Abstract
The invention of the placenta facilitated the evolution of mammals. How the placenta evolved from the simple structure observed in birds and reptiles into the complex organ that sustains human life is one of the great mysteries of evolution. By using a timecourse microarray analysis including the entire lifetime of the placenta, we uncover molecular and genomic changes that underlie placentation and find that two distinct evolutionary mechanisms were utilized during placental evolution in mice and human. Ancient genes involved in growth and metabolism were co-opted for use during early embryogenesis, likely enabling the accelerated development of extraembryonic tissues. Recently duplicated genes are utilized at later stages of placentation to meet the metabolic needs of a diverse range of pregnancy physiologies. Together, these mechanisms served to develop the specialized placenta, a novel structure that led to expansion of the eutherian mammal, including humankind.
Over 120 million years ago, eutherian mammals appeared on the earth. Their appearance was dependent on the evolution of the placenta, which, by mediating nutrient and gas exchange between mother and fetus, serves the central role in bearing live offspring. How the placenta evolved to meet these needs is a long-standing mystery. It is believed that the last common ancestor of mammals, birds, and reptiles had a simple chorion, the outermost of four extraembryonic membranes made in amniotes, as well as a functioning yolk sac, amnion, and allantois. In modern-day birds and in most reptiles, the chorion is a relatively simple membrane that adheres to the egg shell to allow gas exchange between the egg and environment (Gilbert 2006). In some amniote species such as chickens, it joins with the mesodermal component of the allantois to form the chorioallantoic membrane, a highly vascular structure that has the additional function of delivering calcium from the egg shell to the developing embryo (Mossman 1987; Gilbert 2006). The most diverse and striking set of adaptations of the basic chorionic membrane has occurred in the eutherian or placental mammals, in which the chorion is vascularized by the allantois to form the chorioallantoic placenta. In addition, the overall placental structure mediates immune challenges found in no other organ of the body, as it involves two genetically distinct organisms: mother (decidua) and fetus (placental trophoblast).
Despite sharing the same basic role during fetal development, modern eutherian placentas vary dramatically in size, shape, cellular composition, and morphology. Physiological differences between the placentas of eutherians may be adaptations to varied reproductive strategies, or may reflect the current balance in a maternal fetal conflict over nutrient allocation during pregnancy. For example, while all eutherians bear live young, their metabolic needs during pregnancy vary significantly: The mouse carries 12 or more offspring and gestates for 20 d, whereas the elephant has only one offspring and gestates for 660 d. Clearly, these animals require significant differences in a range of physiological adaptations to allow for reproductive success.
The genomic mechanisms that led to the emergence and diversification of the eutherian placenta are unknown. Three distinct mechanisms have been proposed to mediate the evolution of new form, including changes in splicing isoforms, increased transcriptional regulatory complexity, and gene family expansion by duplication and divergence (Carroll 2005). MiRNAs and noncoding regulatory RNAs have also been recently suggested as a mechanism for species diversification (Lee et al. 2007; Niwa and Slack 2007). Several approaches have recently been published that demonstrate how genomic analysis can be mined to answer questions about global events that occurred during evolution to facilitate a particular process (McCarroll et al. 2004; Mitiku and Baker 2007). Therefore, we hypothesized that the mechanism of placental evolution might be deduced by a comprehensive analysis of expressed genes through time during placentation. A close analysis of these placental molecules may reveal the changes, whether regulatory or coding, within the genome that gave rise to such an important and novel form. Indeed, we find that the placenta during early developmental stages disproportionately utilizes evolutionarily ancient genes, suggesting an important role for regulatory element co-option in the evolution of basic placental structures. During the later or mature placental stages, we find that the placenta disproportionately utilizes recently duplicated genes. This strategy is seen in both the human and murine placenta, suggesting that genes were created anew to adapt the eutherian placenta for highly varied reproductive environments.
Results
A global molecular switch during midgestation in the placenta
A comprehensive expression profiling encompassing the full lifetime of the murine placenta from embryonic day 8.5 (e8.5), at the time of chorioallantoic fusion, until postnatal day 0 (P0), just after birth, uncovered a striking molecular transition occurring during mid-gestation (Figs. 1 2). This transition separates two phases of placentation that are distinct on a molecular level and encompasses thousands of genes in both the fetal placenta and maternal decidua (3543 and 1108, respectively). We refer to these different phases as the developing phase and the mature phase (D and M, respectively; Fig. 2). These phases were validated by extensive qRT-PCR (Supplemental Table 1). It is surprising to find such unique molecular signatures for these two phases, given that the placenta at e10.5 is morphologically and histologically very similar to the placenta at late gestation. We investigated whether this transition was due to the major morphological change during this window of time, namely a relative increase in the size of the labyrinth layer, which occurs between e10.5 and e17.5. To this end, we conducted an in situ hybridization screen on e10.5 and e17.5 placentas with 55 genes whose expression changed significantly between the two phases. We found no bias in the staining of any individual cell type between genes expressed highly in the developing versus mature placenta (Supplemental Fig. 1). Therefore, the transition is not simply due to the expansion of an individual cell type, but appears to be a global shift in gene usage in the absence of major morphological change. This gene switch must represent altered function and may reflect a global adaptation of the placenta to metabolic changes occurring during the later stages of pregnancy, including changes in oxygen concentrations and hormonal regulation.
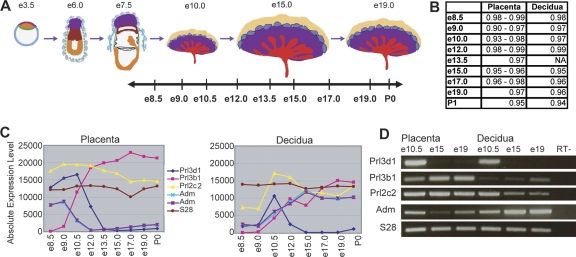
Figure 1.
Genome-wide expression profiling of the fetal and maternal placenta. (A) Samples were taken at nine stages throughout placental development, and dissected into placental and decidual portions. The data set includes biological triplicates for placenta samples at e8.5, e9.0, e10.5, e12.0, e15.0, and e17.0, biological duplicates for placenta samples at e13.5, e19.0, and P0, and biological duplicates for decidual samples at e8.5, e9.0, e10.5, e12.0, e15.0, e17.0, e19.0, and P0. (B) Linear correlation coefficients (r-values) between biological replicates range from 0.90 to 0.99 for fetally derived tissues and from 0.94 to 0.99 for maternally derived tissues, indicating a high degree of correlation between replicate samples at each timepoint. Microarray profiles for known placental hormones correspond with previously published expression patterns (C) and are confirmed by RT–PCR (D) (Linzer et al. 1985; Faria et al. 1991; Yotsumoto et al. 1998; Linzer and Fisher 1999; Soares 2004). Prl3d1 (chorionic somatomammotropin hormone 1); Prl3b1 (chorionic somatomammotropin hormone 2); Prl2c2 (proliferin); Adm (adrenomedullin) (represented by two probe sets). Figure 1A adapted by permission from Macmillan Publishers Ltd.: Nature Reviews Genetics (Rossant and Cross 2001), copyright 2001 (http://www.nature.com/nrg).
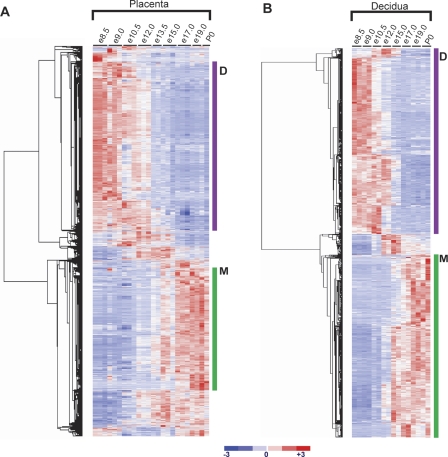
Figure 2.
