Abstract
Drosophila Mad proteins are intracellular signal transducers of decapentaplegic (dpp), the Drosophila transforming growth factor β (TGF-β)/bone morphogenic protein (BMP) homolog. Studies in which the mammalian Smad homologs were transiently overexpressed in cultured cells have implicated Smad2 in TGF-β signaling, but the physiological relevance of the Smad3 protein in signaling by TGF-β receptors has not been established. Here we stably expressed Smad proteins at controlled levels in epithelial cells using a novel approach that combines highly efficient retroviral gene transfer and quantitative cell sorting. We show that upon TGF-β treatment Smad3 becomes rapidly phosphorylated at the SSVS motif at its very C terminus. Either attachment of an epitope tag to the C terminus or replacement of these three serine residues with alanine abolishes TGF-β-induced Smad3 phosphorylation; these proteins act in a dominant-negative fashion to block the antiproliferative effect of TGF-β in mink lung epithelial cells. A Smad3 protein in which the three C-terminal serines have been replaced by aspartic acids is also a dominant inhibitor of TGF-β signaling, but can activate plasminogen activator inhibitor 1 (PAI-1) transcription in a ligand-independent fashion when its nuclear localization is forced by transient overexpression. Phosphorylation of the three C-terminal serine residues of Smad3 by an activated TGF-β receptor complex is an essential step in signal transduction by TGF-β for both inhibition of cell proliferation and activation of the PAI-1 promoter.
Members of the transforming growth factor β (TGF-β) superfamily of cytokines regulate many important cellular processes, including proliferation, differentiation, and development. TGF-β transmits its signals through heteromeric complexes of types I and II transmembrane serine/threonine kinase receptors (reviewed in refs. 1–3). In the absence of TGF-β the type II receptor kinase is active and undergoes autophosphorylation on at least three serine residues that regulate receptor activity (4). Binding of TGF-β1 to the type II receptor induces hetero-oligomerization with type I receptors and trans-phosphorylation of the GS domain of the type I receptor by the type II receptor kinase (5–7).
In Drosophila, genetic screening for genes required for decapentaplegic (dpp) function has led to the identification of Mothers against dpp (Mad) as a downstream signal transduction protein (8, 9). Mad-related proteins have been found in Caenorhabditis elegans, Xenopus, mouse, and humans (10–22). Seven mammalian Mad-related proteins, Smad1–7, have been identified (23–25). Smad proteins contain evolutionary-conserved regions at the N and C termini that are highly similar among all members of this family.
Smad1 is phosphorylated and translocated to the nucleus following bone morphogenic protein (BMP) stimulation (11, 21). Smad2 is phosphorylated directly by TGF-β receptors at its C-terminal serine residues (26) and moves to the nucleus upon ligand addition (22). Xmadr2 (Xenopus Smad2) overexpression resembled the dorsalizing effects of activin in Xenopus (17, 20). Smad2 interacts transiently with the type I receptors for both TGF-β and activin when overexpressed in COS cells and is a direct substrate of both receptors in vitro; phosphorylation of Smad2 is required for its nuclear accumulation in COS cells and induction of transcription (26). Overexpression of Smad2 with Smad4, or Smad3 with Smad4, induces TGF-β-independent activation of the normally TGF-β-responsive plasminogen activator inhibitor 1 (PAI-1) promoter (16, 27–29). Smad4 is essential for the function of Smad1 and Smad2 in pathways that signal mesoderm induction and patterning in Xenopus embryos. Smad4 also associates with Smad1 in response to BMP and with Smad2 in response to activin or TGF-β. Smad4 is therefore a regulated partner of Smads that functions in different signaling pathways of members of the TGF-β family (27, 29).
When fused to the GAL4 DNA binding domain, Smad proteins can activate transcription of a reporter gene (21). Recently the Xenopus winged-helix transcription factor FAST-1 was cloned and shown to complex with activated Xmad2 (Smad2) and participate in transcription of the Mix.2 gene, an immediate-early response gene specific to activin-like members of the TGF-β superfamily (15). The current model, therefore, is that activated type I BMP receptors bind to and phosphorylate Smad1 on serine residues at its very C-terminus, whereas activated type I TGF-β and/or activin receptors phosphorylate Smad2 and/or possibly Smad3. Phosphorylated Smad1, Smad2, or Smad3 then bind with Smad4, move to the nucleus, and function as transcription factors to affect gene expression. The importance of Smad proteins is underscored by discoveries that human Smad2 and Smad4 (DPC4) are frequently mutated in colorectal and pancreatic cancers, respectively (12, 17).
