mutations, while those that mark lineages within a major haplogroup are referred to as “internal” mutations. We also tried to incorporate published markers other than P and M on the tree. The major challenge here was the absence of “positive control” samples (i.e., a DNA sample known to carry the derived state at the polymorphic site), especially for singletons and very-low-frequency mutations. When more than one marker mapped on the same branch of the tree, we tried to identify the order of mutational events by cross-typing positive control samples for each mutation. Large insertion/deletion or simple repeat mutations were not included in this study.
Most of the SNPs reported by Hinds et al. (2005) were discovered (or rediscovered) in the same set of samples. These SNPs have a uniform ascertainment scheme, which makes them useful for estimating the length of particular branches on the tree. However, the number of mutations may be under-represented on some lineages, especially on those branches that are present at low frequency in the ascertainment sample. Because haplogroups R-M269, E-P1, and I-P30 are found at relatively high frequency in the European and African populations used in the ascertainment process (Hinds et al. 2005), we expect the number of SNPs discovered on these lineages to be fairly representative of the relative length of these branches of the tree.
We used a novel method to estimate the relative ages of internal nodes of the tree that relies on a uniform probability distribution for the age of mutations in the ancestry of a lineage. If time is partitioned into k subintervals and mutational events n occur at a constant rate, then the number of events in each subinterval X1, . . . , Xk follows a multinomial distribution with parameters p1, . . . , pk, where pi is the length of the subinterval i relative to the whole interval. To estimate the age of nodes, we partitioned the time interval between the most recent common ancestor (MRCA) of all lineages in branches C through T (denoted MRCA-CT) and the present into two subintervals: one extending from the MRCA-CT to the internal node whose age is estimated, and a second extending from the internal node to the present. The number of mutations in each subinterval follows a binomial distribution with parameter pi equal to the relative length of the subinterval. The relative length of the second subinterval is multiplied by the assumed age of the MRCA-CT to obtain the age in years of the internal node.
To establish confidence intervals, we find the range of relative ages of the internal node such that the distribution of the number of mutations in each subinterval is not too extreme (i.e., neither too many mutations from the MRCA-CT to the internal node nor too many from the internal node to the present). We choose the endpoints of the intervals such that the probability of the observed or more extreme distributions of the mutations in the ancestry is equal to or smaller than 0.05. If the total number of mutations in the lineage (N) equals the observed number of mutations (n) and the observed number of mutations occurring between the node and the present is m, we choose pmax and pmin, the maximum and minimum relative lengths of the recent subinterval, respectively, according to:
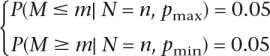 |
where M is the number of mutations in the more recent subinterval. The first equation says that given pmax, it would be very unlikely to observe a smaller number of mutations in the recent subinterval, whereas the second equation indicates that given pmin, it would be very unlikely to observe a larger number of mutations in the recent interval. To find the confidence interval for the age of the subintervals, we multiply the pmin and pmax values by the age of MRCA-CT:
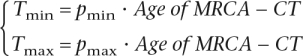 |
See Supplemental material for a worked example, which calculates the time to the most recent common ancestor (TMRCA) for the F clade.
Results and Discussion
Nomenclature systems
The Y Chromosome Consortium (2002) defined a set of rules to label the different lineages within the tree of binary haplogroups. Capital letters (from A to R) were used to identify 18 major clades. Lineages that were not defined on the basis of a derived character represented interior nodes of the tree. Because they were potentially paraphyletic, they were called “paragroups” (indicated by the symbol *). Two complementary nomenclature systems were proposed. The first system used selected aspects of set theory to define hierarchical subclades within each major haplogroup using an alphanumeric system (e.g., E1, E1a, E1a1, etc.). A shorter alternative mutation-based system named haplogroups by the terminal mutation that defined them (e.g., E-M81). Here we continue to follow the rules of these systems, making use of the flexibility inherent in them when new mutations are mapped on the tree.
Major revisions in Y chromosome tree topology
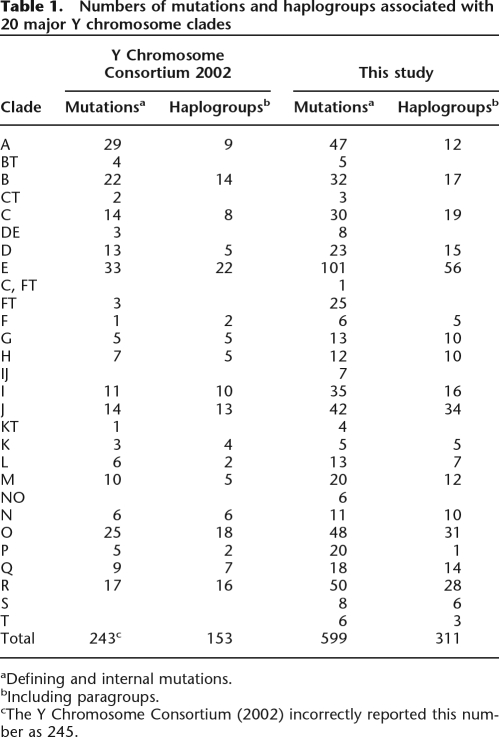
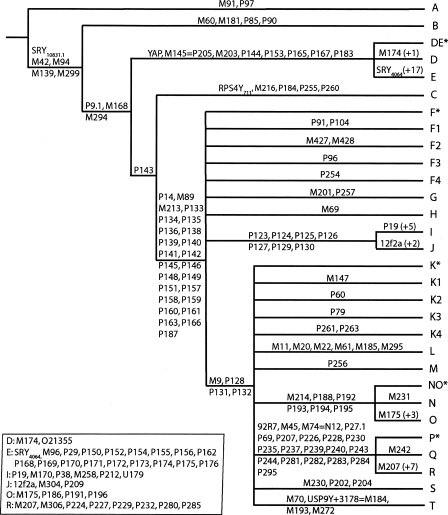
In 2002, 235 mutations mapped to the Y chromosome tree, eight of which were found to occur on different branches (i.e., represented recurrent mutations). Thus, the 243 mutational events gave rise to 153 NRY haplogroups (Table 1). Here, 586 mutations are incorporated into the overall tree and subtrees shown in the accompanying poster enclosed in this issue and Supplemental Figures 1–18, 13 of which are recurrent. These 599 mutational events resulted in 311 distinct haplogroups. Figure 1 shows the backbone of the new Y chromosome binary haplogroup tree and its 20 major clades. Notably, there are several key changes compared with the Y Chromosome Consortium (2002) tree. The deepest polychotomy in the YCC tree has now been resolved by virtue of a new binary marker, P143, which unites haplogroups C and FT (Fig. 1). This supercluster contains lineages that are not typically found in sub-Saharan Africa, suggesting that the ancestral C-FT chromosome may have been carried out of Africa very early in the modern human diaspora (Hammer et al. 1998; Ke et al. 2001; Underhill et al. 2001; Semino et al. 2002; Jobling and Tyler-Smith 2003; Underhill and Kivisild 2007). Seven mutations (P123, P124, P125 = M429, P126, P127, 129, and P130) join haplogroups I and J into the “IJ” clade. Six polymorphisms (M214, P188, P192, P193, P194, and P195) merge the N and O lineages into the “NO” clade. A new marker, P256, merges lineage M with two K haplogroups: K-M353/M387 (Kayser et al. 2006) and K-P117/P118 (Scheinfeldt et al. 2006) into the superclade M. Two K lineages, defined by the M230 and M70 mutations, were newly labeled as haplogroups S and T, respectively. The following sections discuss changes within each of the major 20 clades.
Table 1.
Numbers of mutations and haplogroups associated with 20 major Y chromosome clades
aDefining and internal mutations.
bIncluding paragroups.
cThe Y Chromosome Consortium (2002) incorrectly reported this number as 245.
Figure 1.
An abbreviated form of the Y chromosome parsimony tree shown in the accompanying poster enclosed in this issue. Mutation names are indicated on the branches. The subtrees corresponding to major clades A–T are collapsed in this figure and are shown in Supplemental Figures 1–13, 15–18, and in the accompanying poster enclosed in this issue, wherein the maximum parsimony tree of 311 Y chromosome haplogroups is shown. Haplogroup names are given at the tips of the tree, and major clades are labeled with large capital letters and shaded in color (the entire cladogram is designated “haplogroup Y”). Mutation names are given along the branches; the length of each branch is not proportional to the number of mutations or the age of the mutation. The order of phylogenetically equivalent markers shown on each branch is arbitrary.
Clade A
Clade A is defined by two mutations (M91 and P97) and contains 12 branches marked with 45 (internal) mutations (Supplemental Fig. 1). This compares with a single defining mutation and a total of 28 internal mutations associated with this clade on the Y Chromosome Consortium (2002) tree (Table 1). The newly discovered P97 marker, which defines clade A, is a T → G transversion in the DFFRY AluY50 region. This base substitution is more robust for designing assays than the length polymorphism at M91 (i.e., 9T → 8T), which was the only previously known marker that could be used to define haplogroup A chromosomes. Although 18 new mutations have been identified, there is no major rearrangement in the order of branching of lineages within clade A. This clade, one of the most basal on the entire Y chromosome tree, is almost completely restricted to the African continent (Hammer et al. 2001; Underhill et al. 2001). Haplogroup A chromosomes are most frequent in Khoisan, Ethiopian, and Sudanese populations (Hammer et al. 2001; Underhill et al. 2001; Semino et al. 2002; Jobling and Tyler-Smith 2003; Wood et al. 2005).