A molecular transition revealed at mid-gestation in the fetal placenta. The transition at mid-gestation is global, encompassing 3543 genes in the placenta and 1108 genes in the decidua that change significantly over time (FDR < 0.1% using SAM [Tusher et al. 2001] and having a minimum 1.5-fold change within a 2-d time period). Genes that change significantly during placental development were clustered using dCHIP (Li and Wong 2001a, b). Expression values for each gene across all samples were linearly scaled to have a mean of 0 and standard deviation of 1; red indicates expression greater than the mean; and blue indicates expression below the mean. (A) Clustergram of 3543 genes that change significantly in the placenta. (B) Clustergram of 1108 genes that change significantly in the decidua. Genes in Clusters labeled D are highly expressed in the developing placenta (e8.5–e13.5), while genes in Clusters labeled M are highly expressed in the mature placenta (e13.5–P0).
We investigated the functions of genes in the placental and decidual clusters using Gene Ontology Biological Process term over-representation (Dennis et al. 2003). Based on a conservative Bonferroni correction for multiple testing, we apply a significance threshold of P < 3.45 × 10−6, rather than the standard P < 0.05; categories with P-values above this threshold are included as useful characterizations rather than significant over-representations. The developing placenta cluster is strongly enriched for many growth and metabolism-related processes, including RNA processing and metabolism (54–146 genes, P-value range 2.0 × 10−54–1.2 × 10−24), cellular and molecular metabolism (343–779 genes, P-value range 7.7 × 10−34–1.3 × 10−22), ribosomal and organelle biogenesis (51–142 genes, P-value range 9.5 × 10−27–8.8 × 10−14), and cell division and cell cycle (55–127 genes, P-value range 3.8 × 10−19–6.5 × 10−15). In contrast, the mature placenta cluster is characterized by genes involved in negative regulation of physiological and cellular processes (62–75 genes, P-value range 2.3 × 10−6–2.3 × 10−5), cell growth and morphogenesis (15–37 genes, P-value range 2.2 × 10−4–5.8 × 10−3), and pregnancy and reproductive physiological processes (8–12 genes, P-value range 3.5 × 10−4–2.6 × 10−2). If we perform the same analysis on the decidual clusters, a different set of functions is inferred: The decidual developing cluster demonstrates over-representation of genes involved in steroid metabolism and biosynthesis (9–11 genes, P-value range 5.6 × 10−4–6.5 × 10−3) and angiogenesis (11 genes, P-value 3.9 × 10−3), while the decidual mature cluster is characterized by genes involved in immune/defense response (39–41 genes, P-value range 2.3 × 10−6–3.8 × 10−5), inflammatory response (16 genes, P-value 6.2 × 10−5), pregnancy and reproduction (7–8 genes, P-value range 6.5 × 10−5–1.7 × 10−3), and angiogenesis and blood vessel development (10–13 genes, P-value range 4.3 × 10−3–8.8 × 10−3).
Placental genes have ancient origins
We hypothesized that this comprehensive data set could provide a window into understanding the molecular evolution of the placenta and the mechanisms by which a complex organ forms from a simple membrane. To this end, we aimed to address a simple question: What are the evolutionary origins of genes that play specific or “nonhousekeeping” roles in placentation? Answering this question required two important steps. First, based on expression levels through time and in comparison with other tissues, we identified genes that were likely to have specific roles in placental development and function. Second, we systematically evaluated the evolutionary histories of these placental genes. In combination, these steps give us a glimpse into the genetic processes by which a simple respiratory membrane evolved into the complex placenta found in modern-day eutherians.
To identify genes with specific roles in placentation, we hypothesized that the majority of these genes would be expressed above background levels in the placenta and would demonstrate significant change during development. Our analysis provided 3543 individual placental genes that meet these criteria. We then further restricted this list to include only genes expressed highly in the placenta as compared with other tissues, using the Novartis mouse GeneAtlas data set, which includes gene expression measurements for 36,182 transcripts across 61 different mouse tissues (Su et al. 2002, 2004). To this end, we assigned the 61 tissues to 32 organ- or tissue-type-specific groups and scaled the GeneAtlas placental sample to reflect the peak expression seen in our timecourse, rather than the single day e18 expression level provided in the baseline data set. We found the average expression across all groups for each gene, and calculated a measure (PlacPEM) that is the expression level in the fetal placenta divided by the average expression. We find that 410 of our initial 3542 genes meet a PlacPEM threshold of 4, with the majority of these genes demonstrating this level of tissue specificity only in the placenta. Although we tested other PEM thresholds, PEM4 maximizes the number of probe sets identified with the least sacrifice of specificity. Thus, the 410 genes, referred to here as “preferentially expressed” genes, demonstrate high placental specificity. Following the same procedure for genes expressed in maternal decidua, we find a corresponding set of 347 decidual preferentially expressed genes.
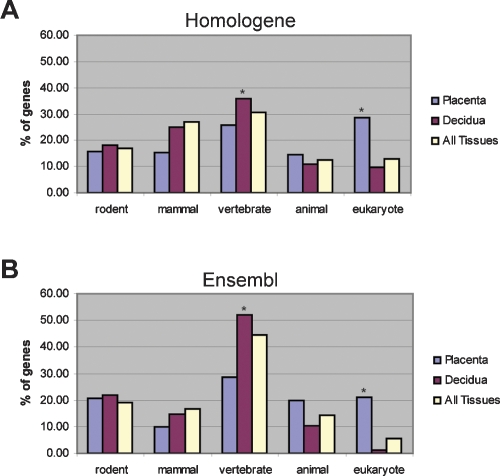
By evaluating the evolutionary origins of the placental and decidual preferentially expressed placental genes using the closest one-to-one, reciprocal best match ortholog, we find the placental preferentially expressed list is significantly enriched for genes with eukaryote orthology according to both Ensembl (P < 1 × 10−29) and Homologene (P < 1 × 10−19), whereas the decidual list is significantly enriched for genes with vertebrate orthology in both datasets (Fig. 3). These enrichments were identified by comparison with a background list of all genes that are expressed in one or more tissues at a minimum of four times the average expression across all tissues. For each gene, orthology is determined based on the most distantly related organism whose genome includes a one-to-one reciprocal best match ortholog, with organisms broadly classified as eukaryote, animal, vertebrate, mammal, or rodent (see Fig. 3). Eukaryotic orthology indicates that the most ancient one-to-one reciprocal best match ortholog is found in an organism that is a eukaryote, but not animal, while animal orthology indicates that such an ortholog is found in an organism that is an animal, but not a vertebrate. Genes with rodent orthology are referred to as “rodent specific,” an estimation based on the available datasets (for details, see Methods). Overall, this data suggests that the development and functions of the placenta were mediated by ancient genes that were co-opted during placental evolution.
Figure 3.
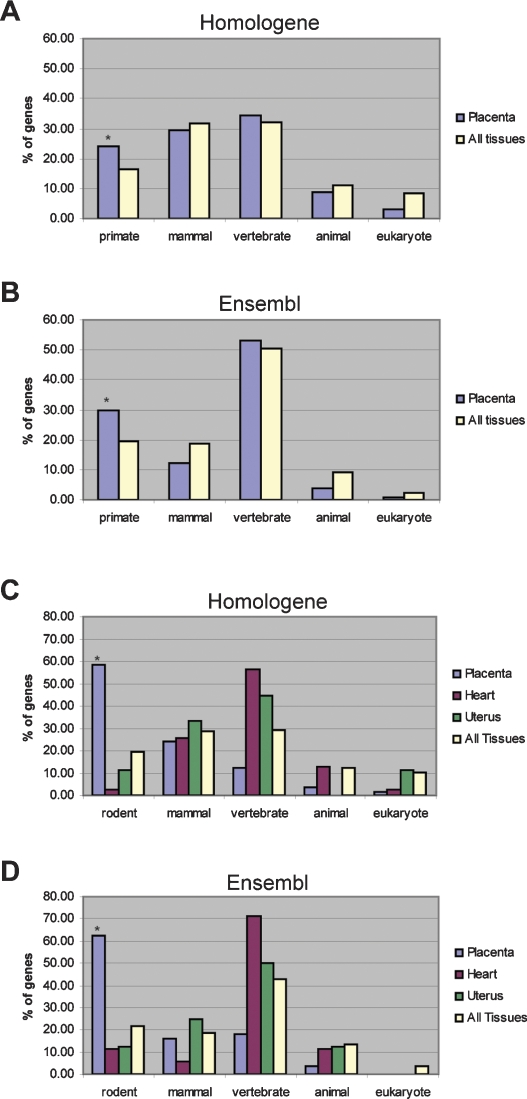
Placental genes have ancient origins. Evolutionary origins of preferentially expressed genes in the placenta (410 genes), decidua (347 genes), or in any tissue (3055 genes). The evolutionary origin of each gene was determined by finding the most distantly related species that has a one-to-one, reciprocal best-match ortholog identified in the Homologene database (A) or the Ensembl database (B). Asterisks (*) indicate significant enrichment over background (All Tissues). The placenta demonstrates significant enrichment for genes with eukaryotic orthology according to both Homologene (P < 1 × 10−19) and Ensembl (P < 1 × 10−29). The decidua demonstrates significant enrichment for genes with vertebrate orthology according to both Homologene (P < 0.05) and Ensembl (P < 0.01).