Despite intensive studies of Smad proteins, there is no definitive evidence implicating Smad3 as an intermediate in TGF-β signaling pathways involving either growth inhibition or induction of expression of genes encoding extracellular matrix proteins such as PAI-1. Although Smad proteins are frequently mutated in cancer cells, these cells often harbor other mutations as well. For this reason, resistance to the growth-inhibiting effects of TGF-β exhibited by these cells cannot be linked definitively to the loss or alteration of Smad proteins. Here we identify dominant-negative forms of Smad3 that enable normally TGF-β-sensitive epithelial cells to escape TGF-β- induced growth inhibition. Furthermore, we show that phosphorylation of a motif at the C terminus of Smad3 is required for TGF-β-dependent nuclear translocation and induction of the PAI-1 promoter. Alterations of this C-terminal region result in the creation of dominant-negative forms of this protein that disrupt TGF-β signaling in vivo. These data provide strong evidence that Smad3 is a signal transduction intermediate both in TGF-β-dependent growth inhibition and transcriptional activation.
MATERIALS AND METHODS
Cell Lines.
Mv1Lu mink lung epithelial cells CCL64 (ATCC) and its derivative L20, in which the murine ecotropic receptor was stably expressed, were grown in minimal essential medium supplemented with nonessential amino acids, penicillin, streptomycin, and 10% fetal bovine serum (GIBCO/BRL). The retroviral packaging cell line BOSC23 (30), was maintained in DMEM supplemented with 10% fetal bovine serum. To generate L20, CCL64 Mv1Lu cells were transfected with 5 μg pM5-EcoRneo (31) and 20 μg carrier DNA using the Ca2+ phosphate precipitation method (Strategene). Stable transfectants were selected in the presence of 800 μg/ml G418 for 2 weeks. Several clones were tested for infectability with the pMX-GFP virus [encoding the green fluorescent protein (GFP)] packaged in BOSC23 cells and analyzed by fluorescent activated cell sorting. One of clones, designated L20, was highly infectable and was expanded and used in subsequent experiments.
Metabolic Labeling of Cells and Two-Dimensional Tryptic Phosphopeptide Analysis.
Five to eight million wild-type Mv1Lu cells and selected pools of cells stably expressing each of the mutant Smad3 proteins were labeled for 2 hr in phosphate-free medium (GIBCO/BRL) containing 1 mCi/ml [32P]orthophosphate (ICN; 1 Ci = 37 GBq), then incubated in the same medium for 30 min in the absence or presence of 1 nM TGF-β. Cells were lysed in 0.2 ml boiled lysis buffer (1% SDS/50 mM Tris⋅HCl, pH 7.4) and centrifuged through a QIAshredder (Qiagen, Chatsworth, CA). The flow-through was diluted to 1 ml with RIPA buffer [1% NP40/50 mM Tris, pH 7.5/150 mM NaCl/2 mM EDTA/50 mM NaF/protease inhibitor mixture (Boehringer Mannheim)]. The lysate was then cleared with 50 μl protein-A beads for 2 hr and 10 μg Flag M2 antibody (Kodak) was added. The lysates were incubated at 4°C for 2 hr followed by addition of 50 μl protein-A beads prebound with 1 μl rabbit anti-mouse antibody (Upstate Biotechnology, Lake Placid, NY). The immunoprecipitates were eluted from beads by boiling for 5 min at 100°C in 50 μl 2× SDS sample buffer (100 mM Tris⋅HCl, pH 6.8/200 mM DTT/4% SDS/0.2% Bromphenol blue/20% glycerol) and electrophoresed through an 8% SDS/PAGE gel, transferred to a nitrocellulose membrane, and autoradiographed. Phosphotryptic peptide analysis was carried out as described (4).
DNA Expression Constructs.
Construction of the retroviral bicistronic expression vector pMX-IRES-GFP (see Fig. 1A) will be described in detail elsewhere. Briefly, this vector was constructed by inserting a GFP gene driven by the encephalomyocarditis virus (EMCV) internal ribosomal entry sequence (IRES) into the retroviral vector pMX (32). pMX-IRES-GFP-Smad3-C-Flag was made by inserting the blunt-ended EcoRI–HindIII fragment of pRK5-Smad3 (16) into the blunt ended and BamHI-digested pMX-IRES-GFP vector. N-terminal Flag-tagged Smad3 (Flag-N-Smad3) and its mutants in which three serines were mutated to alanine (Flag-N-Smad3A) or aspartic acid (Flag-N-Smad3D) were generated by PCR with Pfu polymerase (Strategene) with primers containing the designed mutations and appropriate restriction sites using pRK5-Smad3 as the template for amplification. After digestion with SalI and NotI, these fragments were ligated to SalI–NotI-digested pMX-IRES-GFP-Flag into which a M2-Flag tag had been introduced upstream of the SalI site in the polylinker sequence. For overexpression and luciferase assays, we subcloned Smad3 and its mutant derivatives from the retrovirus vectors into pEGFP-N1 (CLONTECH) to allow high level expression.
