Abstract
Meiosis-specific homologs of RecA protein have been identified in Saccharomyces cerevisiae and higher eukaryotes including mammals, but their enzymatic activities have not been described. We have purified the human protein HsDmc1 produced in Escherichia coli from a cloned copy of the cDNA. The recombinant enzyme had DNA-dependent ATPase activity with an estimated kcat of 1.5 min−1. DNase protection experiments with oligonucleotides as substrates indicated that HsDmc1 protein binds preferentially to single-stranded DNA with a stoichiometry of approximately one molecule of protein per three nucleotide residues. HsDmc1 protein catalyzed the formation of D-loops in superhelical DNA, as well as strand exchange between single-stranded and double-stranded oligonucleotides. The requirements for strand exchange catalyzed by HsDmc1 were similar to those of RecA protein, but exchange caused by HsDmc1 was not supported by ATPγS.
Keywords: homologous recombination, meiotic recombination, RecA homologs, D-loop formation, strand exchange
Eukaryotic homologs of the Escherichia coli recombination protein RecA were first discovered in Saccharomyces cerevisiae where certain members of the RAD52 epistasis group, RAD51 and RAD57 were found to have significant homology to the recA gene (1–4). DMC1, a gene that is specifically expressed in meiosis was also found to encode a homolog of RecA protein (5). Genes that are homologous to recA are now known to be widely distributed in both prokaryotes and eukaryotes, thus constituting a class of protein that plays central roles in DNA recombination and repair (6, 7). Studies of the biological functions of recA homologs in eukaryotes are revealing additional complex roles of these proteins (see below).
The genes RAD51 and DMC1 represent phylogenetically distinct subclasses of the gene family (8). In the mouse, and in man, homologs of both RAD51 and DMC1 have been found. Full-length homologs of DMC1 are expressed only in meiotic tissues, but in the mouse a second shortened form is expressed in somatic tissues (6, 9–11).
In baker’s yeast, RAD51 is important for double-strand break repair in mitotic cells. In meiosis, rad51 and dmc1 mutants have very similar phenotypes: both are essential for meiotic recombination, and both are essential for completion of the meiotic cell cycle leading to production of viable spores (1, 5). In yeast meiosis, Rad51 and Dmc1 proteins colocalize on synaptonemal complexes in an apparently regulated order in which Rad51 is required for the formation of complexes of Dmc1 (12).
Completion of the meiotic cell cycle in yeast is one example of a complex role of recA homologs in eukaryotes. In this case, the failure of meiosis presumably results from a defect in recombination. In S. cerevisiae, rad51 mutants are viable, whereas in the mouse, homozygous rad51 mutants are lethal during embryogenesis (13, 14). The basis for this complex role in viability remains to be determined. In mammalian cells, damage to DNA results in the organization of Rad51 into multiple foci in the nucleus without a detectable increase in net synthesis (15), whereas increased production of Rad51 is associated with the induction of Ig class switching (16).
Previous studies have shown that Rad51 protein from S. cerevisiae and man make nucleoprotein filaments that resemble the filament made by RecA (17–19). Strand exchange activities have been reported for Rad51 from S. cerevisiae, Xenopus laevis, and Homo sapiens (20–23), but none has been reported for Dmc1. Other studies show that both Rad51 and Dmc1 associate with synaptonemal complexes in the mouse; but according to the location and time of appearance of the two proteins during meiosis, Rad51 and Dmc1 appear to play different roles (24, 25). An understanding of the enzymology of Dmc1 is important for the further understanding of its role in meiosis. In the present report, we describe the purification of human Dmc1 protein (denoted HsDmc1 for H. sapiens Dmc1) and demonstrate that it has several recombination activities.
MATERIALS AND METHODS
Enzymes and Other Reagents.
RecA protein was purified as described (26). ATP and adenosine 5′-[γ-thio]triphosphate (ATPγS) were purchased from Sigma; T4 polynucleotide kinase, T4 DNA ligase and restriction enzymes were purchased from New England Biolabs (Beverly, MA); DNase I, DTT, BSA, and an Expand High Fidelity PCR kit were purchased from Boehringer Mannheim. Vector pQE-30 was obtained from Qiagen (Chatsworth, CA). Expression vector pET-29a(+) and E. coli K12 strain NovaBlue(DE3) were from Novagen (Madison, WI).