Genes expressed in mature placenta are rodent specific and recently evolved
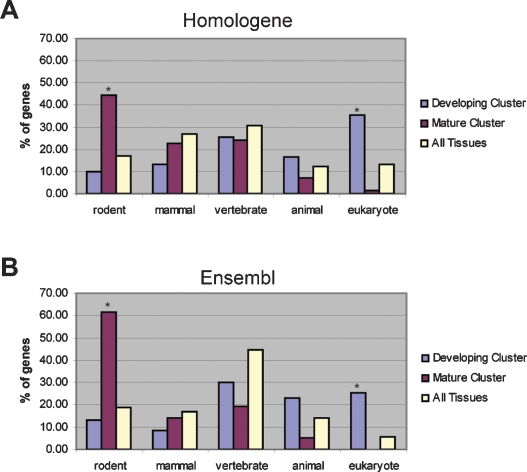
Since the molecular profiles of the developing placenta and the mature placenta are so distinct, we hypothesized that the genes representing these two phases might have unique evolutionary origins that were masked in our initial analysis of the full set (Fig. 3). Hierarchical clustering of the preferentially expressed genes (1.5-fold change and PlacPEM ≥ 4), revealed that—like the entire gene set—these molecules underwent the midgestation transition (see Supplemental Fig. 2). Therefore, we separately assessed the evolutionary histories of the placental preferentially expressed genes in the developing phase versus the mature phase of placentation. We find a striking difference between the two developmental stages (Fig. 4). As found in the full data set, we show that the developing phase is significantly enriched for genes with eukaryotic orthology: 35.3% according to Homologene and 25.3% according to Ensembl, a significant enrichment over the 6%–13% found in the background list (P < 1.0 × 10−27 for Homologene and P < 1.0 × 10−35 for Ensembl) (Fig. 4). In marked contrast to the developing placenta, however, the mature phase has a striking enrichment for rodent-specific genes: 44.3% according to Homologene and 61.4% according to Ensembl, a significant enrichment over the 17%–19% found in the background list (P < 1.0 × 10−7 for Homologene and P < 1.0 × 10−11 for Ensembl) (Fig. 4). The mature phase has no enrichment for genes with eukaryotic orthology.
Figure 4.
Genes representing the mature phase reveal recent evolution by duplication and divergence. Evolutionary origins of placental preferentially expressed genes during the developing (329 genes) and mature (70 genes) phases of placentation. The evolutionary origin of each gene was determined by finding the most distantly related species that has a one-to-one, reciprocal best-match ortholog identified in the Homologene database (A) or the Ensembl database (B). Asterisks (*) indicate significant enrichment over background (All Tissues; 3055 genes). The developing placenta is significantly enriched for genes with eukaryotic orthology according to both Homologene (P < 1.0 × 10−27) and Ensembl (P < 1.0 × 10−35). In contrast, the mature placenta is significantly enriched for genes with rodent orthology according to both Homologene (P < 1.0 × 10−7) and Ensembl (P < 1.0 × 10−11).
Developing and mature genes have distinct functional roles
We next asked whether the difference between genes of the developing and mature clusters extends to gene functions as well as to evolutionary origins. We used DAVID to classify genes by Gene Ontology Biological Process Annotation (Dennis et al. 2003), and again apply a conservative over-representation significance threshold of P < 3.45 × 10−6 based on a Bonferroni correction for multiple testing. The developing placenta cluster is significantly enriched for genes involved in growth and metabolic processes, including cellular and molecular metabolism (P-value range 9.5 × 10−14–5.2 × 10−7), DNA replication (6–25 genes, P-value range 2.5 × 10−14–2.9 × 10−4), RNA processing and metabolism (10–27 genes, P-value range 2.5 × 10−10–1.4 × 10−4) and cell cycle (14–48 genes, P-value range 9.5 × 10−14–5.1 × 10−5). In contrast, the mature cluster demonstrates over-representation (albeit not at the conservative significance threshold) for genes involved in pregnancy and reproduction (P-value range 9.3 × 10−4–7.5 × 10−3). This trend is evident, even given that most of the genes with predicted roles in pregnancy are not annotated; in fact, the only genes annotated to this role are CEA-related cell adhesion molecule 2, CEA-related cell adhesion molecule 3, CEA-related cell adhesion molecule 12, and pregnancy-specific glycoprotein 28. A closer analysis of the mature fetal placenta cluster shows that three large gene families, all of which have specific roles in pregnancy, account for a majority of the rodent-specific genes: the prolactin-like protein family, the carcinoembryonic antigen-related cell adhesion molecule (CEACAM), and pregnancy-specific glycoprotein (PSG) family, and the cathepsin family (Fig. 5). Rodent-specific members of each of these families are believed to have evolved from duplication of one or more ancestral loci following the divergence of primates and rodents ∼85 million years ago (Mason et al. 2002; Sol-Church et al. 2002; Wiemers et al. 2003; Soares 2004; Futuyma 2005; McLellan et al. 2005a, b; Zebhauser et al. 2005; Alam et al. 2006).
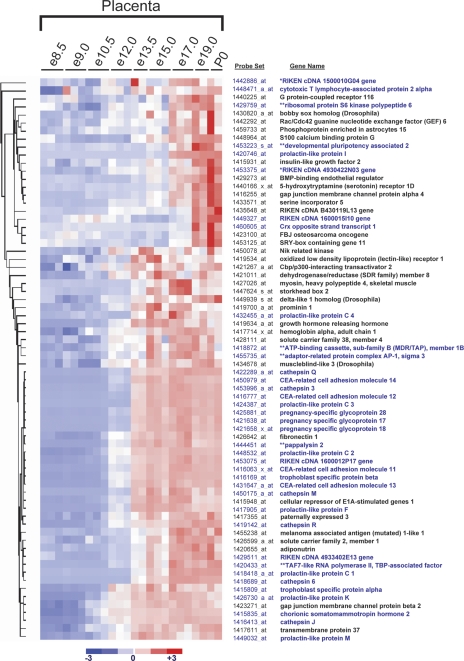
Figure 5.
Newly evolved rodent-specific genes from the mature phase represent three distinct gene families. Among preferentially expressed genes, the mature cluster is highly enriched for genes that have evolved since rodents diverged from primates. Of the 37 rodent specific genes identified by either Homologene or Ensembl (see blue gene names), 29 are called rodent specific by both Homologene and Ensembl, two by Homologene only (*), and six by Ensembl only (**). Three large gene families account for a majority of the rodent-specific genes: the prolactin-like protein family, the CEA-related cellular adhesion molecule, and pregnancy-specific glycoprotein family, and the cathepsin family.