Caveats
Because of the lack of positive control samples, chromosomes carrying the M28 and M59 mutations have not been typed for P289. Thus, there is a possibility that the P289 mutation defines the A3 branch, representing a predecessor of the A-M28 and A-M59 lineages.
Clade B
Clade B is defined by four mutations (M60, M181, P85, and P90) and contains 17 branches with 28 internal markers (Supplemental Fig. 2). This represents an increase of two defining and eight internal mutations since 2002. Like haplogroup A, the B clade is almost entirely restricted to sub-Saharan Africa. B chromosomes are found at their highest frequency among Pygmies, with some lineages being virtually restricted to these ethnic groups (Hammer et al. 2001; Underhill et al. 2001; Cruciani et al. 2002; Semino et al. 2002; Jobling and Tyler-Smith 2003; Wood et al. 2005).
Major rearrangements
The P112 mutation defines a new branch called B2c or B-P112 (Supplemental Fig. 2).
Clade C
A total of 30 mutations mark lineages within haplogroup C, which is defined by five mutations: RPS4Y711, M216, P184, P255, and P260 (Supplemental Fig. 3). Haplogroup C now contains 19 branches with 29 internal markers. This represents an increase in 16 markers (three defining and 13 internal) associated with this haplogroup since 2002. Haplogroup C still has not been detected in sub-Saharan African populations, suggesting an Asian origin after anatomically modern humans migrated out of Africa. The ancestral paragroup of haplogroup C, as well as many downstream lineages, are commonly found among Asian, Australian, and Oceanic populations (Capelli et al. 2001; Hammer et al. 2001; Karafet et al. 2001; Ke et al. 2001; Underhill et al. 2001; Kivisild et al. 2003; Kayser et al. 2006; Scheinfeldt et al. 2006). Recently, a sublineage of haplogroup C (C3b or C-P39) was found to be restricted to Native American populations (Zegura et al. 2004), while branch C4 or C-M347 was observed only in Australia (Hudjashov et al. 2007).
Major rearrangements
While there are no major rearrangements within the C clade, three new subclades—designated C4, C5, and C6—have been defined. These subclades are geographically restricted to Australia, South and Central Asia, and New Guinea, respectively.
Caveats
P33 is a T → C transition within one of three paralogous regions on the Y chromosome mapping to Y chromosome positions 23,397,374 or 25,031,122 or 25,750,056 (Cox et al. 2007). Thus, chromosomes carrying the derived state at P33 also carry additional copies with the ancestral state.
Clade D
In 2002, a single mutation defined haplogroup D and 12 markers defined lineages nested within this clade. Now there are a total of 23 mutations associated with this clade, two defining (M174 and JST021355) (Underhill et al. 2001; Nonaka et al. 2007) and 21 that are internal to this clade (Supplemental Fig. 4). Haplogroup D chromosomes have not been found anywhere outside of Asia, the likely place of origin of this haplogroup. D lineages are most commonly found in Central Asia (Tibet) and in Japan, and are also present at low frequencies in Southeast Asia and among Andaman Islanders (Su et al. 2000; Karafet et al. 2001; Thangaraj et al. 2003; Hammer et al. 2006).
Major rearrangements
The newly discovered P99 polymorphism that defines the D3 branch occurs in Mainland Asia, specifically in Tibet, the Altai, and Mongolia.
Caveats
The N3 mutation (Deng et al. 2004) was not incorporated into the new tree because of uncertainty in its phylogenetic position.
Clade E
Haplogroup E is now defined by 18 mutations (SRY4064, M96, P29, P150, P152, P154, P155, P156, P162, P168, P169, P170, P171, P172, P173, P174, P175, and P176) (Supplemental Fig. 5). There are a total of 83 polymorphic sites that mark lineages within this clade, compared with a total of 30 internal mutations in 2002. This makes haplogroup E by far the most mutationally diverse of all major Y chromosome clades. These polymorphisms define 56 distinct haplogroups, which can be found at high frequencies in Africa, at moderate frequencies in the Middle East and southern Europe, and with occasional occurrence in Central and South Asia (Hammer et al. 1998; Underhill et al. 2001; Cruciani et al. 2002; Jobling and Tyler-Smith 2003; Semino et al. 2004; Sims et al. 2007).
Major rearrangements
In 2002, haplogroup E was characterized by three basal branches: E-M33 (E1), E-M75 (E2), and E-P2 (E3). The newly discovered polymorphism P147 requires major rearrangements within this clade. Subclades E-P147 (E1) and E-M75 (E2) are the most basal haplogroups, with E-P147 (E1) having two subclades: E-M33 (E1a) and E-P177 (E1b). The new polymorphism P177 joins haplogroup E-P2 (E1b1) and recently detected haplogroup E-P75 (E1b2). Haplogroup E-P1 (E1b1a) consists of 19 branches compared with six on the YCC tree.
The most basal paragroup lineage in this clade, E*, was found in a single Bantu-speaking male from South Africa. Haplogroups E-M191 (E1b1a7) and E-U175 (E1b1a8) are widely distributed in Africa. On the other hand, E-P268 (E1b1a9) was detected in a single Gambian, and chromosomes E-M58 (E1b1a1), E-M116.2 (E1b1a2), E-M149 (E1b1a3), E-M154 (E1b1a4), E-M155 (E1b1a5), and E-M10 (E1b1a6) are rarely observed in Africa (Underhill et al. 2001; Sims et al. 2007). The M215 polymorphism is a predecessor of the E-M35 mutation. Haplogroup E-M35 (E1b1b) contains a lineage undefined by a binary marker, as well as six derived sub-branches. Three additional haplogroups have also been added to the tree since 2002: E-M281 (E1b1b1d), E-V6 (E1b1b1e), and E-P72 (E1b1b1f).
Caveats
Two pairs of haplogroups, namely, E-V27 (E1b1b1a2a)/E-P65 (E1b1b1a2b) and E-M148 (E1b1b1a3a)/E-V19 (E1b1b1a3b), are shown as separate branches; however, the positions of these mutations have not been resolved because of a lack of a DNA sample containing the derived state at V27 and V19.
Clade F
A total of 25 mutations define the lineage leading to the FT clade (Fig. 1). Four additional subclades: F-P91/P104 (F1), F-M427/M428 (F2), F-P96 (F3), and F-P254 (F4) were found to have the derived state at P14 and the ancestral state at mutations that define the G-T lineages. F-P91/P104 and F-P254 chromosomes were found in Sri Lanka, F-M427 and M428 chromosomes were present in East Asia (Sengupta et al. 2006), and a single individual with the F-P96 chromosome was observed in the Netherlands. Paragroup F* was observed primarily on the Indian subcontinent at low or moderate frequency (Kivisild et al. 2003; Karafet et al. 2005; Sengupta et al. 2006; Zerjal et al. 2007).
Caveats
The Apt marker, reported as the F1 haplogroup by the Y Chromosome Consortium (2002), has the derived state at M69 and belongs to the H lineage (Supplemental Fig. 7).
Clade G
Clade G, defined by two mutations, contains 11 internal mutations that mark 10 lineages (Supplemental Fig. 6). This haplogroup is divided into two subclades: G1 is defined by M285 and M342, while G2 is defined by P287. The G clade is not widely distributed, being present mostly in the Middle East, the Mediterranean, and the Caucasus Mountains (Jobling and Tyler-Smith 2003; Behar et al. 2004; Cinnioglu et al. 2004; Nasidze et al. 2005; Regueiro et al. 2006; Sengupta et al. 2006). Of the two subclades, G2 is more diverse and has broader geographic distribution (Cinnioglu et al. 2004; Sengupta et al. 2006).
Major rearrangements
The newly discovered polymorphism P287 unites three subclades: G-P15 (G2a), G-M287 (G2b), and G-M377 (G2c) into one branch G2.
Clade H
Haplogroup H is defined by the derived state at a single marker, M69 (Supplemental Fig. 7). Within H, there is an unmarked paragroup (H*) and nine additional lineages marked by 11 mutations. Subclades within H include H-M52 (H1) and H-Apt (H2). The geographic distribution of haplogroup H is almost entirely restricted to the Indian subcontinent (Jobling and Tyler-Smith 2003; Karafet et al. 2005; Sengupta et al. 2006).
Major rearrangements
In 2002, the H lineage was defined on the basis of the derived state at M69 and M52. Newly discovered samples demonstrate that M52 lies downstream from M69. The Apt polymorphism has the derived state at M69 and defines the H-Apt (H2) branch, not the F1 branch as previously reported (Jobling and Tyler-Smith 2003).
Clade I
Clade I is now defined by six mutations, four more than in 2002 (Supplemental Fig. 8). A total of 29 polymorphisms define the topology of the clade compared with a total of nine internal mutations in the Y Chromosome Consortium (2002) report. There is an underived lineage (I*) and 15 lineages that are subdivided into two major subclades: I1, which is defined by the derived state at P30, P40, M253, M307, and M450; and I2, which is defined by P215. The newly discovered polymorphism at P215 joins lineages defined by the P37.2 and M223 mutations into a higher-order cluster. Clade I represents one of two major European Y chromosome haplogroups (Hammer et al. 2001; Jobling and Tyler-Smith 2003; Rootsi et al. 2004), with haplogroup R being the other major group (see below). Unlike R, clade I is widespread in Europe and is virtually absent elsewhere. Subclade I1 is found mostly in Northern Europe, while subclade I2 is the most frequent haplogroup in Eastern Europe and the Balkans (Rootsi et al. 2004).