Five 83-mer oligonucleotides were synthesized on an Applied Biosystems DNA synthesizer. One, W16(−): 5′-TTGATAAGAGGTCATTTTTGCGGATGGCTTAGAGCTTAATTGCTGAATCTGGTGCTGTAGCTCAACATGTTTTAAATATGCAA, is complementary to the viral strand of phage M13 DNA (23). Four others, which had no known sequence homology to M13 DNA contained 84% AT bp: A16(−): 5′-AAATGAACATAAAGTAAATAAGTATAAGGATAATACAAAATAAGTAAATGAATAAACATAGAAAATAAAGTAAAGGATATAAA and its complement, A16(+); ATII(−): 5′-ATGTATATTGATATATTGATTAGTATTAGTTATTGTTATGTTTAGTTTATTT-CTTGATTTGATTATTACTTTGTATTATAGAT, and its complement, ATII(+).
Oligonucleotides were labeled with 32P at their 5′ ends in reactions with T4 polynucleotide kinase, or at their 3′ ends by reactions with terminal transferase (27). Duplex oligonucleotides were prepared by annealing as described (28), and examined by electrophoresis on nondenaturing 12% polyacrylamide gels to confirm complete annealing. All DNA concentrations refer to molecules of nucleotide residues.
Cloning of the HsDmc1 Gene.
The entire coding sequence of HsDmc1 protein was amplified by the PCR from a human testis cDNA library (in bacteriophage λ). Sequences of the upstream and downstream primers were CGCGGATCCATGAAGGAGGATCAAGTTGTG (oligonucleotide EG264) and CGGGGTACCACCTACTCCTTGGCATC (oligonucleotide EG266). Underlined sequences are homologous to the published sequence of the HsDmc1 gene (11). The PCR reaction was carried out by using an Expand High Fidelity PCR kit (Boehringer Mannheim). The reaction mixture was heated at 95°C for 3 min and used in PCR consisting of five cycles at 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min, followed by 30 cycles at 94°C for 1 min and 69°C for 2 min. The resulting DNA fragment was inserted into pQE-30 plasmid by use of BamHI and KpnI restriction sites. The construct was used as a template for a PCR reaction with upstream and downstream primers EG265 (GGAATTCCATATGAAGGAGGATCAAGTTGTC) and EG266 (see above). The second PCR reaction was similar to the first one, but 10 cycles at 74°C for 1 min and 69°C for 2 min were used instead of 30. The amplified DNA fragment was inserted into expression vector pET-29a(+)(see above) by use of restriction sites NdeI and KpnI to make plasmid pEG1019. The choice of primers and restriction enzymes enabled us to make HsDmc1 protein without any tags which are otherwise encoded by the vector. The sequence of our cloned cDNA for HsDmc1 was identical to the sequence published by Habu et al. (11) except at position 110 where there was T in place of A, as in the sequence of Sato et al. (10). This variation in the sequence, which substitutes isoleucine for aspartic acid, is in a nonconserved region of the gene.
Production of HsDmc1 Protein in E. coli.
Plasmid pEG1019 was introduced in E. coli NovaBlue(DE3), which is recA1−, a null mutant. Bacteria from a single colony of the transformants, which were grown no longer than 12 h on Luria–Bertani broth plates containing 25 μg kanamycin/ml, were used to inoculate 50 ml of Luria–Bertani broth containing 125 μg kanamycin/ml, and 12.5 μg/ml tetracycline. Bacteria grown to stationary phase with shaking at 37°C were used to inoculate 12 l of fresh medium. This culture was grown at 37°C to an optical density of 0.5, at 595 nm, in 12 l of Luria–Bertani broth, containing 25 μg kanamycin/ml, and induced by the addition of isopropyl β-d-thiogalactoside, at 1 mM, followed by a further incubation for 2 h at 37°C. Bacteria were harvested by centrifugation and resuspended in 200 ml of 50 mM Tris⋅HCl, (pH 7.5), and 10% sucrose.
Purification of HsDmc1 Protein.
Cells were lysed by the addition of the following reagents at the indicated final concentrations: 0.8 mg lysozyme/ml, 10 mM DTT, 1 mM EDTA and 0.5 mM phenylmethylsulfonyl fluoride. The mixture was stirred for 30 min. Brij 58 at 0.5% and KCl at 500 mM were added, followed by stirring for another 30 min. Cell debris and insoluble materials were removed by centrifugation at 35,000 rpm for 75 min in a Beckman 45Ti rotor.
After precipitation by 50% ammonium sulfate, the pellets obtained were resuspended in 20 ml of P buffer: 20 mM potassium phosphate (pH 7.0), 1 mM EDTA, 10 mM 2-mercaptoethanol, and 10% glycerol. The 38-kDa protein was further purified by successive column chromatography on Q Sepharose, Sephacryl 200, MonoQ, native DNA cellulose, and single-stranded DNA (ssDNA) cellulose.
ATPase Activity.