Human mature placental genes are primate specific
Our data demonstrate that the genes highly and specifically expressed in the mature murine placenta have only recently evolved. Since placental morphology varies significantly between species, we sought to determine whether gene expansion in the mature placenta might be a general evolutionary mechanism in other species. To determine whether the mature human placenta has an over-representation of primate-specific genes, we again used the Novartis GeneAtlas data set (Su et al. 2002, 2004). While no samples from the developing placenta are included in the human data set, it does include a full-term placental sample, which we analyzed. We identified those genes that have a PlacPEM (the placental expression level divided by the average expression across tissues) of ≥4, as described above. Again, as a background list we used the set of genes that are expressed in at least one tissue at fourfold the average expression level across all tissues. In the mature human placenta, 24.0% percent of genes with a PlacPEM ≥ 4 are primate specific according to Homologene and 29.9% using Ensembl, compared with 16.4% and 19.5% in the background lists (P < 0.05 for Homologene and P < 0.01 for Ensembl) (Fig. 6). Among the human mature placenta genes with PlacPEM ≥ 4, we find significant over-representation of genes involved in pregnancy and reproduction (11–15 genes, P-value range 1.4 × 10−8–6.1 × 10−6), again using the Bonferroni corrected significance threshold of P < 3.45 × 10−6. These genes include pregnancy-specific beta-1-glycoproteins 1, 2, 3, 4, 5, and 9, adrenomedullin, chorionic somatomammotropin hormone 1, corticotropin releasing hormone, and at least one member of the chorionic gonadotropin beta polypeptide family (the probe set identified targets chorionic gonadotropin beta polypeptide, chorionic gonadotropin beta polypeptide 5, and chorionic gonadotropin beta polypeptide 7). Overall, this indicates that the trend toward use of newly evolved genes is not unique to rodents, but is also observed in the human lineage.
Figure 6.
Primate-specific genes enriched in human mature placenta, indicating conservation of a gene duplication mechanism. Evolutionary origins of genes with PlacPEM ≥4 in mature placenta samples from the unscaled Novartis GeneAtlas human and mouse datasets using either Homologene (A) or Ensembl (B). Asterisks (*) indicate significant enrichment over background (All Tissues). In human, the GeneAtlas mature placenta gene set demonstrates significant enrichment of primate specific genes according to Homologene (P < 0.05) and Ensembl (P < 0.01). The GeneAtlas mature placenta gene set for the mouse is significantly enriched for rodent-specific genes according to both Homologene (P < 1.0 × 10−11) (C) and Ensembl (P < 1.0 × 10−11) (D). Genes with HeartPEM ≥ 4 (i.e., genes for which the heart expression level divided by the average expression is ≥4), UterusPEM ≥ 4, or PEM ≥ 4 in any tissue are included for comparison.
Mammalian duplication and divergence strategy is unique to the placenta
To further confirm our approach, we evaluated the evolutionary histories of a term placenta, an adult heart, and an adult uterus based exclusively on the mouse GeneAtlas data set (Su et al. 2002, 2004). The uterus and heart were selected as two organs providing contrasting, informative comparisons to the placenta in terms of evolution and function. The uterus was selected as a reproductive organ found only in mammals. The heart, in contrast, exists in peristaltic tubular form in animals such as Drosophila and in a more complex form involving chambers and unidirectional circulation in vertebrates. Therefore, the mouse term placenta serves as a direct comparison with the human results, and the uterus and heart provide additional evidence for a correlation between organ system ages and the evolutionary histories of genes expressed in mature organ tissues. In agreement with our earlier findings, a full 58.6% of genes with PlacPEM ≥ 4 in the mature (e18) mouse placenta of the GeneAtlas data set are rodent specific according to Homologene and 62.5% according to Ensembl, compared with 19.4% and 21.6% in the background list (P < 1.0 × 10−11 for Homologene and P < 1.0 × 10−11 for Ensembl) (Fig. 6). Furthermore, genes with HeartPEM (the heart expression level divided by the average expression) ≥ 4 are enriched, not for rodent-specific genes, but for genes with vertebrate orthology (P < 0.01 for Homologene, P < 0.001 for Ensembl). A similar, but not significant trend is observed for genes with UterusPEM ≥ 4. Taken together, these results demonstrate that the mature placenta in both mouse and human overutilizes newly evolved genes.
Discussion
Rapid growth of extraembyonic tissues is conserved throughout evolution
Our results provide evidence that rapid growth is the dominant facet of the developing murine placenta, and here we hypothesize that this program is conserved within eutherian species. Eutherian mammals must establish a functioning chorioallantoic placenta in order to accommodate the survival of the fetus (Mossman 1987). Given that all eutherian mammals must undergo a phase of rapid extraembryonic growth, to what extent are the genes involved in early placental development conserved between species? While comparable genome-wide expression data from early stages of placenta development in other mammals may allow us to answer this question directly in the future, two interconnected lines of evidence now suggest that at least some aspects of early placental development are likely to be well conserved. First, we have found that the murine-developing placenta relies disproportionately on evolutionarily ancient genes, particularly those involved in basic growth and metabolism, and that genes with rodent and mammalian orthology genes are under-represented. This tells us that rapid growth of the placenta is largely achieved by co-option of previously existing genes for use in this trophoblast-specific function. Second, rapid development and differentiation of extraembryonic tissues, or analogous fetal membranes, is conserved among all amniotes (Mossman 1987). In the sister group of mammals, the sauropsid amniotes (lizards, birds, turtles), timely initiation of rapid absolute fetal growth is dependent on rapid development of chorioallantoic membrane respiratory function (Mossman 1987), indicating that a program of accelerated extraembryonic membrane development was in place prior to the divergence of mammals from birds and reptiles. Thus, while order- or species-specific modifications may have occurred, the rapid trophoblast development and differentiation found in all eutherian mammals is likely based upon a program already established in their last common ancestor.
Genomic response to varied reproductive strategies in mammals
Although placentas of all eutherian mammals must carry out the same basic functions of nutrient, waste, and gas exchange, they display an incredible amount of morphologic diversity (Ramsey 1982; Mossman 1987). This diversity is particularly striking, given that the fetal membranes of noneutherian amniotes are morphologically very similar, even between classes as otherwise diverse as reptiles, birds, monotremes, and marsupials (Mossman 1987). If all mammalian placentas are able to carry out the same basic functions, why have so many variations evolved? One possibility is that this placental diversity has evolved to facilitate the broad range of reproductive strategies found among eutherians. While the basic exchange function of the placenta is constant among all mammals, the modulations of maternal physiology that are required for successful pregnancy are likely to vary dramatically between eutherian species with such highly diverged reproductive strategies.
A comparison of human and mouse provides a good example of the potential interplay between reproductive strategy and placental evolution. The last common ancestor of primates and rodents is believed to have had a hemochorial maternal fetal interface, with labyrinthine interdigitation and a discoid shape (Carter and Mess 2006; Mess and Carter 2006; Wildman et al. 2006). While both humans and rodents have retained a hemochorial interface and a discoid shape, humans have evolved a villous interdigitation, which is associated across species with long gestation times and single offspring, and possibly with reduced metabolic demand on the mother, while rodents have maintained a labyrinthine interdigitation (Wildman et al. 2006). Murine gestation time is ∼19–21 d and average litter sizes are in the range of 12, while humans have a gestation time of ∼9 mo and largely singleton pregnancies. How is the mouse able to provide nutrient, waste, and gas exchange at a sufficient rate to allow full development of 12–15 fetuses within a mere 3-wk period? According to a model in which the placenta adapts to facilitate divergent reproductive strategies, we would expect to observe different outcomes in the mature placenta of mice compared with humans. In fact, this is what we found: The mouse mature placenta is dramatically enriched for rodent-specific genes, including prolactin-like hormones, pregnancy-specific glycoproteins, and CEA-related cellular adhesion molecules and cathepsins, while the human placenta is enriched for primate-specific genes, including several pregnancy-specific glycoproteins (pregnancy specific beta-1 glycoproteins 1, 3–7, and 9), five hormones (CSH1, CSH2, CGB/CGB5/CGB7, GH1, and GH2) and no specific proteases. Given the direct connection between maternal blood and fetal trophoblast cells that is characteristic of the hemochorial placenta, it is not surprising to find recently evolved pregnancy-specific glycoproteins, molecules believed to play a role in modulating the maternal immune response, up-regulated in the mature placenta of both mice and humans. That the expansion of placenta-specific hormones in mouse is more dramatic than that found in humans and that the mouse has a large expansion of placental proteases may reflect an increased range of functions required when metabolic demand on the mother is high.