Major rearrangements
We found that P38 does not define a separate subclade as previously reported; rather, it is one of several mutations that define the I haplogroup. Additionally, we found that the P40 mutation does not mark chromosomes that are a subset of the P30 mutation; together with M253, M307, M450, and P30, the P40 mutation defines I1.
Clade J
Three mutations (12f2a, M304, and P209) define clade J (Supplemental Fig. 9). A total of 39 mutations (compared with 13 in 2002) mark 34 lineages nested within this clade. Along with the A, E, O, and R haplogroups, J is one of five clades with >40 mutations. In addition to the paragroup (J*), there are two major subclades (J1 and J2), which are defined by mutations M267 and M172, respectively. Haplogroup J lineages are found at high frequencies in the Middle East, North Africa, Europe, Central Asia, Pakistan, and India (Hammer et al. 2000, 2001; Underhill et al. 2001; Semino et al. 2002; Behar et al. 2004; Cinnioglu et al. 2004; Sengupta et al. 2006), with haplogroup J-M172 being the most common J haplogroup in Europe, while haplogroup J-M267 predominates in the Middle East, North Africa, and Ethiopia (Semino et al. 2004).
Major rearrangements
The J2 branch is now split into two subclades. J2a, defined by J-M410, contains 17 haplogroups, while J2b, defined by M12, M102, M221, and M314, contains seven haplogroups.
Caveats
The M419 mutation was not tested on P81 and P279 chromosomes because of the absence of positive control DNAs. The topology of P58, M365, and M390 markers has not been completely resolved. The branches defined by mutations M365 and M390 might descend from P58+ chromosomes.
Clade K
As for haplogroups F and P (see below), lineages within haplogroup K are not united by a single mutation. Instead, haplogroup K is characterized by the derived state at four sites (M9, P128, P131, and P132) and the ancestral state at the mutations that define the L, M, NO, P, S, and T lineages (Fig. 1). In addition to paragroup (K*), there are five mutations that define four different K lineages (compared with three mutations and three lineages in 2002). Haplogroup K1 was found at low frequencies in India/Pakistan (Underhill et al. 2001). Lineages K2, K3, and K4 are found in Oceania, Indonesia, and/or Australia. K2 (Y Chromosome Consortium 2002) was renamed as a new haplogroup, T.
Clade L
Haplogroup L is defined by six mutations (M11, M20, M22, M61, M185, and M295). There is an unmarked paragroup (L*) and a total of seven lineages marked by seven mutations (Supplemental Fig. 10). The majority of L haplogroups are found on the Indian subcontinent. Haplogroup L chromosomes are also present in the Middle East, Central Asia, Northern Africa, and Europe along the Mediterranean coast (Underhill et al. 2001; Cruciani et al. 2002; Jobling and Tyler-Smith 2003; Behar et al. 2004; Cinnioglu et al. 2004; Karafet et al. 2005; Sengupta et al. 2006).
Clade M
A newly discovered mutation, P256, combined lineage M with two K haplogroups: K-M353/M387 (Kayser et al. 2006) and K-P117/P118 (Scheinfeldt et al. 2006) into the superclade M (Supplemental Fig. 11). The new haplogroup M contains 19 internal mutations. The lineage M1, previously known as haplogroup M (Y Chromosome Consortium 2002), is defined by seven mutations (M4, M5=P73, M106, M186, M189, M296, and P35). In addition to paragroup M1* there are seven lineages defined by seven mutations within the M1 branch. The M2 branch is marked by the presence of mutations M353 and M387. The M3 branch is defined by mutations P117 and P118. The geographic distribution of haplogroup M is restricted to near and remote Oceania and Eastern Indonesia. The majority of males from Papua New Guinea and Melanesia carry haplogroup M chromosomes (Su et al. 2000; Capelli et al. 2001; Hammer et al. 2001; Hurles et al. 2002; Karafet et al. 2005; Kayser et al. 2006; Scheinfeldt et al. 2006).
Major rearrangements
The recently discovered P87 polymorphism is a precursor of the P22 mutation. The M-P87* (M1b*) branch is almost entirely restricted to Melanesia (Scheinfeldt et al. 2006).
Clade N
Haplogroup N is defined by a newly discovered mutation, M231 (Cinnioglu et al. 2004). In addition to paragroup (N*), 10 mutations define nine N haplogroups (Supplemental Fig. 12). Haplogroup N is mainly found in Northern Eurasia and is absent or only marginally present in other regions of the globe (Karafet et al. 2001; Rootsi et al. 2007). The LLY22g polymorphism, which is characterized by a C → A mutation in one of two amplified units, has the derived state at M231.
Major rearrangements
The Y Chromosome Consortium (2002) reported two markers, M175 and M214, on the lineage leading to haplogroup O. The mutation M214 is now known to combine the N and O lineages into one superclade (NO) (Fig. 1) (Jobling and Tyler-Smith 2003).
Clade O
This highly diverse clade, which is defined by four mutations (M175, P186, P191, and P196), contains 44 internal mutations that mark 30 haplogroups, as well as an unmarked paragroup, O* (Supplemental Fig. 13). This compares with a total of two defining and 23 internal mutations in 2002. O is the major haplogroup in East Asia. Haplogroup O chromosomes are also found in Central Asia and Oceania at moderate or low frequencies (Su et al. 1999; Karafet et al. 2001; Underhill et al. 2001; Deng et al. 2004).
Major rearrangements
The MSY2.2 marker is now a predecessor of the M119 mutation. Two mutations (M122 and P198) now define the large O3 clade, which is subsequently divided into a major subclade (O3a) that is defined by five mutations (M324, P93, P197, P199, and P200) and an underived lineage (O3*), which is found at low frequencies in China, Taiwan, and Indonesia. The L1 retroposon insertion (LINE 1) polymorphism was removed from this tree because of contradictory results with N7 (Xue et al. 2006) and the newly discovered polymorphisms, P201 (=IMS-JST021354) and IMS-JST002611 (Nonaka et al. 2007). This likely reflects multiple deletions of this element and homoplasy on the binary haplogroup tree (Supplemental Fig. 14). P201 joins the O-M159 (O3a3a), O-M7 (O3a3b), and O-M134 (O3a3c) subclades.
Caveats
Six polymorphisms (N6, N7, N8, N9, N10, and N11) (Deng et al. 2004) were not incorporated into this version of the Y chromosome tree because of the absence of positive control DNAs. M164, M300, and M333 were not tested on P201(+) chromosomes because of the absence of the positive control DNAs. Thus, the branches defined by these mutations might descend from P201+ chromosomes. Chromosomes M300(+) and M333(+) were not typed for the IMS-JST002611 marker.
Clade P
Haplogroup P consists of two widely distributed Q and R lineages (Fig. 1).
Clade Q
Clade Q is defined by the M242 mutation (Supplemental Fig. 15). Within this clade, there are 13 haplogroups marked by 17 SNPs, as well as an unmarked paragroup (Q*). Haplogroup Q is distributed widely in North Eurasia and is found at high frequencies in some Siberian groups (Karafet et al. 2002) and at low frequencies in Europe, East Asia, and the Middle East. It is also the major lineage among the Native Americans, with Q-M3 (Q1a3a) being almost completely restricted to the Americas (Zegura et al. 2004). Undifferentiated paragroup Q* is observed at low frequencies in India and Pakistan.
Major rearrangements
The M242 mutation is positioned upstream of P36.2. MEH2 was not typed in a set of our samples; however, there is evidence that MEH2 is a subset of the Q-P36.2 haplogroup (Peter de Knijff, pers. comm.).
Clade R
Haplogroup R is identified by eight mutations: M207, M306, P224, P227, P229, P232, P280, and P285 (Supplemental Fig. 16). A total of 42 mutations identify 28 subclades nested in clade R. This compares with a total of one defining and 16 internal mutations for this clade in 2002. The majority of European Y chromosomes belong to this clade.
Major rearrangements
While the M124 mutation was shown as defining the P1 haplogroup in the Y Chromosome Consortium (2002) report, it is now known to have the derived state downstream from M207 in clade R (Jobling and Tyler-Smith 2003). The newly discovered P297 polymorphism combines M73 and M269 into one R1b1b subcluster. The M269 mutation joins the M37, M65, M153, SRY2627, M222, P66, U106, and U152 lineages into the R1b1b2 subclade. The recently discovered U152 polymorphism (Sims et al. 2007) joins the R-M126 and R-M160 lineages, as well as underived U152 chromosomes, into the R1b1b2h subclade.
Caveats
Additional changes in the topology of the R1a1 subclade may come about as a result of further testing of the M56, M157, M64.2, and PK5 polymorphisms on the P98(+) background. M18 and M335 were not tested on P297(+) chromosomes because of the absence of positive control DNAs. Thus, the branches defined by the M18 and M335 mutations might descend from P297(+) chromosomes. The P25 marker represents a paralogous sequence variant (PSV): three copies of the P25 sequence lie within palindromic repeats, with the mutation at P25 on one copy (Adams et al. 2006). P25 is also known to undergo reversion by gene conversion. It is recommended that this marker be used in conjunction with P297.