Reaction mixtures (20 μl) contained circular single-stranded M13 DNA (50 μM), 30 mM Tris⋅HCl (pH 7.5), 1 mM DTT, 1 mM ATP, 100 μg BSA/ml, 6 mM MgCl2, and 3 μM HsDmc1 protein. Each reaction contained 1 μCi [3H]ATP (1 Ci = 37 GBq). Incubation was at 37°C, and in samples taken at different times, the reaction was stopped by adding 10 mM EDTA and 1 mM of ADP and ATP. Samples of 5 μl were spotted directly onto thin layer chromatography paper (Polygram CEL 300, PEI, Brinkmann). The paper was developed in 1 M formic acid/0.5 M LiCl. The amount of ATP hydrolyzed was determined from the dried paper by scintillation counting.
DNase I Protection.
HsDmc1 protein or RecA protein (1 μM) was incubated at 37°C for 10 min with 3 μM 32P-labeled oligonucleotide A16(−) or duplex oligonucleotide A16(−)/A16(+) in a reaction mixture (20 μl) containing 1 mM MgCl2, 25 mM Pipes (pH 7.0), 1 mM DTT, 2 mM ATP, and 100 μg BSA/ml. The concentration of MgCl2 was brought to 10 mM, and DNase I was then added to the reaction mixture: 0.5 units for A16(−); 0.05 units for A16(−)/A16(+). (Units were as defined by the supplier, Boehringer Mannheim.) Incubation was continued at 37°C for another 5 min. The reaction was immediately quenched by addition of final concentrations of SDS at 0.5% and EDTA at 25 mM. Labeled DNA was precipitated by adding unlabeled carrier DNA at 0.1 mg/ml and cold trichloroacetic acid at 10% final concentrations. Acid-soluble radioactive material in the supernatant was measured by scintillation counting.
D-Loop Assay.
HsDmc1 protein or RecA protein (1 μM) was preincubated at 37°C for 10 min with 3 μM 32P-labeled 83-mer W16(−) in a reaction mixture (20 μl) containing 1 mM MgCl2, 25 mM Pipes (pH 7.0), 1 mM DTT, 2 mM ATP or ATPγS, and 100 μg BSA/ml. After preincubation, the concentration of MgCl2 was increased to 10 mM and M13 superhelical DNA was added at 50 μM. (Thus, W16 83-mer was present in 5-fold excess over homologous sites in M13 DNA.) At various times, aliquots were deproteinized at 37°C for 10 min in 0.1 mg proteinase K/ml and 0.005 mg SDS/ml. Samples were loaded on an 0.8% agarose gel and run at 3.3 V/cm for 1 h at room temperature in TBE buffer (45 mM Tris/45 mM boric acid, pH 8/0.001 M EDTA). The gel was dried and the reactions were quantitated by use of a PhosphorImager (Molecular Dynamics).
Strand Exchange.
Preincubation of HsDmc1 or RecA protein with 83-mer A16(−) was done as just described under D-loop assay. After 10 min, the concentration of MgCl2 was increased to 10 mM, followed by addition of homologous duplex oligonucleotide 5′-32P-A16(−)/A16(+) or 3′-32P-A16(−)/A16(+) at 3 μM final concentration. (Thus the single-stranded 83-mer was present in 2-fold molar excess over molecules of duplex 83-mer.) Incubation was continued at 37°C and stopped as described under “D-loop assay”. Oligonucleotide W16(−) served as a heterologous control. Samples were analyzed on a nondenaturing 12% polyacrylamide gel run at 8 V/cm for 2 h at room temperature in TBE buffer. Quantitation was done as described under D-loop assay.
RESULTS
Purified HsDmc1 Protein.
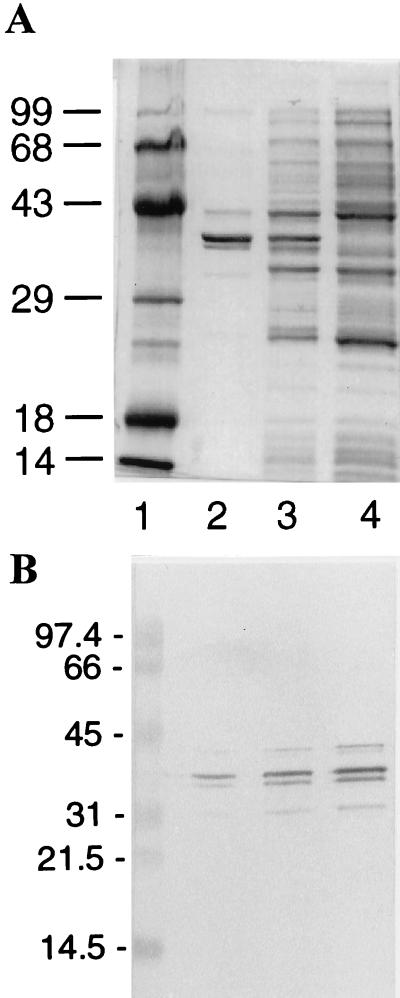
The sequence of the cloned cDNA from which we produced HsDmc1 was confirmed as described in Materials and Methods. Analysis by SDS/PAGE of fractions from ssDNA cellulose that contained about 1.3 mg of protein/ml revealed one major component corresponding to a protein with an Mr of 38 kDa (Fig. 1A). Western blot analysis indicated that two faint bands below the major band may be degradation products of Dmc1 protein (Fig. 1B). Antibodies raised against a peptide in the HsDmc1 sequence, from amino acids 186–196 inclusive, reacted with the purified protein by a blot test. Determination of the first 11 amino acid residues by microsequencing confirmed the identity of the purified protein with HsDmc1.