Duplicated and diverged rodent specific placental gene families
The majority of the rodent-specific genes that we have found to be highly expressed in the mature placenta are members of three gene families that have undergone extensive rodent or mouse-specific expansion. These include the prolactin-like proteins C1, C2, C3, C4, F, I, K, and M, pregnancy-specific glycoproteins 17, 18, and 28, CEA-related cellular adhesion molecules 11–14, and cathepsins 3, 6, J, M, Q, and R. The prolactin-like proteins are part of a rodent-specific expansion of the prolactin (Prl) locus, on Chromosome 13, which contains 26 related genes (for review, see Soares 2004). The mouse Prl gene family members encode a broad range of hormones and cytokines believed to be involved in regulation of adaptations to pregnancy, with primary expression in the placenta and uterus (Ain et al. 2004; for review, see Soares 2004). While some of the Prl gene family members appear to mimic the biological actions of prolactin itself, others, including many of those we have found to be characteristic of the mature placenta, are believed to possess novel biological actions (for review, see Soares 2004). Multiple copies of the Prl family have been found in the mouse, rat, and cow. While the mouse and rat loci are similarly organized, with orthologous members similarly positioned in the two species (Alam et al. 2006), members of the expanded cow locus are not orthologous with genes of the mouse and rat Prl family (for review, see Soares 2004). In human and dog there has been no expansion, and Prl is the single gene at the locus. The human genome, however, has undergone moderate expansion of the related growth hormone (GH) locus (Alam et al. 2006; for review, see Soares 2004).
The CEA-related cellular adhesion molecules (CEACAMs) and pregnancy-specific glycoproteins (PSGs) are members of the carcinoembryonic antigen (CEA) family of glycoproteins, a subgroup of the Immunoglobulin (Ig) superfamily. The CEA family has undergone extensive expansion in both rodents and humans; however, only five of 31 known murine CEA-related genes are found in primates; the other 26 are unique to rodents (Zebhauser et al. 2005). The majority of the rodent-specific genes, including Psg 17,18, 28, and Ceacam 11–14, are expressed only in the trophoblast (Beauchemin et al. 1999; for review, see Zebhauser et al. 2005). PSGs are believed to play a role in modulating the maternal immune system to prevent rejection of the fetus, possibly by binding to macrophage and monocyte receptors to stimulate production of anti-inflammatory cytokines and shifting the immune system from a Th1 bias to the Th2 bias characteristic of pregnancy. It has been proposed that CEACAM 11–14 may share similar functions, despite their structural divergence from the PSGs, since they demonstrate a similar spongiotrophoblast-specific expression pattern with increases in expression levels during pregnancy (discussed in Zebhauser et al. 2005).
The mouse placentally expressed cathepsin (PEC) family is composed of a minimum of eight members, Cathepsins 3, 6, 7, 8, M, P/J, Q, and R, all of which are located in a dense cluster on chromosome 13 (Deussing et al. 2002). In contrast to the closely related Cathepsin L, which is found in a broad variety of eukaryotic species and is ubiquitously expressed, the PECs are expressed only in the placenta and are found only in rodents (for review, see Mason et al. 2002). Of the Cathepsins in our mature placenta cluster, J/P, M, Q, and R are found in both rats and mice, while 3 and 6 are believed to exist in mice alone (Deussing et al. 2002; for reviews, see Mason et al. 2002; Sol-Church et al. 2002). The PECs are cysteine proteases that are believed to have arisen via gene duplication, then evolved to have greater substrate specificity compared with the related Cathepsins L and K. Based on their exclusive expression in the placenta, it has been suggested that PECs may contribute to embryonic nutrition or may have evolved to more efficiently process conserved or novel placental hormones and proteins (for review, see Mason et al. 2002). While there is no expansion of placentally specific proteases in humans, an analogous expansion of aspartic proteases has occurred in artiodactyls (cloven-hoofed animals). This family also demonstrates placental-specific expression, suggesting important roles for proteases in placentation across species (for reviews, see Mason et al. 2002; Sol-Church et al. 2002).
Conclusion
We began this study by asking how the genome adapted over evolutionary time to yield a new and important organ system. We have found evidence that the basic placental structure relied largely on co-option of ancient genes (True and Carroll 2002; Carroll 2005), particularly those involved in growth and metabolism. We further propose that once the basic placenta was formed, duplication and divergence allowed for new and highly varied reproductive strategies among the rapidly diversifying eutherian mammals.
Methods
Placental sample collection and dissection
Three timed pregnant female Swiss Webster mice from Charles River Laboratories were sacrificed at each timepoint used in our analysis, generating two to three biological replicates at each stage. For each pregnancy, the entire litter of fetuses and placentas was dissected. Fetal tissues were used to confirm staging of the embryos using Theiler’s morphological criteria (Theiler 1989). Placentas were manually dissected into fetal and decidual (maternal) portions, using fine forceps to separate the decidua from the spongiotrophoblast and trophoblast giant cells. Fetally derived placenta samples and maternally derived placenta samples were pooled separately for each litter, then flash frozen in liquid nitrogen and stored at –80°C until RNA isolation. Tissues from two to 10 placentas from a single litter were combined for each sample except for two of the e8 samples, which were pooled from multiple litters after staging.
Array sample preparation and hybridization
RNA was isolated from each of the pooled samples. Each sample was homogenized in TRIzol with a rotor homogenizer. RNA was isolated by chloroform extraction from the TRIzol sample, purified using a Qiagen RNeasy column, then ethanol precipitated using glycogen as a carrier. Due to limited tissue availability, two e8.5 fetally derived placenta samples were homogenized in Ambion lysis buffer rather than TRIzol. For these samples, RNA was isolated using the Ambion RNAqueous Micro kit prior to ethanol precipitation, again with glycogen as a carrier. A total of 10 μg of starting RNA was used for each sample and labeled cRNA was prepared according to Affymetrix protocols. Briefly, RNA was reverse transcribed and the resulting double-stranded cDNA was purified by phenolchloroform extraction followed by ethanol precipitation. The cDNA was used as a template for a biotin labeling in vitro transcription. The resulting cRNA was purified using an RNeasy column, then quantified. For each sample, 20 μg of cRNA was fragmented using KOAc RNA fragmentation buffer, then hybridized to an Affymetrix mouse 430 2.0 microarray using standard hybridization procedures (www.affymetrix.com).
Microarray data analysis
In order to control for differences in overall strength of hybridization, all arrays were normalized to the same median intensity using the dCHIP invariant set method (Li and Wong 2001a). Modeling was performed using the dCHIP Perfect Match (PM) modeling algorithm. Prior to conducting comparative analysis, the data set was filtered to remove probe sets that are not expressed above background levels. This was done separately for fetal and maternal placenta sample groups; for each group, probe sets were filtered to include only those for which at least two arrays had an absolute expression value over 200 and at least two arrays were called “Present” by the dCHIP software. All expression values were log2 transformed prior to further analysis.
Statistical Analysis of Microarrays (SAM) (Tusher et al. 2001) “multiclass” function was used to identify probe sets for which there is a statistically significant difference between one or more timepoints. SAM identifies 8237 probe sets in the fetal placenta and 1660 probe sets in the maternal placenta for which there is a significant difference between one or more timepoints, with less than one out of every 1000 genes predicted to be a false positive. Because the SAM uses a random permutation-based approach, the gene lists generated for a given FDR threshold can vary somewhat between individual analyses. The lists used here, however, were generated using 1000 permutations of the data, allowing for near perfect convergence of gene lists given different random number seeds. The dataset is available from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo/).