Clade S
Clade S, previously known as haplogroup K-M230, is identified by three mutations (M230, P202, and P204) (Supplemental Fig. 17). A total of eight mutations distinguish five haplogroups and paragroup S*. Haplogroup S chromosomes are predominantly found in Oceania and Indonesia (Kayser et al. 2006; Scheinfeldt et al. 2006).
Clade T
Clade T, formerly haplogroup K2 (K-M70) (Y Chromosome Consortium 2002), is defined by four markers (M70, M193, M272, and USP9Y + 3178 = M184) (Supplemental Fig. 18). Six mutations identify two subclades and one paragroup nested in clade T. Clade T was observed at low frequencies in the Middle East, Africa, and Europe (Underhill et al. 2001; King et al. 2007).
Caveats
M320 chromosomes were not typed for the P77 mutation because of the absence of a positive DNA control for M320. Therefore, it is possible that T-M320 (T1) might be a sub-branch of T-P77 (T2).
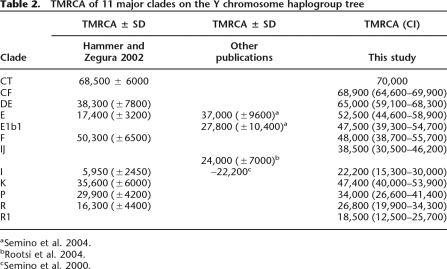
Age estimates for 11 major clades of the Y chromosome tree
The time to the most recent common ancestral Y chromosome and the estimated ages of 11 major clades are presented in Table 2. To provide estimates of the age of the nodes, we chose to fix the time to the most recent common ancestor of CT (defined by P9.1, M168, and M294) at 70 thousand years ago (Kya), which is consistent with previous estimates from genetic and archaeological data (Lahr and Foley 1998; Hammer and Zegura 2002; Macaulay et al. 2005), and is the chronological approximation given in Jobling et al. (2004) (p250) for the first major human out-of-Africa dispersals. We estimated the times for intermediate nodes by using a linear interpolation. The age estimates in years should be viewed with caution because we do not know if the calibration date chosen above is accurate.
Table 2.
TMRCA of 11 major clades on the Y chromosome haplogroup tree
A single lineage was used for the estimation of the ages of the MRCAs of DE, E, and E1b1. Also, the ages of the MRCAs of IJ, I, K, P, R, and R1 were estimated using a single lineage. The ages of the MRCAs of lineages in CF and of lineage F were estimated using the ancestry of R-M269 and the ancestry of I-P30. Each estimation and confidence interval was obtained independently for R-M269 and I-P30. For the cases where the age of an internal node was estimated using different lineages, we report the average of the estimates and the union of the confidence intervals. We show the distribution of the number of counts in Supplemental Figure 19 and details on the SNPs used in Supplemental Table 2. We note that although the number of mutations in each lineage is not the same, the difference is not significant (the largest χ2 for homogeneity is 1.12; P = 0.29), which is consistent with the assumption of homogeneous ascertainment of mutations in these lineages.
Dating estimates for the majority of lineages show earlier coalescence times than those in previous studies (Table 2). Many of the previous estimates were based on a much smaller number of SNPs and a specific demographic model (Hammer and Zegura 2002). We present estimates that are free from assumptions about population history and rely only on the assumption of uniform ascertainment of SNPs. Additional previous estimates were based on microsatellite variation within clades (Semino et al. 2000, 2004; Rootsi et al. 2004). Limited knowledge of the appropriate mutation rate and model, as well as demography, makes it difficult to assess the accuracy of these estimates. Importantly, our model provides an independent method to estimate relative TMRCAs that does not rely on a specific demographic model (Tang et al. 2002).
Future directions
With decreasing costs for genome sequencing and SNP typing (Service 2006), we anticipate the discovery of many more Y chromosome markers in the near future. While we have attempted to retain previously utilized nomenclature as much as possible, some of the branch names may change in future iterations of the tree (Calafell et al. 2002). Indeed, there are known mutations that are not yet incorporated into this version of the tree. For example, a recent survey of sequence variation within an 80-kb region on 47 Y chromosomes identified 94 SNPs (Repping et al. 2006), some of which have been typed in additional population samples (Sun et al. 1999; Shen et al. 2000; Underhill et al. 2000, 2001; Cinnioglu et al. 2004). It would be extremely useful to incorporate this set of SNPs in the next version of the YCC tree. The Y chromosome research community would also benefit from a set of publicly accessible cell line DNAs that could be used as positive controls for all the known markers. While the original set of 74 Y Chromosome Consortium (2002) cell lines is no longer available for this purpose, we suggest that typing of the current markers on the widely available CEPH-HGDP panel (Cann et al. 2002) would be a highly desirable goal for a future study.
Acknowledgments
We thank Chris Tyler-Smith for helpful comments, Peter de Knijff for unpublished information, and A. Richard Diebold and the Salus Mundi Foundation for funding.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7172008.
References
- Adams S.M., King T.E., Bosch E., Jobling M.A., King T.E., Bosch E., Jobling M.A., Bosch E., Jobling M.A., Jobling M.A. The case of the unreliable SNP: Recurrent back-mutation of Y-chromosomal marker P25 through gene conversion. Forensic Sci. Int. 2006;159:14–20. doi: 10.1016/j.forsciint.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Behar D.M., Garrigan D., Kaplan M.E., Mobasher Z., Rosengarten D., Karafet T.M., Quintana-Murci L., Ostrer H., Skorecki K., Hammer M.F., Garrigan D., Kaplan M.E., Mobasher Z., Rosengarten D., Karafet T.M., Quintana-Murci L., Ostrer H., Skorecki K., Hammer M.F., Kaplan M.E., Mobasher Z., Rosengarten D., Karafet T.M., Quintana-Murci L., Ostrer H., Skorecki K., Hammer M.F., Mobasher Z., Rosengarten D., Karafet T.M., Quintana-Murci L., Ostrer H., Skorecki K., Hammer M.F., Rosengarten D., Karafet T.M., Quintana-Murci L., Ostrer H., Skorecki K., Hammer M.F., Karafet T.M., Quintana-Murci L., Ostrer H., Skorecki K., Hammer M.F., Quintana-Murci L., Ostrer H., Skorecki K., Hammer M.F., Ostrer H., Skorecki K., Hammer M.F., Skorecki K., Hammer M.F., Hammer M.F. Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations. Hum. Genet. 2004;114:354–365. doi: 10.1007/s00439-003-1073-7. [DOI] [PubMed] [Google Scholar]
- Calafell F., Comas D., Bertranpetit J., Comas D., Bertranpetit J., Bertranpetit J. Why names? Genome Res. 2002;12:219–221. doi: 10.1101/gr.226502. [DOI] [PubMed] [Google Scholar]
- Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Bonne-Tamir B., Cambon-Thomsen A., Cambon-Thomsen A., et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Capelli C., Wilson J.F., Richards M., Stumpf M.P., Gratrix F., Oppenheimer S., Underhill P., Pascali V.L., Ko T.M., Goldstein D.B., Wilson J.F., Richards M., Stumpf M.P., Gratrix F., Oppenheimer S., Underhill P., Pascali V.L., Ko T.M., Goldstein D.B., Richards M., Stumpf M.P., Gratrix F., Oppenheimer S., Underhill P., Pascali V.L., Ko T.M., Goldstein D.B., Stumpf M.P., Gratrix F., Oppenheimer S., Underhill P., Pascali V.L., Ko T.M., Goldstein D.B., Gratrix F., Oppenheimer S., Underhill P., Pascali V.L., Ko T.M., Goldstein D.B., Oppenheimer S., Underhill P., Pascali V.L., Ko T.M., Goldstein D.B., Underhill P., Pascali V.L., Ko T.M., Goldstein D.B., Pascali V.L., Ko T.M., Goldstein D.B., Ko T.M., Goldstein D.B., Goldstein D.B. A predominantly indigenous paternal heritage for the Austronesian-speaking peoples of insular Southeast Asia and Oceania. Am. J. Hum. Genet. 2001;68:432–443. doi: 10.1086/318205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinnioglu C., King R., Kivisild T., Kalfoglu E., Atasoy S., Cavalleri G.L., Lillie A.S., Roseman C.C., Lin A.A., Prince K., King R., Kivisild T., Kalfoglu E., Atasoy S., Cavalleri G.L., Lillie A.S., Roseman C.C., Lin A.A., Prince K., Kivisild T., Kalfoglu E., Atasoy S., Cavalleri G.L., Lillie A.S., Roseman C.C., Lin A.A., Prince K., Kalfoglu E., Atasoy S., Cavalleri G.L., Lillie A.S., Roseman C.C., Lin A.A., Prince K., Atasoy S., Cavalleri G.L., Lillie A.S., Roseman C.C., Lin A.A., Prince K., Cavalleri G.L., Lillie A.S., Roseman C.C., Lin A.A., Prince K., Lillie A.S., Roseman C.C., Lin A.A., Prince K., Roseman C.C., Lin A.A., Prince K., Lin A.A., Prince K., Prince K., et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum. Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- Cox M.P., Redd A., Karafet T.M., Ponder C.A.,, Sudoyo H., Hammer M.F., Redd A., Karafet T.M., Ponder C.A.,, Sudoyo H., Hammer M.F., Karafet T.M., Ponder C.A.,, Sudoyo H., Hammer M.F., Ponder C.A.,, Sudoyo H., Hammer M.F., Sudoyo H., Hammer M.F., Hammer M.F. A Polynesian motif on the Y chromosome: Population structure in remote Oceania. Hum. Biol. 2007;79:525–535. doi: 10.1353/hub.2008.0004. [DOI] [PubMed] [Google Scholar]
- Cruciani F., Santolamazza P., Shen P., Macaulay V., Moral P., Olckers A., Modiano D., Holmes S., Destro-Bisol G., Coia V., Santolamazza P., Shen P., Macaulay V., Moral P., Olckers A., Modiano D., Holmes S., Destro-Bisol G., Coia V., Shen P., Macaulay V., Moral P., Olckers A., Modiano D., Holmes S., Destro-Bisol G., Coia V., Macaulay V., Moral P., Olckers A., Modiano D., Holmes S., Destro-Bisol G., Coia V., Moral P., Olckers A., Modiano D., Holmes S., Destro-Bisol G., Coia V., Olckers A., Modiano D., Holmes S., Destro-Bisol G., Coia V., Modiano D., Holmes S., Destro-Bisol G., Coia V., Holmes S., Destro-Bisol G., Coia V., Destro-Bisol G., Coia V., Coia V., et al. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am. J. Hum. Genet. 2002;70:1197–1214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Shi B., He X., Zhang Z., Xu J., Li B., Yang J., Ling L., Dai C., Qiang B., Shi B., He X., Zhang Z., Xu J., Li B., Yang J., Ling L., Dai C., Qiang B., He X., Zhang Z., Xu J., Li B., Yang J., Ling L., Dai C., Qiang B., Zhang Z., Xu J., Li B., Yang J., Ling L., Dai C., Qiang B., Xu J., Li B., Yang J., Ling L., Dai C., Qiang B., Li B., Yang J., Ling L., Dai C., Qiang B., Yang J., Ling L., Dai C., Qiang B., Ling L., Dai C., Qiang B., Dai C., Qiang B., Qiang B., et al. Evolution and migration history of the Chinese population inferred from Chinese Y-chromosome evidence. J. Hum. Genet. 2004;49:339–348. doi: 10.1007/s10038-004-0154-3. [DOI] [PubMed] [Google Scholar]
- Hammer M.F., Zegura S.L., Zegura S.L. The human Y chromosome haplogroup tree: Nomenclature and phylogeography of its major divisions. Annu. Rev. Anthropol. 2002;31:303–321. [Google Scholar]
- Hammer M.F., Karafet T.M., Rasanayagam A., Wood E.T., Altheide T.K., Jenkins T., Griffiths R.C., Templeton A.R., Zegura S.L., Karafet T.M., Rasanayagam A., Wood E.T., Altheide T.K., Jenkins T., Griffiths R.C., Templeton A.R., Zegura S.L., Rasanayagam A., Wood E.T., Altheide T.K., Jenkins T., Griffiths R.C., Templeton A.R., Zegura S.L., Wood E.T., Altheide T.K., Jenkins T., Griffiths R.C., Templeton A.R., Zegura S.L., Altheide T.K., Jenkins T., Griffiths R.C., Templeton A.R., Zegura S.L., Jenkins T., Griffiths R.C., Templeton A.R., Zegura S.L., Griffiths R.C., Templeton A.R., Zegura S.L., Templeton A.R., Zegura S.L., Zegura S.L. Out of Africa and back again: Nested cladistic analysis of human Y chromosome variation. Mol. Biol. Evol. 1998;15:427–441. doi: 10.1093/oxfordjournals.molbev.a025939. [DOI] [PubMed] [Google Scholar]
- Hammer M.F., Redd A.J., Wood E.T., Bonner M.R., Jarjanazi H., Karafet T., Santachiara-Benerecetti S., Oppenheim A., Jobling M.A., Jenkins T., Redd A.J., Wood E.T., Bonner M.R., Jarjanazi H., Karafet T., Santachiara-Benerecetti S., Oppenheim A., Jobling M.A., Jenkins T., Wood E.T., Bonner M.R., Jarjanazi H., Karafet T., Santachiara-Benerecetti S., Oppenheim A., Jobling M.A., Jenkins T., Bonner M.R., Jarjanazi H., Karafet T., Santachiara-Benerecetti S., Oppenheim A., Jobling M.A., Jenkins T., Jarjanazi H., Karafet T., Santachiara-Benerecetti S., Oppenheim A., Jobling M.A., Jenkins T., Karafet T., Santachiara-Benerecetti S., Oppenheim A., Jobling M.A., Jenkins T., Santachiara-Benerecetti S., Oppenheim A., Jobling M.A., Jenkins T., Oppenheim A., Jobling M.A., Jenkins T., Jobling M.A., Jenkins T., Jenkins T., et al. Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc. Natl. Acad. Sci. 2000;97:6769–6774. doi: 10.1073/pnas.100115997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M.F., Karafet T.M., Redd A.J., Jarjanazi H., Santachiara-Benerecetti S., Soodyall H., Zegura S.L., Karafet T.M., Redd A.J., Jarjanazi H., Santachiara-Benerecetti S., Soodyall H., Zegura S.L., Redd A.J., Jarjanazi H., Santachiara-Benerecetti S., Soodyall H., Zegura S.L., Jarjanazi H., Santachiara-Benerecetti S., Soodyall H., Zegura S.L., Santachiara-Benerecetti S., Soodyall H., Zegura S.L., Soodyall H., Zegura S.L., Zegura S.L. Hierarchical patterns of global human Y-chromosome diversity. Mol. Biol. Evol. 2001;18:1189–1203. doi: 10.1093/oxfordjournals.molbev.a003906. [DOI] [PubMed] [Google Scholar]
- Hammer M.F., Blackmer F., Garrigan D., Nachman M.W., Wilder J.A., Blackmer F., Garrigan D., Nachman M.W., Wilder J.A., Garrigan D., Nachman M.W., Wilder J.A., Nachman M.W., Wilder J.A., Wilder J.A. Human population structure and its effects on sampling Y chromosome sequence variation. Genetics. 2003;164:1495–1509. doi: 10.1093/genetics/164.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M.F., Karafet T.M., Park H., Omoto K., Harihara S., Stoneking M., Horai S., Karafet T.M., Park H., Omoto K., Harihara S., Stoneking M., Horai S., Park H., Omoto K., Harihara S., Stoneking M., Horai S., Omoto K., Harihara S., Stoneking M., Horai S., Harihara S., Stoneking M., Horai S., Stoneking M., Horai S., Horai S. Dual origins of the Japanese: Common ground for hunter-gatherer and farmer Y chromosomes. J. Hum. Genet. 2006;51:47–58. doi: 10.1007/s10038-005-0322-0. [DOI] [PubMed] [Google Scholar]
- Hinds D.A., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Ballinger D.G., Frazer K.A., Cox D.R., Frazer K.A., Cox D.R., Cox D.R. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Hudjashov G., Kivisild T., Underhill P.A., Endicott P., Sanchez J.J., Lin A.A., Shen P., Oefner P., Renfrew C., Villems R., Kivisild T., Underhill P.A., Endicott P., Sanchez J.J., Lin A.A., Shen P., Oefner P., Renfrew C., Villems R., Underhill P.A., Endicott P., Sanchez J.J., Lin A.A., Shen P., Oefner P., Renfrew C., Villems R., Endicott P., Sanchez J.J., Lin A.A., Shen P., Oefner P., Renfrew C., Villems R., Sanchez J.J., Lin A.A., Shen P., Oefner P., Renfrew C., Villems R., Lin A.A., Shen P., Oefner P., Renfrew C., Villems R., Shen P., Oefner P., Renfrew C., Villems R., Oefner P., Renfrew C., Villems R., Renfrew C., Villems R., Villems R., et al. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc. Natl. Acad. Sci. 2007;104:8726–8730. doi: 10.1073/pnas.0702928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles M.E., Nicholson J., Bosch E., Renfrew C., Sykes B.C., Jobling M.A., Nicholson J., Bosch E., Renfrew C., Sykes B.C., Jobling M.A., Bosch E., Renfrew C., Sykes B.C., Jobling M.A., Renfrew C., Sykes B.C., Jobling M.A., Sykes B.C., Jobling M.A., Jobling M.A. Y chromosomal evidence for the origins of Oceanic-speaking peoples. Genetics. 2002;160:289–303. doi: 10.1093/genetics/160.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling M.A., Tyler-Smith C., Tyler-Smith C. The human Y chromosome: An evolutionary marker comes of age. Nat. Rev. Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Jobling M.A., Hurles M.E., Tyler-Smith C., Hurles M.E., Tyler-Smith C., Tyler-Smith C. Human evolutionary genetics: Origins, peoples & disease. Garland Publishing; New York: 2004. [Google Scholar]