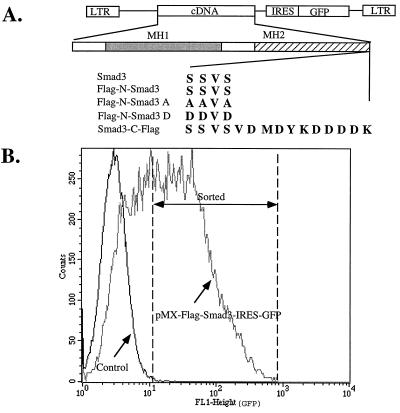
Figure 1.
Stable expression of Smad3 proteins in Mv1Lu L20 cells using the pMX-IRES-GFP retroviral vector. (A) Diagram of the expression vector and the wild-type and mutant Smad3 genes used in this study. (B) Mink lung L20 cells are a derivative of mink lung CCL64 cells that stably express a transfected murine receptor for retroviruses containing the murine ecotropic envelope glycoprotein; L20 cells are highly infectable by the ecotropic retroviruses generated by from BOSC 23 cells. A population of 5 × 105 L20 cells was infected with pMX-IRES-GFP-Smad3 retroviruses encoding the wild-type or mutant Smad3 proteins; shown here is a typical profile of cells infected with pMX-Flag-Smad3-IRES-GFP. After 3 days growth 105 infected GFP-positive cells were sorted by flow cytometry. Indicated by the dotted lines is the population of GFP-positive cells that were selected and used for all subsequent experiments. As expected, cells infected by the same virus but lacking the GFP gene did not exhibit any green fluorescence.
Transfection, Infection, and Cell Sorting.
The pMX-Smad3-IRES-GFP1.0 and various mutant DNA constructs were transfected into BOSC23 cells as described (30). Forty-eight hours after transfection, virus supernatants were collected. The titer of virus is generally 5 × 105/ml as determined by infecting 105 of target cells with a series of dilutions of the pMX-GFP virus. The number of GFP-positive cells was scored by FACScan (Becton Dickinson), and the virus titer was calculated based on the average of the number of cells infected at different dilutions. Then 5 × 105 Mv1Lu L20 cells were infected with 1.5 ml virus supernatant in the presence of 4 μg/ml of Polybrene for 4 hr and fresh medium were added afterwards. Forty-eight hours after the infection, cells were sorted by FACstar or FACSvantage cell sorter (Becton Dickinson) according to their GFP levels.
Thymidine Incorporation Assay.
Twenty thousand wild-type Mv1Lu cells or Mv1Lu cells expressing various Smad3 derivatives were seeded in triplicate in each well of 24-well culture plate. Twenty-four hours later, varying concentrations of TGF-β1 were added and the cells were incubated for 20 hr. Cells were labeled with 1 μCi per well of [3H]thymidine for 3 hr and fixed in 5% trichloroacetic acid for 30 min followed by several washes with trichloroacetic acid and water. Acid insoluble materials were dissolved in 0.5 M NaOH and counted in a scintillation counter.
Luciferase Assay.
Twenty-four hours before transfection Mv1Lu cells were seeded in triplicate at 2 × 105 cells per well in a 6-well plate and grown for 24 hr. DEAE dextran-mediated transfections (33) were performed with 0.75 μg of p3TP-Lux together with a total of 1 μg of the vector (see above) alone or expressing the indicated Smad3 variant. 0.5 μg of pCH110 or pSVβ (CLONTECH), encoding β-galactosidase, was included in each sample as a control for the efficiency of transfection. After 20 hr incubation in the absence or presence of 240 pM TGF-β1, luciferase activity was determined using the luciferase assay system (Promega) and β-galactosidase activity was measured using the Luminescent β-galactosidase detection kit (CLONTECH) with an Analytical Luminescent Laboratory luminometer.
RESULTS
Quantitative and Stable Expression of Smad3 Genes in Mink Lung Epithelial Cells.
To address the physiological relevance of the Smad3 protein in cellular responses to TGF-β, we introduced the wild-type and various mutant Smad3 genes into a novel retroviral vector. In this vector the left LTR promoted Smad3 expression, followed by an IRES (34) linked to GFP. Thus both genes resided in a common bicistronic transcriptional unit (Fig. 1A). We used transient transfection of BOSC23 cells to produce a very high titer of recombinant retroviruses. As these retroviruses express the ecotropic envelope glycoprotein, they will not infect mink or other nonmurine cell lines. Thus, we generated a line of CCL64 mink lung epithelial cells (Mv1Lu), called L20, that stably express the murine ecotropic receptor (31) and showed that >80% could be infected by the high titer recombinant virus. In control experiments in which the cDNA encoding the cell surface protein CD2 was used instead of Smad3, we showed that the level of CD2 expressed by infected cells was proportional to the amount of GFP (data not shown). Thus, we used flow cytometry to sort infected cells expressing both Smad3 and GFP for those with a particular level of GFP fluorescence (Fig. 1B) and we could be assured that the same amount of wild-type or mutant Smad3 protein was expressed in the several populations of infected cells we studied. As detailed in Fig. 1, we generated populations of infected L20 cells that indeed expressed similar amounts (Fig. 2 Lower) of versions of wild-type Smad3 with a Flag tag at the N terminus (Flag-N-Smad3) or mutant derivatives of Flag-N-Smad3 in which the three C-terminal serine residues were changed either to alanine (Smad3A) or aspartic acid (Smad3D).