The stage-to-stage fold change requirement, in which a fold change threshold of at least 1.5-fold within a 2-d time window was applied, was conducted using dCHIP (Li and Wong 2001b). Genes could pass this threshold by changing 1.5-fold from e8.5–e10.5, e10.5–12.0, e12.0–e13.5, e13.5–e15.0, e15.0–e17.0, e17.0–e19.0, or e19.0–P0 in the fetal placenta timecourse. Because there are no sample arrays at e13.5 for the maternal timecourse, a 1.5-fold change was allowed from e12.0 to e15.0 for the maternal timecourse only. Because two of the e8 RNA samples were isolated with the Ambion column rather than TRIzol/RNeasy, only the single e8.5 sample that was isolated with TRIzol/RNeasy was used in determining the stage-to-stage requirements, rather than using an average of the three replicates. This ensures that any change identified between e8.5 and e9.0 or e10.5 is not due to the difference in RNA isolation protocol.
Hierarchical clustering, using Pearson’s correlation and centroid linkage, was performed using dCHIP (Li and Wong 2001b). Before clustering, dCHIP linearly scales the expression values for a gene across all samples so that they have a mean of 0 and a standard deviation of 1. These standardized values are then used to calculate the Pearson correlations between genes and to guide the merging of nodes during the clustering process. The standardized values are also used in generating the clustergrams.
Correlation analysis, RT–PCR, and qRT–PCR:
Linear correlation coefficients (r-values) were calculated in Excel using log2-transformed expression values for the background-filtered gene lists described above. Conventional RT–PCR analysis of placental tissue samples was conducted using previously published primers for Prl3d1 (chorionic somatomammotropin hormone 1), Prl3b1 (chorionic somatomammotropin hormone 2), Prl2c2 (proliferin) (Hemberger et al. 2004), and Adm (adrenomedullin) (Mantuenga et al. 1998), and using custom primers for S28 (F: 5′-TAGGGTAACCAAAGTGCTGGGCAG-3′; R: 5′-GACATTTCGGATGATAGAGCGG-3′). All analyses were done using 27 cycles. Quantitative RT–PCR was conducted using custom-designed primers for chorionic somatomammotropin hormone 1, adrenomedullin, ribonucleotide reductase M2, placental specific protein 1, prolactin-like protein A, inner centromere protein, tubulin, beta 6, cyclin E1, CEA-related cell adhesion molecule 9, galactokinase 1, chorionic somatomammotropin hormone 2, prolactin-like protein I, cathepsin Q, CEA-related cell adhesion molecule 12, pregnancy specific glycoprotein 17, cathepsin M, prolactin-like protein C 1, and cathepsin 6, with beta-actin serving as a reaction control. All primer sets were validated using conventional nonquantitative RT-PCR and agarose gel electrophoresis.
In situ hybridization:
Placental samples for in situ hybridization were collected from pregnant Swiss Webster females at e10.0 and e17.0. Placentas were fixed in 4% paraformaldehyde, infused with sucrose, then embedded in OCT and stored at −80°C until sectioning. Digoxigenin-labeled riboprobes were generated from linearized plasmids (Dr. Mark Krasnow’s laboratory at Stanford University). In situ hybridization was conducted at 55°C overnight on e17.0 and e10.0 placental sections. Digoxigenin signal was detected using a Roche anti-digoxigenin alkaline phosphatase conjugated antibody. Staining was performed for 4–6 h using Roche NBT and BCIP; counterstaining was done with Biomeda nuclear fast red.
Determination of tissue specificity
The GeneAtlas datasets used for determining tissue specificity of mouse and human genes were provided by Novartis (Su et al. 2004). In order to control for differences in overall strength of hybridization, all arrays in the mouse data set were normalized to the same median intensity using the dCHIP invariant set method (Li and Wong 2001b). Modeling was performed using the dCHIP Perfect Match (PM) modeling algorithm. The mouse GeneAtlas data set includes 36,182 probe sets and 61 tissues. Of the 45,037 probe sets on the Affymetrix mouse 430 2.0 array, 35,587 were mapped to corresponding GNF IDs using Entrez Gene annotations provided by Novartis and Affymetrix; the 9450 probe sets that were not mapped to GNF IDs either were not annotated to Entrez Gene IDs themselves, or had no corresponding gene in the GNF array. Each GeneAtlas tissue sample has one replicate, meaning that each tissue is represented by two arrays. For each tissue, dCHIP was used to calculate a weighted average expression value that takes into account the measurement error associated with each replicate (Li and Wong 2001a); all further analyses were conducted using these average expression values.
In order to assess the specificity of expression of genes up-regulated throughout the lifetime of the placenta, rather than only at the single e18 timepoint provided by GeneAtlas, expression levels from our timecourse were mapped onto the GeneAtlas data set. The two GeneAtlas placenta sample replicates are from C57BL/6 mice, taken at e18.0. E17.0 placentas were collected from a C57Bl/6 mouse, RNA was isolated using TRIzol and RNeasy, and 10 μg of RNA was prepared for microarray hybridization using standard Affymetrix protocols, as described above. The C57BL/6 mouse used in the analysis was a phenotypically normal heterozygote for the imprinted gene p57/kip2; the placentas used in the analysis were wild type (+/+). Affymetrix mouse 430 2.0 arrays for the two e17.0 C57BL/6 placental samples were normalized and modeled with the full set of arrays for our placenta timecourse. These arrays provide a standard with which to scale the placenta timecourse data to be comparable to the GeneAtlas data. For each gene, the ratio of the peak expression in the fetal timecourse to the expression in the e17.0 C57BL/6 placenta samples was calculated. This scaling factor indicates how the peak expression in the timecourse compares with the e17.0 C57BL/6 placentas when expression is measured using the same preparation protocol and the same microarray platform. The GeneAtlas e18.0 placenta value was then multiplied by the scaling factor, so that the GeneAtlas placenta datapoint now reflects the peak placental expression of the gene, rather than the expression at e18.0. This was done for the fetal placenta and maternal placenta timecourses separately, so that the modified GeneAtlas data set includes a peak fetal placenta expression level for each gene and a peak maternal placenta expression level for each gene.
The measure of expression specificity (PlacPEM) used in these analyses is based on the average expression across all tissue types for each gene. In order to avoid weighting of the average by heavily represented tissue types, such as brain (which is represented by 12 different samples in the mouse GeneAtlas data set), the 61 tissue types were grouped according to organ or tissue type of origin. In addition, samples without clear tissue type categories were eliminated. This yielded a total of 56 samples pooled into 32 groups for the scaled mouse data set (including the scaled fetal and maternal placental samples) and 55 samples pooled into 31 groups for the unscaled, original mouse data set (including the single e18.0 GeneAtlas placental sample). For each of these groups, a group expression value was calculated by taking the average expression within that group. The average expression level for each gene was calculated by taking the average of the 32 group expression values. The PlacPEM for a gene is the placenta expression value divided by this average expression value.
The human GeneAtlas data set was normalized and modeled using dCHIP (Li and Wong 2001b) as described above for the mouse data. The human data set includes 44,775 probe sets and 79 tissues. To maintain consistency with the procedure used for the mouse, this GeneAtlas data was mapped onto the Affymetrix human whole genome microarray (HG-U133 Plus 2.0). Of the 54,613 probe sets on the HG-U133 Plus 2.0 array, 40,741 were mapped to corresponding GNF IDs using Entrez Gene annotations provided by Novartis and Affymetrix; the 13,872 probe sets that were not mapped to GNF IDs either were not annotated to Entrez Gene IDs themselves, or had no corresponding gene in the GeneAtlas data set. As with the mouse data, each human GeneAtlas tissue sample has one replicate, meaning that each tissue is represented by two arrays. Again, all analyses were conducted using the dCHIP weighted average expression values for each tissue. Expression specificity was calculated as described for the mouse data set above, using a similar grouping strategy to avoid weighting the average toward over-represented tissues types. After eliminating pathological samples, cell line samples, and other samples without clear tissue-type categories, the final data set contained a total of 50 samples pooled into 27 groups. The placenta sample in this data set is the original human GeneAtlas sample, representing normal term human placenta. In mapping from the human GNF data set to the Affymetrix GH-U133 Plus 2.0 IDs, the association of some GH-U133 Plus 2.0 with multiple Entrez IDs allowed the possibility of redundancy among output genes. To ensure that this did not affect the results, the three possibly redundant probe sets that have primate origins were removed from the human list and primate-specific enrichment was reassessed. Based on this modified list, the trend toward increased primate-specific genes in the mature human placenta is maintained (data not shown).