- Karafet T., Xu L., Du R., Wang W., Feng S., Wells R.S., Redd A.J., Zegura S.L., Hammer M.F., Xu L., Du R., Wang W., Feng S., Wells R.S., Redd A.J., Zegura S.L., Hammer M.F., Du R., Wang W., Feng S., Wells R.S., Redd A.J., Zegura S.L., Hammer M.F., Wang W., Feng S., Wells R.S., Redd A.J., Zegura S.L., Hammer M.F., Feng S., Wells R.S., Redd A.J., Zegura S.L., Hammer M.F., Wells R.S., Redd A.J., Zegura S.L., Hammer M.F., Redd A.J., Zegura S.L., Hammer M.F., Zegura S.L., Hammer M.F., Hammer M.F. Paternal population history of East Asia: Sources, patterns, and microevolutionary processes. Am. J. Hum. Genet. 2001;69:615–628. doi: 10.1086/323299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet T.M., Osipova L.P., Gubina M.A., Posukh O.L., Zegura S.L., Hammer M.F., Osipova L.P., Gubina M.A., Posukh O.L., Zegura S.L., Hammer M.F., Gubina M.A., Posukh O.L., Zegura S.L., Hammer M.F., Posukh O.L., Zegura S.L., Hammer M.F., Zegura S.L., Hammer M.F., Hammer M.F. High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum. Biol. 2002;74:761–789. doi: 10.1353/hub.2003.0006. [DOI] [PubMed] [Google Scholar]
- Karafet T.M., Lansing J.S., Redd A.J., Reznikova S., Watkins J.C., Surata S.P., Arthawiguna W.A., Mayer L., Bamshad M., Jorde L.B., Lansing J.S., Redd A.J., Reznikova S., Watkins J.C., Surata S.P., Arthawiguna W.A., Mayer L., Bamshad M., Jorde L.B., Redd A.J., Reznikova S., Watkins J.C., Surata S.P., Arthawiguna W.A., Mayer L., Bamshad M., Jorde L.B., Reznikova S., Watkins J.C., Surata S.P., Arthawiguna W.A., Mayer L., Bamshad M., Jorde L.B., Watkins J.C., Surata S.P., Arthawiguna W.A., Mayer L., Bamshad M., Jorde L.B., Surata S.P., Arthawiguna W.A., Mayer L., Bamshad M., Jorde L.B., Arthawiguna W.A., Mayer L., Bamshad M., Jorde L.B., Mayer L., Bamshad M., Jorde L.B., Bamshad M., Jorde L.B., Jorde L.B., et al. Balinese Y-chromosome perspective on the peopling of Indonesia: Genetic contributions from pre-Neolithic hunter-gatherers, Austronesian farmers, and Indian traders. Hum. Biol. 2005;77:93–114. doi: 10.1353/hub.2005.0030. [DOI] [PubMed] [Google Scholar]
- Kayser M., Brauer S., Cordaux R., Casto A., Lao O., Zhivotovsky L.A., Moyse-Faurie C., Rutledge R.B., Schiefenhoevel W., Gil D., Brauer S., Cordaux R., Casto A., Lao O., Zhivotovsky L.A., Moyse-Faurie C., Rutledge R.B., Schiefenhoevel W., Gil D., Cordaux R., Casto A., Lao O., Zhivotovsky L.A., Moyse-Faurie C., Rutledge R.B., Schiefenhoevel W., Gil D., Casto A., Lao O., Zhivotovsky L.A., Moyse-Faurie C., Rutledge R.B., Schiefenhoevel W., Gil D., Lao O., Zhivotovsky L.A., Moyse-Faurie C., Rutledge R.B., Schiefenhoevel W., Gil D., Zhivotovsky L.A., Moyse-Faurie C., Rutledge R.B., Schiefenhoevel W., Gil D., Moyse-Faurie C., Rutledge R.B., Schiefenhoevel W., Gil D., Rutledge R.B., Schiefenhoevel W., Gil D., Schiefenhoevel W., Gil D., Gil D., et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol. Biol. Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- Ke Y., Su B., Song X., Lu D., Chen L., Li H., Qi C., Marzuki S., Deka R., Underhill P., Su B., Song X., Lu D., Chen L., Li H., Qi C., Marzuki S., Deka R., Underhill P., Song X., Lu D., Chen L., Li H., Qi C., Marzuki S., Deka R., Underhill P., Lu D., Chen L., Li H., Qi C., Marzuki S., Deka R., Underhill P., Chen L., Li H., Qi C., Marzuki S., Deka R., Underhill P., Li H., Qi C., Marzuki S., Deka R., Underhill P., Qi C., Marzuki S., Deka R., Underhill P., Marzuki S., Deka R., Underhill P., Deka R., Underhill P., Underhill P., et al. African origin of modern humans in East Asia: A tale of 12,000 Y chromosomes. Science. 2001;292:1151–1153. doi: 10.1126/science.1060011. [DOI] [PubMed] [Google Scholar]
- King T.E., Bowden G.R., Belaresque P.L., Adams S.M., Shanks M.E., Jobling M.A., Bowden G.R., Belaresque P.L., Adams S.M., Shanks M.E., Jobling M.A., Belaresque P.L., Adams S.M., Shanks M.E., Jobling M.A., Adams S.M., Shanks M.E., Jobling M.A., Shanks M.E., Jobling M.A., Jobling M.A. Thomas Jefferson’s Y chromosome belongs to a rare European lineage. Am. J. Phys. Anthropol. 2007;132:583–589. doi: 10.1002/ajpa.20557. [DOI] [PubMed] [Google Scholar]
- Kivisild T., Rootsi S., Metspalu M., Mastana S., Kaldma K., Parik J., Metspalu E., Adojaan M., Tolk H.V., Stepanov V., Rootsi S., Metspalu M., Mastana S., Kaldma K., Parik J., Metspalu E., Adojaan M., Tolk H.V., Stepanov V., Metspalu M., Mastana S., Kaldma K., Parik J., Metspalu E., Adojaan M., Tolk H.V., Stepanov V., Mastana S., Kaldma K., Parik J., Metspalu E., Adojaan M., Tolk H.V., Stepanov V., Kaldma K., Parik J., Metspalu E., Adojaan M., Tolk H.V., Stepanov V., Parik J., Metspalu E., Adojaan M., Tolk H.V., Stepanov V., Metspalu E., Adojaan M., Tolk H.V., Stepanov V., Adojaan M., Tolk H.V., Stepanov V., Tolk H.V., Stepanov V., Stepanov V., et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am. J. Hum. Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr M.M., Foley R.A., Foley R.A. Towards a theory of modern human origins: Geography, demography, and diversity in recent human evolution. Am. J. Phys Anthropol. 1998;41:137–176. doi: 10.1002/(sici)1096-8644(1998)107:27+<137::aid-ajpa6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Macaulay V., Hill C., Achilli A., Rengo C., Clarke D., Meehan W., Blackburn J., Semino O., Scozzari R., Cruciani F., Hill C., Achilli A., Rengo C., Clarke D., Meehan W., Blackburn J., Semino O., Scozzari R., Cruciani F., Achilli A., Rengo C., Clarke D., Meehan W., Blackburn J., Semino O., Scozzari R., Cruciani F., Rengo C., Clarke D., Meehan W., Blackburn J., Semino O., Scozzari R., Cruciani F., Clarke D., Meehan W., Blackburn J., Semino O., Scozzari R., Cruciani F., Meehan W., Blackburn J., Semino O., Scozzari R., Cruciani F., Blackburn J., Semino O., Scozzari R., Cruciani F., Semino O., Scozzari R., Cruciani F., Scozzari R., Cruciani F., Cruciani F., et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- Nasidze I., Quinque D., Ozturk M., Bendukidze N., Stoneking M., Quinque D., Ozturk M., Bendukidze N., Stoneking M., Ozturk M., Bendukidze N., Stoneking M., Bendukidze N., Stoneking M., Stoneking M. MtDNA and Y-chromosome variation in Kurdish groups. Ann. Hum. Genet. 2005;69:401–412. doi: 10.1046/j.1529-8817.2005.00174.x. [DOI] [PubMed] [Google Scholar]
- Nonaka I., Minaguchi K., Takezaki N., Minaguchi K., Takezaki N., Takezaki N. Y-chromosomal binary haplogroups in the Japanese population and their relationship to 16 Y-STR polymorphisms. Ann. Hum. Genet. 2007;71:480–495. doi: 10.1111/j.1469-1809.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- Regueiro M., Cadenas A.M., Gayden T., Underhill P.A., Herrera R.J., Cadenas A.M., Gayden T., Underhill P.A., Herrera R.J., Gayden T., Underhill P.A., Herrera R.J., Underhill P.A., Herrera R.J., Herrera R.J. Iran: Tricontinental nexus for Y-chromosome driven migration. Hum. Hered. 2006;61:132–143. doi: 10.1159/000093774. [DOI] [PubMed] [Google Scholar]