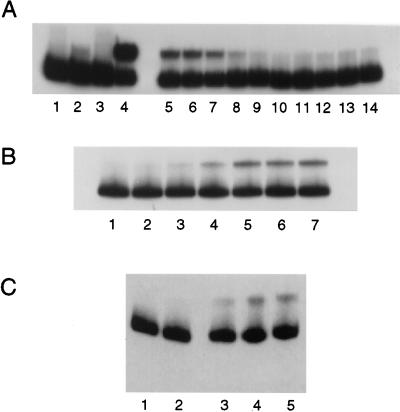
Figure 1.
(A) SDS/PAGE showing the overexpression and purification of HsDmc1 protein from E. coli. Lane 1, Mr standards with sizes indicated in kDa; lane 2, purified HsDmc1 protein; lanes 3 and 4, respectively, cellular proteins after and before induction with isopropyl-β-d-thiogalactoside. The 12% polyacrylaminde gel was stained with Coomassie blue. (B) Western blot analysis of purified protein. Dmc1 protein was visualized by using rabbit polyclonal anti-HsRad51 antibody that cross-reacts with Dmc1 protein but not with RecA protein. Lane 1, protein standards with sizes indicated in kDa. Lanes 2–4 were loaded, respectively, with 1.2, 5.0, and 12.0 ng of purified protein.
HsDmc1 was purified from a strain of E. coli bearing recA1, a null mutation (29). Separate endpoint titrations of antibody to RecA were done with purified RecA protein and with our preparation of purified Dmc1 protein. The titrations indicated that any contamination of HsDmc1 protein by inactive RecA1 protein did not exceed 3%, at most (data not shown).
In the preparation of Dmc1, exonuclease or endonuclease activity on ssDNA and double-stranded DNA digested <2% of DNA after incubations for 1 h at 37°C, and no separation of the strands of a duplex oligonucleotide was detected.
DNA-Dependent ATPase Activity.
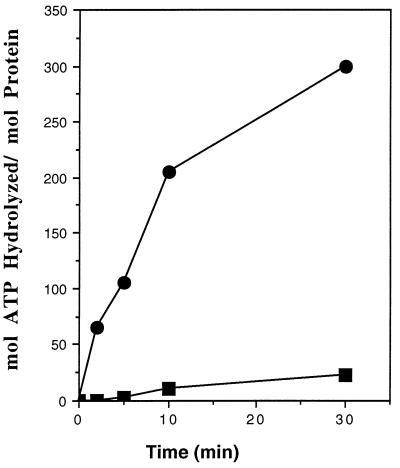
The ATPase activity of HsDmc1 protein compared with that of RecA protein is shown in Fig. 2. In the absence of DNA, HsDmc1 protein showed less than 15% ATPase activity compared with that in the presence of DNA. The turnover numbers (kcat) calculated from the initial slopes of the plots in Fig. 2 were ≈25 min−1 for RecA protein, and 1.5 min−1 for HsDmc1. A similarly low kcat has been observed for Rad51 protein from both S. cerevisiae and H. sapiens. (18, 19, 23).
Figure 2.
ssDNA-dependent ATPase activity of HsDmc1 compared with that of RecA. Reactions were carried out as described. ▪, HsDmc1 protein; •, RecA protein.
Binding to DNA.
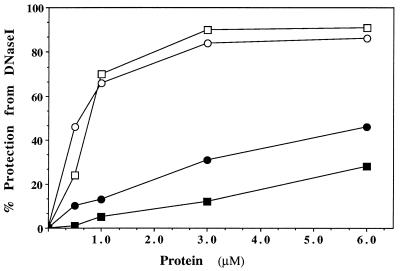
To assess the binding of HsDmc1 protein to ssDNA and double-stranded DNA, we examined the protection of oligonucleotides against digestion by DNase I (Fig. 3). In the same experiment, we compared the protection afforded by RecA protein. HsDmc1 and RecA protein were similarly effective in protecting a single-stranded 83-mer against DNase I, both in terms of the maximal protection afforded and the molar ratios of protein to DNA required for that degree of protection. By extrapolation, we estimate that both proteins gave maximal protection at one molecule of protein per two or three molecules of nucleotide residues, which compares well with the established 1:3 ratio for the binding of RecA protein to ssDNA. Neither HsDmc1 nor RecA protein protected a double-stranded oligonucleotide very well, but HsDmc1 was somewhat more effective than RecA. We conclude that HsDmc1 binds preferentially to ssDNA, with a stoichiometry of binding similar to that of RecA protein.