Determining closest one-to-one ortholog with Homologene and Ensembl
Evolutionary origins were determined using (1) Homologene build 49.1 (mouse and human) and (2) Ensembl version 39 (mouse) and 40 (human). In both cases, only one-to-one, reciprocal best-match orthologs were considered. For each gene, the one-to-one, reciprocal best-match ortholog found in the most distantly related species was used to classify the gene by evolutionary origin. For the mouse, using Homologene, genes with no one-to-one best-reciprocal ortholog or with the most distant one-to-one best-reciprocal ortholog in Mus musculus or Rattus norvegicus were classified as having rodent orthology. Similarly, genes with one-to-one best reciprocal orthologs found in Canis familiaris, Pan troglodytes, or Homo sapiens were classified as having mammalian orthology; Galluss gallus as vertebrate orthology; Caenorhabditis elegans, Anopheles gambiae, or Drosophila melanogaster as animal orthology; and Oryza sativa, Arabidopsis thaliana, Plasmodium falciparum, Neurospora crassa, Eremothecium gossypii, Magnaporthe grisea, Schizosaccharomyces pombe, Kluyveromyces lactis, or Saccharomyces cerevisiae as eukaryotic orthology. Using Ensembl, genes with a one to one best ortholog in M. musculus or R. norvegicus were classified as having rodent orthology; H. sapiens, P. troglodytes, Macaca mulatta, C. familiaris, or Bos taurus as mammalian orthology; G. gallus, Xenopus tropicalis, Danio rerio, Tetraodon nigroviridis, or Takifugu rubripes as vertebrate orthology; C. elegans, A. gambiae, D. melanogaster, Ciona savignyi, or Ciona intestinalis as animal orthology; and S. cerevisiae as eukaryotic orthology. In determining human gene orthologs, the same classifications were used except that (1) in Homologene, genes with one-to-one best-reciprocal orthologs found in P. troglodytes (chimp) and H. sapiens were classified as having primate orthology and R. norvegicus and M. musculus as mammalian orthology and (2) in Ensembl, genes with one-to-one best reciprocal orthologs found in H. sapiens, P. troglodytes (chimp), and M. mulatta (rhesus) were classified as having primate orthology and R. norvegicus and M. musculus as mammalian orthology. Here “rodent specific” and “primate specific” are estimations based on the available data, referring to genes classified as having rodent orthology and primate orthology, respectively.
Gene Ontology term enrichment
Over-representation of Gene Ontology Biological Process terms was conducted using the NIH DAVID tool (Dennis et al. 2003). Over-representation was calculated using custom background lists. For the fetal and maternal placenta 1.5-fold clusters, the background list included all genes present on the Affymetrix mouse 430 2.0 array, with duplicates removed. For each of the clusters of genes requiring PlacPEM ≥ 4 (scaled mouse, mouse GeneAtlas only, and human GeneAtlas only), the background list included all genes for which any tissue has a PEM ≥ 4, with duplicates removed. The P-values reported for over-representation were adjusted using the EASE score method, which is a more conservative variation of the Fisher exact score. In addition, a highly conservative significance threshold (P < 3.45 × 10–6) was calculated based on a Bonferroni correction for multiple testing (accounting for testing of ∼14,500 Gene Ontology Biological Process terms). Given the conservative nature of the Bonferroni correction and the utility of the classifications in characterizing the gene sets, several categories not achieving this high threshold for significance are included in the text, with corresponding P-values.
Acknowledgments
We thank Mark Krasnow and Hernan Espinoza for providing the cDNA clones used in assessing cell type-specific expression patterns of placental genes. We also thank Robert Tibshirani for helping us with statistical and technical issues related to the SAM program, and Si Wan Kim and Rakhi Gupta for helping with qPCR verification. We thank Greg Barsh, Josh Stuart, Gill Berjerano, and Eric Chiao for support, advice, and helpful discussions. This work was supported by NIH (R01 HD41557), the March of Dimes, and the Stanford University Medical Scientist Training Program.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.071407.107.
References
- Ain R., Dai G., Dunmore J.H., Godwin A.R., Soares M.J., Dai G., Dunmore J.H., Godwin A.R., Soares M.J., Dunmore J.H., Godwin A.R., Soares M.J., Godwin A.R., Soares M.J., Soares M.J. A prolactin family paralog regulates reproductive adaptations to a physiological stressor. Proc. Natl. Acad. Sci. 2004;101:16543–16548. doi: 10.1073/pnas.0406185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S.M., Ain R., Konno T., Ho-Chen J.K., Soares M.J., Ain R., Konno T., Ho-Chen J.K., Soares M.J., Konno T., Ho-Chen J.K., Soares M.J., Ho-Chen J.K., Soares M.J., Soares M.J. The rat prolactin gene family locus: Species-specific gene family expansion. Mamm. Genome. 2006;17:858–877. doi: 10.1007/s00335-006-0010-1. [DOI] [PubMed] [Google Scholar]
- Beauchemin N., Draber P., Dveksler G., Gold P., Gray-Owen S., Grunert F., Hammarstrom S., Holmes K.V., Karlsson A., Kuroki M., Draber P., Dveksler G., Gold P., Gray-Owen S., Grunert F., Hammarstrom S., Holmes K.V., Karlsson A., Kuroki M., Dveksler G., Gold P., Gray-Owen S., Grunert F., Hammarstrom S., Holmes K.V., Karlsson A., Kuroki M., Gold P., Gray-Owen S., Grunert F., Hammarstrom S., Holmes K.V., Karlsson A., Kuroki M., Gray-Owen S., Grunert F., Hammarstrom S., Holmes K.V., Karlsson A., Kuroki M., Grunert F., Hammarstrom S., Holmes K.V., Karlsson A., Kuroki M., Hammarstrom S., Holmes K.V., Karlsson A., Kuroki M., Holmes K.V., Karlsson A., Kuroki M., Karlsson A., Kuroki M., Kuroki M., et al. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 1999;252:243–249. doi: 10.1006/excr.1999.4610. [DOI] [PubMed] [Google Scholar]
- Carroll S.B. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.M., Mess A., Mess A. Evolution of the placenta in eutherian mammals. Placenta. 2006;28:259–262. doi: 10.1016/j.placenta.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Gao W., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Deussing J., Kouadio M., Rehman S., Werber I., Schwinde A., Peters C., Kouadio M., Rehman S., Werber I., Schwinde A., Peters C., Rehman S., Werber I., Schwinde A., Peters C., Werber I., Schwinde A., Peters C., Schwinde A., Peters C., Peters C. Identification and characterization of a dense cluster of placenta-specific cysteine peptidase genes and related genes on mouse chromosome 13. Genomics. 2002;79:225–240. doi: 10.1006/geno.2002.6696. [DOI] [PubMed] [Google Scholar]
- Faria T.N., Ogren L., Talamantes F., Linzer D.I., Soares M.J., Ogren L., Talamantes F., Linzer D.I., Soares M.J., Talamantes F., Linzer D.I., Soares M.J., Linzer D.I., Soares M.J., Soares M.J. Localization of placental lactogen-I in trophoblast giant cells of the mouse placenta. Biol. Reprod. 1991;44:327–331. doi: 10.1095/biolreprod44.2.327. [DOI] [PubMed] [Google Scholar]
- Futuyma D.J. Evolution. Sinauer Associates; Sunderland, MA: 2005. [Google Scholar]
- Gilbert S.F. Developmental biology. Sinauer Associates; Sunderland, MA: 2006. [Google Scholar]