- Repping S., van Daalen S.K., Brown L.G., Korver C.M., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C., van Daalen S.K., Brown L.G., Korver C.M., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C., Brown L.G., Korver C.M., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C., Korver C.M., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C., Pyntikova T., van der Veen F., Skaletsky H., Page D.C., van der Veen F., Skaletsky H., Page D.C., Skaletsky H., Page D.C., Page D.C., et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- Rootsi S., Magri C., Kivisild T., Benuzzi G., Help H., Bermisheva M., Kutuev I., Barac L., Pericic M., Balanovsky O., Magri C., Kivisild T., Benuzzi G., Help H., Bermisheva M., Kutuev I., Barac L., Pericic M., Balanovsky O., Kivisild T., Benuzzi G., Help H., Bermisheva M., Kutuev I., Barac L., Pericic M., Balanovsky O., Benuzzi G., Help H., Bermisheva M., Kutuev I., Barac L., Pericic M., Balanovsky O., Help H., Bermisheva M., Kutuev I., Barac L., Pericic M., Balanovsky O., Bermisheva M., Kutuev I., Barac L., Pericic M., Balanovsky O., Kutuev I., Barac L., Pericic M., Balanovsky O., Barac L., Pericic M., Balanovsky O., Pericic M., Balanovsky O., Balanovsky O., et al. Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in Europe. Am. J. Hum. Genet. 2004;75:128–137. doi: 10.1086/422196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rootsi S., Zhivotovsky L.A., Baldovic M., Kayser M., Kutuev I.A., Khusainova R., Bermisheva M.A., Gubina M., Fedorova S.A., Ilumae A.M., Zhivotovsky L.A., Baldovic M., Kayser M., Kutuev I.A., Khusainova R., Bermisheva M.A., Gubina M., Fedorova S.A., Ilumae A.M., Baldovic M., Kayser M., Kutuev I.A., Khusainova R., Bermisheva M.A., Gubina M., Fedorova S.A., Ilumae A.M., Kayser M., Kutuev I.A., Khusainova R., Bermisheva M.A., Gubina M., Fedorova S.A., Ilumae A.M., Kutuev I.A., Khusainova R., Bermisheva M.A., Gubina M., Fedorova S.A., Ilumae A.M., Khusainova R., Bermisheva M.A., Gubina M., Fedorova S.A., Ilumae A.M., Bermisheva M.A., Gubina M., Fedorova S.A., Ilumae A.M., Gubina M., Fedorova S.A., Ilumae A.M., Fedorova S.A., Ilumae A.M., Ilumae A.M., et al. A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur. J. Hum. Genet. 2007;15:204–211. doi: 10.1038/sj.ejhg.5201748. [DOI] [PubMed] [Google Scholar]
- Scheinfeldt L., Friedlaender F., Friedlaender J., Latham K., Koki G., Karafet T., Hammer M., Lorenz J., Friedlaender F., Friedlaender J., Latham K., Koki G., Karafet T., Hammer M., Lorenz J., Friedlaender J., Latham K., Koki G., Karafet T., Hammer M., Lorenz J., Latham K., Koki G., Karafet T., Hammer M., Lorenz J., Koki G., Karafet T., Hammer M., Lorenz J., Karafet T., Hammer M., Lorenz J., Hammer M., Lorenz J., Lorenz J. Unexpected NRY chromosome variation in Northern Island Melanesia. Mol. Biol. Evol. 2006;23:1628–1641. doi: 10.1093/molbev/msl028. [DOI] [PubMed] [Google Scholar]
- Semino O., Passarino G., Oefner P.J., Lin A.A., Arbuzova S., Beckman L.E., De Benedictis G., Francalacci P., Kouvatsi A., Limborska S., Passarino G., Oefner P.J., Lin A.A., Arbuzova S., Beckman L.E., De Benedictis G., Francalacci P., Kouvatsi A., Limborska S., Oefner P.J., Lin A.A., Arbuzova S., Beckman L.E., De Benedictis G., Francalacci P., Kouvatsi A., Limborska S., Lin A.A., Arbuzova S., Beckman L.E., De Benedictis G., Francalacci P., Kouvatsi A., Limborska S., Arbuzova S., Beckman L.E., De Benedictis G., Francalacci P., Kouvatsi A., Limborska S., Beckman L.E., De Benedictis G., Francalacci P., Kouvatsi A., Limborska S., De Benedictis G., Francalacci P., Kouvatsi A., Limborska S., Francalacci P., Kouvatsi A., Limborska S., Kouvatsi A., Limborska S., Limborska S., et al. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: A Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- Semino O., Santachiara-Benerecetti A.S., Falaschi F., Cavalli-Sforza L.L., Underhill P.A., Santachiara-Benerecetti A.S., Falaschi F., Cavalli-Sforza L.L., Underhill P.A., Falaschi F., Cavalli-Sforza L.L., Underhill P.A., Cavalli-Sforza L.L., Underhill P.A., Underhill P.A. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am. J. Hum. Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O., Magri C., Benuzzi G., Lin A.A., Al-Zahery N., Battaglia V., Maccioni L., Triantaphyllidis C., Shen P., Oefner P.J., Magri C., Benuzzi G., Lin A.A., Al-Zahery N., Battaglia V., Maccioni L., Triantaphyllidis C., Shen P., Oefner P.J., Benuzzi G., Lin A.A., Al-Zahery N., Battaglia V., Maccioni L., Triantaphyllidis C., Shen P., Oefner P.J., Lin A.A., Al-Zahery N., Battaglia V., Maccioni L., Triantaphyllidis C., Shen P., Oefner P.J., Al-Zahery N., Battaglia V., Maccioni L., Triantaphyllidis C., Shen P., Oefner P.J., Battaglia V., Maccioni L., Triantaphyllidis C., Shen P., Oefner P.J., Maccioni L., Triantaphyllidis C., Shen P., Oefner P.J., Triantaphyllidis C., Shen P., Oefner P.J., Shen P., Oefner P.J., Oefner P.J., et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: Inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 2004;74:1023–1034. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S., Zhivotovsky L.A., King R., Mehdi S.Q., Edmonds C.A., Chow C.E., Lin A.A., Mitra M., Sil S.K., Ramesh A., Zhivotovsky L.A., King R., Mehdi S.Q., Edmonds C.A., Chow C.E., Lin A.A., Mitra M., Sil S.K., Ramesh A., King R., Mehdi S.Q., Edmonds C.A., Chow C.E., Lin A.A., Mitra M., Sil S.K., Ramesh A., Mehdi S.Q., Edmonds C.A., Chow C.E., Lin A.A., Mitra M., Sil S.K., Ramesh A., Edmonds C.A., Chow C.E., Lin A.A., Mitra M., Sil S.K., Ramesh A., Chow C.E., Lin A.A., Mitra M., Sil S.K., Ramesh A., Lin A.A., Mitra M., Sil S.K., Ramesh A., Mitra M., Sil S.K., Ramesh A., Sil S.K., Ramesh A., Ramesh A., et al. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am. J. Hum. Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service R.F. Gene sequencing. The race for the $1000 genome. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- Shen P., Wang F., Underhill P.A., Franco C., Yang W.H., Roxas A., Sung R., Lin A.A., Hyman R.W., Vollrath D., Wang F., Underhill P.A., Franco C., Yang W.H., Roxas A., Sung R., Lin A.A., Hyman R.W., Vollrath D., Underhill P.A., Franco C., Yang W.H., Roxas A., Sung R., Lin A.A., Hyman R.W., Vollrath D., Franco C., Yang W.H., Roxas A., Sung R., Lin A.A., Hyman R.W., Vollrath D., Yang W.H., Roxas A., Sung R., Lin A.A., Hyman R.W., Vollrath D., Roxas A., Sung R., Lin A.A., Hyman R.W., Vollrath D., Sung R., Lin A.A., Hyman R.W., Vollrath D., Lin A.A., Hyman R.W., Vollrath D., Hyman R.W., Vollrath D., Vollrath D., et al. Population genetic implications from sequence variation in four Y chromosome genes. Proc. Natl. Acad. Sci. 2000;97:7354–7359. doi: 10.1073/pnas.97.13.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Dong Y.L., Wen B., Xiao C.J., Underhill P.A., Shen P.D., Chakraborty R., Jin L., Su B., Dong Y.L., Wen B., Xiao C.J., Underhill P.A., Shen P.D., Chakraborty R., Jin L., Su B., Wen B., Xiao C.J., Underhill P.A., Shen P.D., Chakraborty R., Jin L., Su B., Xiao C.J., Underhill P.A., Shen P.D., Chakraborty R., Jin L., Su B., Underhill P.A., Shen P.D., Chakraborty R., Jin L., Su B., Shen P.D., Chakraborty R., Jin L., Su B., Chakraborty R., Jin L., Su B., Jin L., Su B., Su B. Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am. J. Hum. Genet. 2005;77:408–419. doi: 10.1086/444436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims L.M., Garvey D., Ballantyne J., Garvey D., Ballantyne J., Ballantyne J. Sub-populations within the major European and African derived haplogroups R1b3 and E3a are differentiated by previously phylogenetically undefined Y-SNPs. Hum. Mutat. 2007;28:97. doi: 10.1002/humu.9469. [DOI] [PubMed] [Google Scholar]