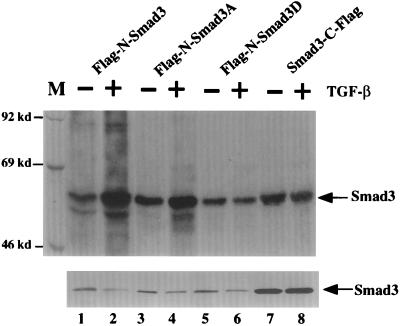
Figure 2.
TGF-β-induced phosphorylation of Smad3. (Upper) Ligand-dependent phosphorylation of Smad3 in response to TGF-β treatment. Stable pools of L20 cells expressing various Smad3 mutants were metabolically labeled with [32P]orthophosphate for 2 hr and subsequently incubated in the presence or absence of 500 pM TGF-β1 for 30 min. Cells were lysed in lysis buffer and the cleared supernatants were subjected to immunoprecipitation with the M2 Flag antibody and analyzed by 8% SDS/PAGE. (Lower) Western blot analysis of Smad3 protein levels in the transfected cells demonstrate equivalent expression of the wild-type and mutant Smad3 proteins. Lysates prepared from unlabeled cells were subjected to Western blot analysis using the Flag-M2 antibody (Kodak). The steady-state level of Smad3-C-Flag is higher than that of the N-terminal Flag-tagged Smad3s; this difference could be due to a positional effect of the Flag tag on stability of the Smad3 protein or because higher levels of expression of this form of the protein are well tolerated by these cells.
TGF-β-Induced Phosphorylation of Smad3.
To determine whether the TGF-β signaling pathway stimulates phosphorylation of Smad3, both N-terminal and C-terminal Flag-tagged Smad3 proteins were stably expressed in mink lung cells by infection with the appropriate viral vector. Pools of cells were metabolically labeled with [32P]orthophosphate and subsequently incubated in the presence or absence of TGF-β. As shown in Fig. 2 Upper (lanes 1, 3, 5, 7, and 9), a basal level of phosphorylation of Smad3 and its derivatives was observed in the absence of ligand. However, upon TGF-β addition the level of phosphorylation of the N-Flag-tagged Smad3 was dramatically increased (lane 2), whereas that of the C-terminal-tagged Smad3 remained unchanged (lane 8).
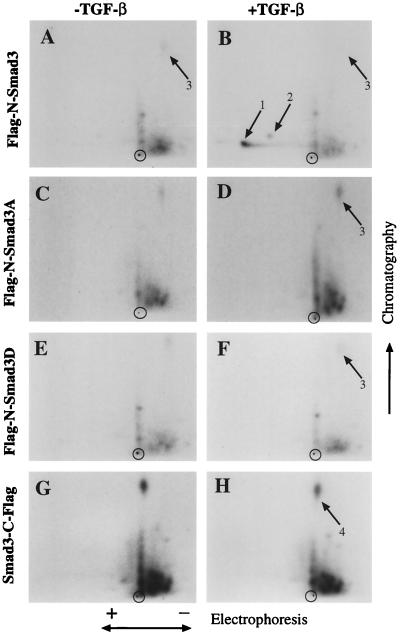
Two-dimensional tryptic phosphopeptide maps of phosphorylated Flag-N-Smad3 from TGF-β-treated or untreated Mv1Lu cells indicated that only one major (spot 1) and one minor peptide (spot 2) showed a substantial increase in phosphorylation in response to ligand addition; these phosphopeptides migrated toward the anode. Several additional minor peptides exhibited only modest increases in phosphorylation under these conditions; we do not know the identity of these peptides, but some may result from partial trypsin digestion.