- Hemberger M., Hughes M., Cross J.C., Hughes M., Cross J.C., Cross J.C. Trophoblast stem cells differentiate in vitro into invasive trophoblast giant cells. Dev. Biol. 2004;271:362–371. doi: 10.1016/j.ydbio.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Lee C.T., Risom T., Strauss W.M., Risom T., Strauss W.M., Strauss W.M. Evolutionary conservation of microRNA regulatory circuits: An examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007;26:209–218. doi: 10.1089/dna.2006.0545. [DOI] [PubMed] [Google Scholar]
- Li C., Wong W.H., Wong W.H. Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biol. 2001a;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wong W.H., Wong W.H. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. 2001b;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzer D.I., Fisher S.J., Fisher S.J. The placenta and the prolactin family of hormones: Regulation of the physiology of pregnancy. Mol. Endocrinol. 1999;13:837–840. doi: 10.1210/mend.13.6.0286. [DOI] [PubMed] [Google Scholar]
- Linzer D.I., Lee S.J., Ogren L., Talamantes F., Nathans D., Lee S.J., Ogren L., Talamantes F., Nathans D., Ogren L., Talamantes F., Nathans D., Talamantes F., Nathans D., Nathans D. Identification of proliferin mRNA and protein in mouse placenta. Proc. Natl. Acad. Sci. 1985;82:4356–4359. doi: 10.1073/pnas.82.13.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R.W., Stabley D.L., Picerno G.N., Frenck J., Xing S., Bertenshaw G.P., Sol-Church K., Stabley D.L., Picerno G.N., Frenck J., Xing S., Bertenshaw G.P., Sol-Church K., Picerno G.N., Frenck J., Xing S., Bertenshaw G.P., Sol-Church K., Frenck J., Xing S., Bertenshaw G.P., Sol-Church K., Xing S., Bertenshaw G.P., Sol-Church K., Bertenshaw G.P., Sol-Church K., Sol-Church K. Evolution of placental proteases. Biol. Chem. 2002;383:1113–1118. doi: 10.1515/BC.2002.120. [DOI] [PubMed] [Google Scholar]
- McCarroll S.A., Murphy C.T., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Murphy C.T., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Kenyon C., Bargmann C.I., Li H., Bargmann C.I., Li H., Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- McLellan A.S., Fischer B., Dveksler G., Hori T., Wynne F., Ball M., Okumura K., Moore T., Zimmermann W., Fischer B., Dveksler G., Hori T., Wynne F., Ball M., Okumura K., Moore T., Zimmermann W., Dveksler G., Hori T., Wynne F., Ball M., Okumura K., Moore T., Zimmermann W., Hori T., Wynne F., Ball M., Okumura K., Moore T., Zimmermann W., Wynne F., Ball M., Okumura K., Moore T., Zimmermann W., Ball M., Okumura K., Moore T., Zimmermann W., Okumura K., Moore T., Zimmermann W., Moore T., Zimmermann W., Zimmermann W. Structure and evolution of the mouse pregnancy-specific glycoprotein (Psg) gene locus. BMC Genomics. 2005a;6:4. doi: 10.1186/1471-2164-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan A.S., Zimmermann W., Moore T., Zimmermann W., Moore T., Moore T.2005bConservation of pregnancy-specific glycoprotein (PSG) N domains following independent expansions of the gene families in rodents and primates BMC Evol. Biol. 539 . 10.1186/1471-2148-5-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mess A., Carter A.M., Carter A.M. Evolutionary transformations of fetal membrane characters in Eutheria with special reference to Afrotheria. J. Exp. Zoolog. B Mol. Dev. Evol. 2006;306:140–163. doi: 10.1002/jez.b.21079. [DOI] [PubMed] [Google Scholar]
- Mitiku N., Baker J.C., Baker J.C. Genomic analysis of gastrulation and organogenesis in the mouse. Dev. Cell. 2007;13:897–907. doi: 10.1016/j.devcel.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Montuenga L.M., Mariano J.M., Prentice M.A., Cuttitta F., Jakowlew S.B., Mariano J.M., Prentice M.A., Cuttitta F., Jakowlew S.B., Prentice M.A., Cuttitta F., Jakowlew S.B., Cuttitta F., Jakowlew S.B., Jakowlew S.B. Coordinate expression of transforming growth factor-1 and adrenomedullin in rodent embryogenesis. Endocrinology. 1998;139:3946–3957. doi: 10.1210/endo.139.9.6205. [DOI] [PubMed] [Google Scholar]
- Mossman H.W. Vertebrate fetal membranes: Comparative ontogeny and morphology, evolution, phylogenetic significance, basic functions, research opportunities. Rutgers University Press; New Brunswick, NJ: 1987. [Google Scholar]
- Niwa R., Slack F.J., Slack F.J. The evolution of animal microRNA function. Curr. Opin. Genet. Dev. 2007;17:145–150. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Ramsey E.M. The placenta: Human and animal. Praeger; New York: 1982. [Google Scholar]
- Rossant J., Cross J.C., Cross J.C. Placental development: Lessons from mouse mutants. Nat. Rev. Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Soares M.J. The prolactin and growth hormone families: Pregnancy-specific hormones/cytokines at the maternal-fetal interface. Reprod. Biol. Endocrinol. 2004;2:51. doi: 10.1186/1477-78267-2-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sol-Church K., Picerno G.N., Stabley D.L., Frenck J., Xing S., Bertenshaw G.P., Mason R.W., Picerno G.N., Stabley D.L., Frenck J., Xing S., Bertenshaw G.P., Mason R.W., Stabley D.L., Frenck J., Xing S., Bertenshaw G.P., Mason R.W., Frenck J., Xing S., Bertenshaw G.P., Mason R.W., Xing S., Bertenshaw G.P., Mason R.W., Bertenshaw G.P., Mason R.W., Mason R.W. Evolution of placentally expressed cathepsins. Biochem. Biophys. Res. Commun. 2002;293:23–29. doi: 10.1016/S0006-291X(02)00167-5. [DOI] [PubMed] [Google Scholar]
- Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Vega R.G., Sapinoso L.M., Moqrich A., Sapinoso L.M., Moqrich A., Moqrich A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiler K. The house mouse: Atlas of embryonic development. Springer-Verlag; New York: 1989. [Google Scholar]
- True J.R., Carroll S.B., Carroll S.B. Gene co-option in physiological and morphological evolution. Annu. Rev. Cell Dev. Biol. 2002;18:53–80. doi: 10.1146/annurev.cellbio.18.020402.140619. [DOI] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G., Tibshirani R., Chu G., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemers D.O., Shao L.J., Ain R., Dai G., Soares M.J., Shao L.J., Ain R., Dai G., Soares M.J., Ain R., Dai G., Soares M.J., Dai G., Soares M.J., Soares M.J. The mouse prolactin gene family locus. Endocrinology. 2003;144:313–325. doi: 10.1210/en.2002-220724. [DOI] [PubMed] [Google Scholar]
- Wildman D.E., Chen C., Erez O., Grossman L.I., Goodman M., Romero R., Chen C., Erez O., Grossman L.I., Goodman M., Romero R., Erez O., Grossman L.I., Goodman M., Romero R., Grossman L.I., Goodman M., Romero R., Goodman M., Romero R., Romero R. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc. Natl. Acad. Sci. 2006;103:3203–3208. doi: 10.1073/pnas.0511344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotsumoto S., Shimada T., Cui C.Y., Nakashima H., Fujiwara H., Ko M.S., Shimada T., Cui C.Y., Nakashima H., Fujiwara H., Ko M.S., Cui C.Y., Nakashima H., Fujiwara H., Ko M.S., Nakashima H., Fujiwara H., Ko M.S., Fujiwara H., Ko M.S., Ko M.S. Expression of adrenomedullin, a hypotensive peptide, in the trophoblast giant cells at the embryo implantation site in mouse. Dev. Biol. 1998;203:264–275. doi: 10.1006/dbio.1998.9073. [DOI] [PubMed] [Google Scholar]
- Zebhauser R., Kammerer R., Eisenried A., McLellan A., Moore T., Zimmermann W., Kammerer R., Eisenried A., McLellan A., Moore T., Zimmermann W., Eisenried A., McLellan A., Moore T., Zimmermann W., McLellan A., Moore T., Zimmermann W., Moore T., Zimmermann W., Zimmermann W. Identification of a novel group of evolutionarily conserved members within the rapidly diverging murine Cea family. Genomics. 2005;86:566–580. doi: 10.1016/j.ygeno.2005.07.008. [DOI] [PubMed] [Google Scholar]