- Su B., Xiao J., Underhill P., Deka R., Zhang W., Akey J., Huang W., Shen D., Lu D., Luo J., Xiao J., Underhill P., Deka R., Zhang W., Akey J., Huang W., Shen D., Lu D., Luo J., Underhill P., Deka R., Zhang W., Akey J., Huang W., Shen D., Lu D., Luo J., Deka R., Zhang W., Akey J., Huang W., Shen D., Lu D., Luo J., Zhang W., Akey J., Huang W., Shen D., Lu D., Luo J., Akey J., Huang W., Shen D., Lu D., Luo J., Huang W., Shen D., Lu D., Luo J., Shen D., Lu D., Luo J., Lu D., Luo J., Luo J., et al. Y-chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am. J. Hum. Genet. 1999;65:1718–1724. doi: 10.1086/302680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B., Jin L., Underhill P., Martinson J., Saha N., McGarvey S.T., Shriver M.D., Chu J., Oefner P., Chakraborty R., Jin L., Underhill P., Martinson J., Saha N., McGarvey S.T., Shriver M.D., Chu J., Oefner P., Chakraborty R., Underhill P., Martinson J., Saha N., McGarvey S.T., Shriver M.D., Chu J., Oefner P., Chakraborty R., Martinson J., Saha N., McGarvey S.T., Shriver M.D., Chu J., Oefner P., Chakraborty R., Saha N., McGarvey S.T., Shriver M.D., Chu J., Oefner P., Chakraborty R., McGarvey S.T., Shriver M.D., Chu J., Oefner P., Chakraborty R., Shriver M.D., Chu J., Oefner P., Chakraborty R., Chu J., Oefner P., Chakraborty R., Oefner P., Chakraborty R., Chakraborty R., et al. Polynesian origins: Insights from the Y chromosome. Proc. Natl. Acad. Sci. 2000;97:8225–8228. doi: 10.1073/pnas.97.15.8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Skaletsky H., Birren B., Devon K., Tang Z., Silber S., Oates R., Page D.C., Skaletsky H., Birren B., Devon K., Tang Z., Silber S., Oates R., Page D.C., Birren B., Devon K., Tang Z., Silber S., Oates R., Page D.C., Devon K., Tang Z., Silber S., Oates R., Page D.C., Tang Z., Silber S., Oates R., Page D.C., Silber S., Oates R., Page D.C., Oates R., Page D.C., Page D.C. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat. Genet. 1999;23:429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- Tang H., Siegmund D.O., Shen P., Oefner P.J., Feldman M.W., Siegmund D.O., Shen P., Oefner P.J., Feldman M.W., Shen P., Oefner P.J., Feldman M.W., Oefner P.J., Feldman M.W., Feldman M.W. Frequentist estimation of coalescence times from nucleotide sequence data using a tree-based partition. Genetics. 2002;161:447–459. doi: 10.1093/genetics/161.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K., Singh L., Reddy A.G., Rao V.R., Sehgal S.C., Underhill P.A., Pierson M., Frame I.G., Hagelberg E., Singh L., Reddy A.G., Rao V.R., Sehgal S.C., Underhill P.A., Pierson M., Frame I.G., Hagelberg E., Reddy A.G., Rao V.R., Sehgal S.C., Underhill P.A., Pierson M., Frame I.G., Hagelberg E., Rao V.R., Sehgal S.C., Underhill P.A., Pierson M., Frame I.G., Hagelberg E., Sehgal S.C., Underhill P.A., Pierson M., Frame I.G., Hagelberg E., Underhill P.A., Pierson M., Frame I.G., Hagelberg E., Pierson M., Frame I.G., Hagelberg E., Frame I.G., Hagelberg E., Hagelberg E. Genetic affinities of the Andaman Islanders, a vanishing human population. Curr. Biol. 2003;13:86–93. doi: 10.1016/s0960-9822(02)01336-2. [DOI] [PubMed] [Google Scholar]
- Underhill P., Kivisild T., Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu. Rev. Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- Underhill P.A., Shen P., Lin A.A., Jin L., Passarino G., Yang W.H., Kauffman E., Bonne-Tamir B., Bertranpetit J., Francalacci P., Shen P., Lin A.A., Jin L., Passarino G., Yang W.H., Kauffman E., Bonne-Tamir B., Bertranpetit J., Francalacci P., Lin A.A., Jin L., Passarino G., Yang W.H., Kauffman E., Bonne-Tamir B., Bertranpetit J., Francalacci P., Jin L., Passarino G., Yang W.H., Kauffman E., Bonne-Tamir B., Bertranpetit J., Francalacci P., Passarino G., Yang W.H., Kauffman E., Bonne-Tamir B., Bertranpetit J., Francalacci P., Yang W.H., Kauffman E., Bonne-Tamir B., Bertranpetit J., Francalacci P., Kauffman E., Bonne-Tamir B., Bertranpetit J., Francalacci P., Bonne-Tamir B., Bertranpetit J., Francalacci P., Bertranpetit J., Francalacci P., Francalacci P., et al. Y chromosome sequence variation and the history of human populations. Nat. Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- Underhill P.A., Passarino G., Lin A.A., Shen P., Mirazon Lahr M., Foley R.A., Oefner P.J., Cavalli-Sforza L.L., Passarino G., Lin A.A., Shen P., Mirazon Lahr M., Foley R.A., Oefner P.J., Cavalli-Sforza L.L., Lin A.A., Shen P., Mirazon Lahr M., Foley R.A., Oefner P.J., Cavalli-Sforza L.L., Shen P., Mirazon Lahr M., Foley R.A., Oefner P.J., Cavalli-Sforza L.L., Mirazon Lahr M., Foley R.A., Oefner P.J., Cavalli-Sforza L.L., Foley R.A., Oefner P.J., Cavalli-Sforza L.L., Oefner P.J., Cavalli-Sforza L.L., Cavalli-Sforza L.L. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann. Hum. Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- Wilder J.A., Kingan S.B., Mobasher Z., Pilkington M.M., Hammer M.F., Kingan S.B., Mobasher Z., Pilkington M.M., Hammer M.F., Mobasher Z., Pilkington M.M., Hammer M.F., Pilkington M.M., Hammer M.F., Hammer M.F. Global patterns of human mitochondrial DNA and Y-chromosome structure are not influenced by higher migration rates of females versus males. Nat. Genet. 2004a;36:1122–1125. doi: 10.1038/ng1428. [DOI] [PubMed] [Google Scholar]
- Wilder J.A., Mobasher Z., Hammer M.F., Mobasher Z., Hammer M.F., Hammer M.F. Genetic evidence for unequal effective population sizes of human females and males. Mol. Biol. Evol. 2004b;21:2047–2057. doi: 10.1093/molbev/msh214. [DOI] [PubMed] [Google Scholar]
- Wood E.T., Stover D.A., Ehret C., Destro-Bisol G., Spedini G., McLeod H., Louie L., Bamshad M., Strassmann B.I., Soodyall H., Stover D.A., Ehret C., Destro-Bisol G., Spedini G., McLeod H., Louie L., Bamshad M., Strassmann B.I., Soodyall H., Ehret C., Destro-Bisol G., Spedini G., McLeod H., Louie L., Bamshad M., Strassmann B.I., Soodyall H., Destro-Bisol G., Spedini G., McLeod H., Louie L., Bamshad M., Strassmann B.I., Soodyall H., Spedini G., McLeod H., Louie L., Bamshad M., Strassmann B.I., Soodyall H., McLeod H., Louie L., Bamshad M., Strassmann B.I., Soodyall H., Louie L., Bamshad M., Strassmann B.I., Soodyall H., Bamshad M., Strassmann B.I., Soodyall H., Strassmann B.I., Soodyall H., Soodyall H., et al. Contrasting patterns of Y chromosome and mtDNA variation in Africa: Evidence for sex-biased demographic processes. Eur. J. Hum. Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- Xue Y.L., Zerjal T., Bao W.D., Zhu S.L., Shu Q.F., Xu J.J., Du R.F., Fu S.B., Li P., Hurles M.E., Zerjal T., Bao W.D., Zhu S.L., Shu Q.F., Xu J.J., Du R.F., Fu S.B., Li P., Hurles M.E., Bao W.D., Zhu S.L., Shu Q.F., Xu J.J., Du R.F., Fu S.B., Li P., Hurles M.E., Zhu S.L., Shu Q.F., Xu J.J., Du R.F., Fu S.B., Li P., Hurles M.E., Shu Q.F., Xu J.J., Du R.F., Fu S.B., Li P., Hurles M.E., Xu J.J., Du R.F., Fu S.B., Li P., Hurles M.E., Du R.F., Fu S.B., Li P., Hurles M.E., Fu S.B., Li P., Hurles M.E., Li P., Hurles M.E., Hurles M.E., et al. Male demography in East Asia: A north–south contrast in human population expansion times. Genetics. 2006;172:2431–2439. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegura S.L., Karafet T.M., Zhivotovsky L.A., Hammer M.F., Karafet T.M., Zhivotovsky L.A., Hammer M.F., Zhivotovsky L.A., Hammer M.F., Hammer M.F. High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol. Biol. Evol. 2004;21:164–175. doi: 10.1093/molbev/msh009. [DOI] [PubMed] [Google Scholar]
- Zerjal T., Pandya A., Thangaraj K., Ling E.Y., Kearley J., Bertoneri S., Paracchini S., Singh L., Tyler-Smith C., Pandya A., Thangaraj K., Ling E.Y., Kearley J., Bertoneri S., Paracchini S., Singh L., Tyler-Smith C., Thangaraj K., Ling E.Y., Kearley J., Bertoneri S., Paracchini S., Singh L., Tyler-Smith C., Ling E.Y., Kearley J., Bertoneri S., Paracchini S., Singh L., Tyler-Smith C., Kearley J., Bertoneri S., Paracchini S., Singh L., Tyler-Smith C., Bertoneri S., Paracchini S., Singh L., Tyler-Smith C., Paracchini S., Singh L., Tyler-Smith C., Singh L., Tyler-Smith C., Tyler-Smith C. Y-chromosomal insights into the genetic impact of the caste system in India. Hum. Genet. 2007;121:137–144. doi: 10.1007/s00439-006-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]