To investigate whether the observed phosphorylation of Smad3 occurred at the C-terminal serine residues, two Flag-N-Smad3 mutants were made in which the three C-terminal serine residues, corresponding to amino acids acid 422, 423, and 425, were replaced with either alanine or aspartic acid residues (Smad3A and Smad3D), respectively. Smad3A did not exhibit TGF-β-induced increase in phosphorylation at spots 1 and 2 (Fig. 3), although there was a slight increase in the overall phosphorylation level of the protein (Fig. 2 Upper, lane 4). Replacement of serine residues 422, 423, and 425 with aspartic acid also resulted in the loss of TGF-β-dependent phosphorylation at spots 1 and 2 (Fig. 3). Interestingly, placing a Flag epitope tag at the C terminus of the wild-type Smad3 protein also disrupted TGF-β-induced phosphorylation at spots 1 and 2 (Fig. 3), and there were no additional phosphorylated peptides in the sample from TGF-β-stimulated cells. In particular, there were none that could derive from the extended C terminus of the protein even through serine residues 422, 423, and 425 are present. The mobility of spot 1 in both dimensions is similar to that of the analogous peptide derived from phosphorylated Smad2, which derives from its very C terminus (data not shown), and spot 2 is likely a version of spot 1 in which only two of the three serine residues are phosphorylated. We conclude that TGF-β induces rapid phosphorylation of Smad3 at its C terminus. Mutation of the C-terminal SSVS motif of Smad3 or addition of an epitope tag to the C terminus results in loss of ligand-dependent phosphorylation.
Figure 3.
TGF-β induced phosphorylation of Smad3 occurs at the three C-terminal serine residues, and mutation of the C-terminal serines or addition of a Flag tag at the C terminus blocks TGF-β-induced phosphorylation of Smad3. Cells expressing Flag-N-Smad3, Smad3-C-Flag, or mutants of Flag-N-Smad3 in which the three C-terminal serine residues was changed to alanine (Smad3A) or aspartate (Smad3D) were labeled with [32P]orthophosphate for 2 hr and subsequently incubated in the presence or absence of 500 pM of TGF-β1 for 30 min. Anti-Flag immunoprecipitates were digested overnight with trypsin, oxidized, and resolved in two dimensions by electrophoresis in pH 1.9 buffer followed by chromatography. Spots 1 and 2 in the resultant autoradiogram derive from the C-terminal tryptic peptide. Spot 3 is probably identical to spot 4 and likely corresponds to a peptide from the N terminus of the protein, because the migration pattern of this peptide depends on whether the Flag tag is attached to the N or C terminus of the Smad3 protein.
Expression of Dominant-Negative Smad3 and Abrogation of TGF-β- Triggered Growth Inhibition.
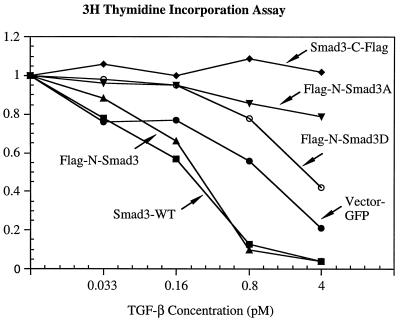
TGF-β strongly inhibits the growth of mink lung epithelial cells. To test the effects of Smad3 and its mutant derivatives on cell proliferation, pools of L20 cells stably expressing similar amounts of Smad3 or its derivatives were incubated in the presence or absence of various concentrations of TGF-β, then subjected to a [3H]thymidine incorporation assay (Fig. 4). Control cells expressing GFP alone were very sensitive to TGF-β growth inhibition; when incubated with 4 pM TGF-β there was an 80% decrease in thymidine incorporation. Cells stably expressing either wild-type Smad3 or the N-terminal-tagged Smad3 (Flag-N-Smad3) were more sensitive to TGF-β than were the control cells; an inhibition of thymidine incorporation by >80% was achieved at a TGF-β concentration of only 0.8 pM.
Figure 4.
Expression of Smad3 variants in mink lung epithelial cells affects the antiproliferative effects of TGF-β. Pools of L20 cells stably expressing similar amounts of Flag-N-Smad3 (▴) or Flag-N-Smad3A (▾) or Flag-N-Smad3D (○) or Smad3-C-Flag (⧫) or wild-type Smad3 (▪) or only GFP (vector; •) were incubated with the indicated concentration of TGF-β1 for 20 hr, then labeled with [3H]thymidine for 3 hr, fixed, and counted. Results for each pool of cells are normalized to the amount of thymidine uptake in cells incubated without TGF-β, and each data point represents a average from three independent wells. Variation is less than 10% for each data point.
Quite different results were observed with cells stably expressing the C-terminal-tagged Smad3 (Smad3-C-Flag) or the Flag-N-Smad3A or Flag-N-Smad3D mutants in which the C-terminal serines were altered; these cells exhibited significant resistance to growth inhibition by TGF-β (Fig. 4). Cells expressing Smad3-C-Flag showed no inhibition of growth at a TGF-β concentration (4 pM) that reduced the growth of control cells by 80%, and the growth of cells expressing Flag-N-Smad3A or Flag-N-Smad3D was inhibited only 18% and 60%, respectively.