Figure 3.
DNase I protection of DNA bound to HsDmc1 or RecA. Reactions were done as described. The amount of DNase I, as determined by prior titration, was the least amount that would render acid-soluble all of the 32P label in 5 min in the absence of either HsDmc1 protein or RecA protein. □, protection of single-stranded 83-mer by RecA; ○, protection of single-stranded 83-mer by Dmc1; ▪, protection of double-stranded 83-mer by RecA; •, protection of double-stranded 83-mer by HsDmc1.
Formation of D-Loops in Superhelical DNA.
The nonenzymic uptake of a homologous single strand by superhelical DNA leads to the formation of a D-loop (30). Superhelicity is not required when the reaction is catalyzed by E. coli RecA protein. This reaction is the prototype of strand invasion, the invasion of duplex DNA by a homologous single strand (31, 32, 33). The ability of a single strand to invade duplex DNA is an essential feature of one of the two major pathways of homologous recombination (34). With the exception of E. coli RecO protein (35), the class of protein represented by RecA and its homologs remains the only one that can catalyze the formation of D-loops.
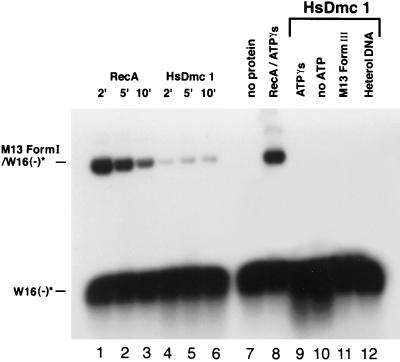
To explore the recombination activities of HsDmc1, we tested its ability to catalyze the formation of D-loops (Fig. 4). The duplex substrate was M13 superhelical DNA and the single-stranded substrate was a homologous 83-mer. For comparison, we examined the formation of D-loops from the same substrates by RecA protein. In 2 min, RecA formed D-loops in 60% of superhelical DNA molecules, after which the yield decreased as observed before (36). The yield of D-loops made by HsDmc1 was 12% at its maximum but showed no tendency to decrease during the 10 min of the reaction, unlike the pseudoreversible reaction promoted by RecA protein. In another experiment, the yield of D-loops made by Dmc1 increased up to 10 min, as in Fig. 4, and at 10 min reached a yield of 23% (data not shown).
Figure 4.
Formation of D-loops in superhelical DNA. HsDmc1 or RecA were present at a ratio of one molecule of protein per three nucleotide residues of single-stranded oligonucleotide. The latter, 32P-labeled W16(−) 83-mer, was present in 5-fold excess over homologous sites in M13 DNA. Other details of the reactions are described in Materials and Methods. Lanes 1–3, RecA at 2, 5, and 10 min, respectively; lanes 4–6, HsDmc1 at 2, 5, and 10 min, respectively; lane 7, no protein; lane 8, RecA with ATPγS; lane 9, Dmc1 with ATPγS; lane 10, HsDmc1 without ATP; lane 11, HsDmc1 with M13 linear duplex DNA (form III); lane 12, HsDmc1 with 83-mer A16(−) substituted for W16(−) as a heterologous control. A16(−), the oligonucleotide used as a heterologous control in this experiment was shown to be active as substrate when it was used with homologous duplex oligonucleotide in the strand exchange reaction shown in Fig. 5.
The formation of D-loops by Dmc1 required superhelical DNA, homology, and ATP. Unlike the reaction promoted by RecA protein, no formation of D-loops was detectable when ATPγS was substituted for ATP (Fig. 4).
Strand Exchange Promoted by HsDmc1.
We readily detected strand exchange by HsDmc1 when we used the A+T-rich oligonucleotide, A16(−), which contained 84% A+T base pairs (see Materials and Methods). The single-stranded oligonucleotide was reacted with the corresponding labeled homologous duplex oligonucleotide. Exchange was detected as the displacement of a labeled strand from a duplex oligonucleotide to the position of a single-stranded oligonucleotide (Fig. 5). Under the best conditions found for HsDmc1, the yield of exchanged product was only ≈20% of the maximum possible, compared with 61% for RecA protein in the same experiment.