We also conducted clonal growth assays in the presence of 50 pM TGF-β (data not shown). No colonies were visible with control L20 cultures or with L20 cells expressing wild-type Smad3 or Flag-N-Smad3. However, a large number of TGF-β-resistant colonies was obtained from the pools of L20 cells expressing Smad3-C-Flag or Flag-N-Smad3A, and a lower but significant number of colonies was obtained from L20 pools expressing Flag-N-Smad3D. Taken together, we conclude that Smad3A, Smad3-C-Flag, and Smad3D function as dominant-negative proteins when stably expressed in mink lung epithelial cells, in that they block TGF-β-induced growth inhibition. Importantly, this dominant-negative effect is correlated with the inability of these proteins to undergo TGF-β-induced phosphorylation at the C-terminal serine residues (Fig. 3).
Regulation of TGF-β Induced Transcriptional Response by Smad3.
To assess the effect of expressing Flag-N-Smad3, Flag-N-Smad3A, Flag-N-Smad3D, and Smad3-C-Flag on the transcriptional activation of TGF-β-responsive promoter, we transiently transfected plasmids expressing these mutants with a 3TP-Lux reporter construct, in which the PAI-1 promoter drives expression of a luciferase gene (33).
In cells cotransfected with empty vector expressing GFP together with the p3TP-Lux reporter construct, TGF-β treatment resulted in a almost 100-fold increase in luciferase activity (Fig. 5). Cotransfection of wild-type Smad3 (data not shown) or Flag-N-Smad3 (Fig. 5) with the p3TP-Lux reporter construct increased luciferase expression both in the presence and absence of TGF-β. However, when the Flag-N-Smad3A or Smad3-C-Flag vector was cotransfected with the reporter gene, the ability of TGF-β to induce transcription of the p3TP-Lux reporter construct was reduced over 5-fold. Expression of Flag-N-Smad3D had a lesser inhibitory effect on the ability of TGF-β to induce the p3TP-Lux reporter.
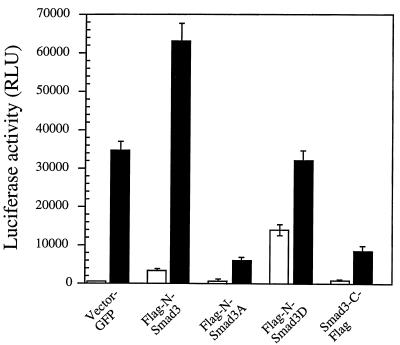
Figure 5.
Effect of Smad3 mutants on transcriptional activation of the TGF-β-responsive PAI-1 promoter (p3TP-Lux) in the absence or presence of 240 pM TGF-β. One million Mv1Lu cells were transiently transfected with 1 μg of vector DNA encoding the indicated Smad3 variants. Open bars, expression of the luciferase reporter gene in the absence of TGF-β; filled bars, in the presence of TGF-β. The data are normalized to β-galactosidase activity to control for transfection efficiency.
Neither transfection of wild-type Smad3 (data not shown), Flag-N-Smad3, Flag-N-Smad3A, or Smad3-C-Flag had a significant effect on expression of the p3TP-Lux reporter in the absence of TGF-β. Strikingly, expression of the Smad3D mutant caused significant induction of the p3TP-Lux reporter even in the absence of TGF-β; addition of TGF-β only caused a further 2-fold increase in luciferase expression. We conclude that Smad3D can activate transcription of the p3TP promoter in the absence of ligand; apparently the presence of the three aspartate residues at the C terminus of the Smad3D can mimic phosphorylation of the normal serine residues and act as a constitutive activator of transcription of the PAI-1 promoter. Perhaps surprisingly, Smad3D also dominantly inhibits the ability of TGF-β to fully activate this promoter. Thus, consistent with their dominant-negative effects on TGF-β-induced growth inhibition, both Smad3A and Smad3-C-Flag exert dominant-negative effects on p3TP-Lux activation in the presence of TGF-β but have little effect in the absence of TGF-β.
DISCUSSION
Genetic analysis of Mad and Mad-related proteins in Drosophila and C. elegans has provided crucial evidence that this protein family is involved in signaling pathways triggered by TGF-β and related ligands (8, 13). Humans express at least seven Mad-like proteins, termed Smads, but ligand-dependent regulation of Smad activity has only been established for Smad1, Smad2, and Smad4 (11, 17, 25, 26, 29). Smad3, which exhibits 91% amino acid identity to Smad2, has been shown to cooperate with Smad4 in inducing ligand-independent activation of the PAI-1 promoter (16). However, it remained unclear whether Smad3 activity is directly regulated by TGF-β, because Zhang et al. (16) were unable to detect any effects of TGF-β addition on Smad3 phosphorylation or function. Our principal result is that TGF-β addition to Mv1Lu cells induces phosphorylation of Smad3 on serine residues near its C terminus. Stable expression of wild-type Smad3 renders Mv1Lu cells hypersensitive to the antiproliferative effects of TGF-β, whereas expression of dominant-negative Smad3 mutants that cannot undergo TGF-β-induced phosphorylation abrogates TGF-β-induced growth inhibition. In particular, the C-terminal epitope-tagged Smad3 (Smad3-C-Flag) used by Zhang et al. (16) in all their experiments is dominant negative for Smad3 function. Therefore, Smad3 is a key regulator that mediates TGF-β-dependent growth inhibition and transcriptional activation.
Ligand-Induced Phosphorylation of Smad3 on C-Terminal Serine Residues.
Previous work showed that in mink lung epithelial cells TGF-β induces phosphorylation of Smad2 at the serine residues in the sequence SSMS found at the very C terminus (26). Smad3, like Smad2, becomes phosphorylated in the absence of TGF-β addition when overexpressed in COS or 293 cells and can be phosphorylated in vitro by the TGF-β type I receptor kinase domain expressed in bacteria (16). It was not clear whether such phosphorylation of Smad3 occurred at the physiologically relevant residues that are phosphorylated in vivo upon ligand addition. The C-terminal sequence SSVS in Smad3 differs in only one position from that of Smad2 (SSMS). Here we showed that in mink lung cells SSVS is the major Smad3 phosphorylation site induced by TGF-β. Smad3 is likely a direct substrate of the activated type I receptor kinase and we showed that the SSVS sequence in Smad3 can be phosphorylated in vitro by the type I receptor kinase (data not shown). Recognition of the C terminus of Smad3 by the type I TGF-β receptor is highly specific; addition of a Flag epitope tag to the C terminus of Smad3 (16) abolishes the ability of the C-terminal serine residues to become phosphorylated. Presumably the extra amino acids at this location affect the local protein structure, possibly masking the serine residues and preventing their phosphorylation by the type I receptor kinase.
Importantly, the in vitro kinase assay using the type I receptor, like many in vitro kinase assays, may not represent the normal substrate specificities of the kinase in the living cell. Smads1, -2, -3, and -5 can be phosphorylated in vitro by the constitutively active type I TGF-β receptor, yet TGF-β does not induce phosphorylation of Smad1 or Smad5 in vivo (data not shown). Similarly, Smad1 becomes phosphorylated when it is overexpressed in COS cells together with the types I and II TGF-β receptors, yet Smad1 is not phosphorylated in cells following TGF-β addition (22). Therefore, results obtained from overexpression of Smad proteins in COS or 293 cells are not always physiologically relevant. Were it not for our evidence that Smad3 is phosphorylated by the type I TGF-β receptor at the same site in vivo and in vitro, as evidenced by the same tryptic phosphopeptides, we could not conclude that phosphorylation of Smad3 by this receptor in vitro is physiologically relevant.
Smad3 Is an Intracellular Mediator of TGF-β-Induced Growth Inhibition.
Although Smad proteins have been intensively characterized, direct evidence that they function in the regulation of cell proliferation by TGF-β has been lacking. Transient overexpression of Smad2, -3, -4, and -5 proteins all induce growth inhibition (16, 18, 29). However, it has been unclear whether this growth inhibition is caused by the same regulatory pathway normally activated by TGF-β or is simply due to nonspecific cytostatic effects associated with overexpression of these proteins. We showed here that stable expression of Smad3 at quasi-physiologic levels sensitizes the response of Mv1Lu cells to growth inhibition by TGF-β, whereas stable expression at the same level of dominant-negative, nonphosphorylatable, Smad3 proteins blocks TGF-β-induced growth inhibition. These data strongly support the notion that the Smad3 protein is a physiologically important downstream mediator of the TGF-β signaling pathway.
Genes encoding Smad proteins are frequently mutated in many cancer cells. In particular, Smad4/DPC4 is frequently deleted in pancreatic tumors, and Smad2 mutations can be detected in many colorectal tumors (12, 17). Loss of Smad proteins may contribute, in part, to the resistance of many tumor cell types to growth inhibition by TGF-β. Our data show that dominant-negative variants of Smad3 can also confer resistance to TGF-β-induced growth inhibition in cells that are normally highly sensitive to this growth inhibitor. We speculate that similar mutant Smad3 proteins could occur in certain tumor cells.
Mechanism of Dominant-Negative Action of Mutant Smad3 Proteins in Growth Inhibition and Transcriptional Regulation.
We showed here that transient overexpression of wild-type Smad3 in mink lung epithelial cells slightly potentiates the activation of the PAI-1 promoter by TGF-β. In contrast, transient overexpression of a mutant Smad3 protein in which the three C-terminal serine residues are mutated to alanine (Smad3A) or of Smad3-C-Flag (Smad3 with a C-terminal Flag-tag) inhibits the ability of TGF-β to activate the PAI-1 promoter—the proteins function as dominant-negative inhibitors, as they do in the growth-inhibition assays.