Figure 5.
Strand exchange mediated by HsDmc1 protein. (A and B) Reactions were done with single-stranded 83-mer A16(−) and duplex 83-mer 5′-32P-A16(−)/A16(+), each at 3 μM. (C) The duplex 83-mer was 3′-32P-A16(−)/A16(+). (A) Requirements: lane 1, HsDmc1 but no ssDNA; lane 2, HsDmc1 without ATP; lane 3, HsDmc1 with W16(−) substituted for A16(−) as a heterologous control; lane 4, RecA substituted for HsDmc1 in a complete reaction; lanes 5–9, HsDmc1 with 5, 10, 20, 30, and 50 mM MgCl2, respectively; lanes 10–14, in the absence of HsDmc1 with 5, 10, 20, 30, and 50 mM MgCl2, respectively. W16(−), used as a heterologous control in this experiment, was shown to be active as substrate when it was used with homologous M13 DNA in the formation of D-loops as shown in Fig. 4. The bands are darker in A than in B and C below because the film for the autoradiogram was exposed for a longer time. (B) Titrations of Dmc1. As above, reactions contained 3 μM each of single-stranded and double-stranded oligonucleotides; other conditions were as described in Materials and Methods. Lane 1, no protein; lanes 2–7, with 0.125 μM, 0.25 μM, 0.5 μM, 1 μM, 2 μM, and 3 μM of HsDmc1 protein, respectively. (C) Time course and 3′-labeled substrate. In this experiment, the concentration of HsDmc1 was 1.3 μM and the concentration of 83-mer A16(−) was 3 μM. Lane 1, HsDmc1 omitted; Lanes 2–5, time course, zero time, 5 min, 20 min, and 60 min, respectively.
As indicated above, characterization of the purified preparation of HsDmc1 showed that contaminating exonuclease or endonuclease activity on ssDNA and double-stranded DNA digested <2% of DNA after incubations for 1 h at 37°C. In addition, in the absence of a homologous single strand, or in the presence of a heterologous single strand no separation of the strands of labeled duplex oligonucleotide was detected. To exclude further the possibility of apparent strand exchange due to contaminating exonuclease activity, we labeled the noncomplementary strand in the duplex oligonucleotide at its 5′ end, as in Fig. 5 A and B, or at its 3′ end (Fig. 5C). The yield of the reaction was similar whether the 5′ or 3′ end of the displaced strand was labeled, indicating further that conversion of the labeled strand from a duplex to a single-stranded form was not attributable to exonucleolytic digestion from either end of the duplex.
The time course of strand exchange is shown in Fig. 5C. Products first appeared at around 5 min, and the reaction reached completion at 20 min, with a yield of ≈20% at 20 and 60 min. By contrast, under similar conditions, RecA protein pushed strand exchange to completion in 1–2 min (23). Exchange promoted by HsDmc1 required homologous substrates and ATP. The reaction was optimal at 5–10 mM MgCl2 (Fig. 5A). In the experiment shown in Fig. 5B, exchange was optimal at a ratio of one molecule of HsDmc1 per three nucleotide residues of single-stranded oligonucleotide, but in later experiments optimal strand exchange required a 2-fold higher ratio of HsDmc1 protein to DNA. No exchange was detectable in the presence of ATPγS (data not shown).
In a previous study, we demonstrated the ability of HsRad51, another human homolog of RecA protein, to promote strand exchange with the same A+T-rich substrate as described here, oligonucleotide A16 (23). In the case of HsRad51 protein, exchange was also detectable with substrates that had 40% GC content (R.G. and C.M.R., unpublished observations), but we were unable to detect strand exchange catalyzed by HsDmc1 when the content of GC base pairs was 40% (data not shown). In the strand exchange reaction catalyzed by HsDmc1, a second, independently derived A+T-rich oligonucleotide, ATII(−) (see Materials and Methods) gave yields comparable to A16(−) (data not shown).
DISCUSSION
This report describes the partial purification and characterization of meiosis-specific human Dmc1 that was overproduced in E. coli. The purified protein, which was identified by microsequencing of its N terminus, and by immunological tests binds more readily to ssDNA than to double-stranded DNA. The complete protection of ssDNA from digestion by DNase I was optimal at a ratio of about one molecule of protein per three nucleotide residues, which corresponds to the stoichiometry for other proteins in the RecA class of proteins, and suggests that the preparation of recombinant HsDmc1 consists principally of protein molecules that were active in binding to DNA. The catalytic rate constant for the hydrolysis of ATP by HsDmc1 was at least an order of magnitude smaller than that of RecA protein, which is also true for the other eukaryotic members of the RecA family of proteins.
HsDmc1 promoted the formation of D-loops in superhelical DNA, a reaction that is fundamental to one of the major pathways of genetic recombination in which a single-stranded end of one molecule invades an intact homologous DNA molecule. The formation of D-loops in vitro is unlikely to be an artifact resulting from contaminating exonuclease activity since such an activity should only diminish the formation of D-loops (See below). The formation of D-loops by HsDmc1 differed in several important respects from the same reaction promoted by RecA protein. In addition to a much lower yield, the reaction promoted by HsDmc1 was much slower and failed to exhibit the pseudoreversibility characteristic of the RecA reaction that has been attributed to “processive unwinding” of superhelical DNA (37). Thus HsDmc1 may lack a similar ability to unwind DNA. Benson et al. (18) described evidence of a reduced ability of HsRad51 to unwind DNA.
Strand exchange was also promoted by HsDmc1. In this case, especially with oligonucleotides as substrates, the possibility of an artifact due to any contaminating exonuclease activity is a serious issue. Nibbling of an unlabeled strand of the duplex DNA from one end could result in creation of a single-stranded site from which annealing and spontaneous branch migration might be initiated. We showed, however, that the yield of product produced by strand exchange was the same whether the duplex substrate was labeled at the 5′ or 3′ end of the strand that is displaced, indicating that apparent strand exchange cannot be attributed to the action of a single contaminating exonuclease with a defined polarity of action. Direct assays for nuclease contamination showed further that the level of contamination was much lower than would be required to explain the observed degree of apparent strand exchange. The lack of apparent strand exchange in the absence of a homologous single strand argues against artifacts that might be created by unrelated combined exonuclease and helicase activity, which might conceivably lead to the conversion of some labeled duplex oligonucleotides into labeled single-stranded oligonucleotides.
The overall reaction leading to strand exchange was a slow reaction (data not shown), and under our conditions, neither the formation of D-loops, nor strand exchange was supported by the ATP analog ATPγS. A prominent characteristic of strand exchange promoted by HsDmc1 is its sensitivity to base composition. We detected strand exchange with two different oligonucleotide substrates that had only 16% GC content, but a substrate that had 40% GC content yielded no detectable product. This observation and the lack of processive unwinding of superhelical DNA (see above) may indicate a general decreased ability of HsDmc1 to unwind DNA, an aspect of its activity that is under further study.
In the case of RecA protein, the formation of D-loops is very rapid, and presumably reflects an early step in the overall reaction (Fig. 4 and ref. 37). The formation of D-loops by HsDmc1, by contrast, is much slower. In previous studies, observations on the interaction of fluorescent probes revealed that homologous pairing, the initial formation of homologously aligned joint molecules is much slower when catalyzed by human Rad51 protein than by RecA protein. Strand exchange itself, as detected specifically by the separation of contiguous fluorescent probes, is also very slow in the case of HsRad51 (23, 38), as are the overall reactions promoted by ScRad51 (20), HsRad51 (22), Xrad51 (21), and HsDmc1 (this paper, see above).
By comparison with RecA protein, the catalytic rate constant for the hydrolysis of ATP is also reduced by more than an order of magnitude for these four eukaryotic members of the RecA family (see above). Bedale and Cox (39) have recently presented data that support the idea that the hydrolysis of ATP by RecA protein is specifically linked to a late step in strand exchange. In the case of the eukaryotic homologs that have been studied, there appears to be a correlation between low rates of ATP hydrolysis and low rates of homologous pairing and strand exchange. The significance of this correlation, however, is challenged by the recent report that in the presence of spermidine and excess ScRad51 protein, homologous pairing and extensive strand exchange occur without ATP hydrolysis (40). It is conceivable that strand exchange in the absence of hydrolysis of ATP proceeds by a different mechanism.
The findings described here show that human Dmc1 protein has homologous pairing and strand exchange activities similar to those of the other eukaryotic homologs of RecA. Solutions to the quandaries about the meiosis-specific role of HsDmc1, and its relation to the role of Rad51, must be sought in future studies of interactions with other proteins and with particular DNA sequences. The strong dependence of the strand exchange activity of Dmc1 on base composition, for example, could make it dependent upon the functioning of other specific proteins, or could limit its function to special sites in chromosomal DNA. Such studies are essential to our understanding of meiosis-specific enzymic functions.
Acknowledgments
We are grateful to Jan Zulkeski for data processing. This research was sponsored by National Institutes of Health Grant 5R37 GM55304.
ABBREVIATIONS
- HsDmc1
human Dmc1 protein
- ATPγS
adenosine 5′-[γ-thio]triphosphate
- ssDNA
single-stranded DNA
References
- 1.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 2.Basile G, Aker M, Mortimer R K. Mol Cell Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kans J A, Mortimer R K. Gene. 1991;105:139–140. doi: 10.1016/0378-1119(91)90527-i. [DOI] [PubMed] [Google Scholar]
- 4.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 6.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 7.Bezzubova O, Shinohara A, Mueller R G, Ogawa H, Buerstedde J. Nucleic Acids Res. 1993;21:1577–1580. doi: 10.1093/nar/21.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stassen N Y, Logsdon J M, Jr, Vora G J, Offenberg H H, Palmer J D, Zolan M E. Curr Genet. 1997;31:144–157. doi: 10.1007/s002940050189. [DOI] [PubMed] [Google Scholar]
- 9.Sato S, Kobayashi T, Hotta Y, Tabata S. DNA Res. 1995;2:147–150. doi: 10.1093/dnares/2.3.147. [DOI] [PubMed] [Google Scholar]
- 10.Sato S, Seki N, Hotta Y, Tabata S. DNA Res. 1995;2:183–186. doi: 10.1093/dnares/2.4.183. [DOI] [PubMed] [Google Scholar]
- 11.Habu T, Taki T, West A, Nishimune Y, Morita T. Nucleic Acids Res. 1996;24:470–477. doi: 10.1093/nar/24.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop D K. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 13.Lim D, Hasty P. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haaf T, Golub E I, Reddy G, Radding C M, Ward D C. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M-J, Peakman M-C, Golub E I, Reddy G, Ward D C, Radding C M, Maizels N. Proc Natl Acad Sci USA. 1996;93:10222–10227. doi: 10.1073/pnas.93.19.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa T, Yu X, Shinohara A, Egelman E H. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 18.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung P, Robberson D L. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 20.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 21.Maeshima K, Morimatsu K, Horii T. Genes Cells. 1996;1:1057–1068. doi: 10.1046/j.1365-2443.1996.d01-224.x. [DOI] [PubMed] [Google Scholar]
- 22.Baumann P, Benson F, West S. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 23.Gupta R, Bazemore L R, Golub E I, Radding C M. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashley T, Plug A, Xu J, Solari A J, Reddy G, Golub E I, Ward D C. Chromosoma. 1995;104:19–28. doi: 10.1007/BF00352222. [DOI] [PubMed] [Google Scholar]
- 25.Terasawa M, Shinohara A, Hotta Y, Ogawa H, Ogawa T. Genes Dev. 1995;9:925–934. doi: 10.1101/gad.9.8.925. [DOI] [PubMed] [Google Scholar]
- 26.Shibata T, Cunningham R P, Radding C M. J Biol Chem. 1981;256:7557–7564. [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Rao B J, Chiu S K, Radding C M. J Mol Biol. 1993;229:328–343. doi: 10.1006/jmbi.1993.1038. [DOI] [PubMed] [Google Scholar]
- 29.Kowalczykowski S C. Biochimie. 1991;72:289–304. doi: 10.1016/0300-9084(91)90216-n. [DOI] [PubMed] [Google Scholar]
- 30.Holloman W K, Wiegand R, Hoessli C, Radding C M. Proc Natl Acad Sci USA. 1975;72:2394–2398. doi: 10.1073/pnas.72.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata T, DasGupta C, Cunningham R P, Radding C M. Proc Natl Acad Sci USA. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McEntee K, Weinstock G M, Lehman I R. Proc Nat Acad Sci USA. 1979;76:2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata T, DasGupta C, Cunningham R P, Radding C M. Proc Natl Acad Sci USA. 1980;77:2606–2610. doi: 10.1073/pnas.77.5.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haber J E. Curr Opin Cell Biol. 1992;4:401–412. doi: 10.1016/0955-0674(92)90005-w. [DOI] [PubMed] [Google Scholar]
- 35.Luisi-DeLuca C. J Bacteriol. 1995;177:566–572. doi: 10.1128/jb.177.3.566-572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata T, Ohtani T, Iwabuchi M, Ando T. J Biol Chem. 1982;257:13981–13986. [PubMed] [Google Scholar]
- 37.Shibata T, Makino O, Ikawa S, Ohtani T, Iwabuchi M, Shibata Y, Maeda H, Ando T. Cold Spring Harbor Symp Quant Biol. 1984;49:541–551. doi: 10.1101/sqb.1984.049.01.061. [DOI] [PubMed] [Google Scholar]
- 38.Bazemore L R, Takahashi M, Radding C M. J Biol Chem. 1997;272:14672–14682. doi: 10.1074/jbc.272.23.14672. [DOI] [PubMed] [Google Scholar]
- 39.Bedale W A, Cox M M. J Biol Chem. 1996;271:5725–5732. doi: 10.1074/jbc.271.10.5725. [DOI] [PubMed] [Google Scholar]
- 40.Sung P, Stratton S A. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]