Interestingly, mutation to aspartic acid of the three C-terminal serine residues in Smad3 results in a constitutively active protein that can stimulate PAI-1 transcription independent of TGF-β addition. Due to its negative charge at neutral pH, aspartic acid presumably can partially mimic a phosphorylated serine or threonine. Our results on the Smad3D mutant indicate that phosphorylation of the C-terminal serine residues is not absolutely required for transcriptional activation but probably required for ligand-induced nuclear accumulation under physiological concentration. Importantly, when expressed at physiologic levels in Mv1Lu cells, the Smad3D mutant has no dramatic effect on cell proliferation in the absence of TGF-β; it is apparently not inducing genes required for growth inhibition. In these stably transfected cells Smad3D does act as a weak dominant negative inhibitor of TGF-β-induced growth inhibition. The complex phenotypes of this mutant suggests that the structural requirements for Smad3 nuclear translocation and transcriptional activation may be separable.
How do mutant forms of Smad3 act in a dominant-negative fashion to block TGF-β-induced cellular responses? Others have previously shown that, when overexpressed in COS cells, a mutant Smad2 having serine-to-alanine substitutions at its C terminus can stably associate with TGF-β receptors (26). When overexpressed in 293 cells, the Smad3-C-Flag protein also becomes stably associated with TGF-β receptor complexes (16). This finding suggests that, when overexpressed, these mutant Smad2 or Smad3 proteins bind to the TGF-β receptor complex but, because they are unable to undergo phosphorylation, remain stably associated and in this fashion block receptor signaling. However, the nonphosphorylatable mutant Smad3 proteins could also associate with endogenous, wild-type Smad proteins or with other essential signal transduction proteins and in this way inhibit the ability of the TGF-β receptors to signal gene activation or growth inhibition.
Smad3 and Smad2 have very similar sequences, suggesting that they act in similar fashions. Which Smad mediates TGF-β signaling in vivo? We suggest that nonphosphorylatable mutant Smad3 proteins could act not only to negate the function of endogenous Smad3 but also might affect the function of the endogenous Smad2 proteins. Indeed, Smad3 and Smad2 may well forms hetero-oligomers and as such may mediate certain TGF-β responses. In any case, our data clearly point to an important role played by Smad3 in the regulation of cell proliferation and transcriptional activation by the TGF-β receptors.
Acknowledgments
We are grateful to Drs. K. Luo, X. Hua, and J. Bogan for helpful discussions. We thank Drs. Warren Pear, Ying Zhang, Rik Derynck, Joan Massague, and Toshio Kitamura for reagents, and Glen Paradis and Mike Jennings for help with cell sorting. We also thank R&D Systems for kindly providing the TGF-β1. This work was supported in part by National Institutes of Health Grant R01 CA-63260 to H.F.L.; by Grant CDR 88-03014 from the National Science Foundation to the Massachusetts Institute of Technology Biotechnology Process Engineering Center and to H.F.L., and by a National Institutes of Health grant (to R.A.W.). R.A.W is a research professor of the American Cancer Society. X.L. was supported by a postdoctoral fellowship from the National Institutes of Health. Y.S. was supported by a postdoctoral fellowship of Robert Steel Foundation for Pediatric Cancer Research, and S.N.C. was supported by a fellowship from the Medical Foundation.
ABBREVIATIONS
- TGF-β
transforming growth factor beta
- GFP
green fluorescent protein
- PAI-1
plasminogen activator inhibitor 1
- IRES
internal ribosomal entry sequence
- BMP
bone morphogenic protein
References
- 1.Kingsley D M. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 2.Massague J, Weis-Garcia F. Cancer Surv. 1996;27:41–64. [PubMed] [Google Scholar]
- 3.Roberts A B, Sporn M B. In: The Transforming Growth Factor Betas. Roberts M B S a A B., editor. Heidelberg: Spring; 1990. pp. 421–472. [Google Scholar]
- 4.Luo K, Lodish H F. EMBO J. 1997;16:1970–1981. doi: 10.1093/emboj/16.8.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen F, Weinberg R A. Proc Natl Acad Sci USA. 1995;92:1565–1569. doi: 10.1073/pnas.92.5.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo K, Lodish H F. EMBO J. 1996;15:4485–4496. [PMC free article] [PubMed] [Google Scholar]
- 7.Wrana J L, Attisano L, Wieser R, Ventura F, Massague J. Nature (London) 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 8.Sekelsky J J, Newfeld S J, Raftery L A, Chartoff E H, Gelbart W M. Genetics. 1995;139:1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newfeld S J, Chartoff E H, Graff J M, Melton D A, Gelbart W M. Development (Cambridge, UK) 1996;122:2099–2108. doi: 10.1242/dev.122.7.2099. [DOI] [PubMed] [Google Scholar]
- 10.Lechleider R J, de Caestecker M P, Dehejia A, Polymeropoulos M H, Roberts A B. J Biol Chem. 1996;271:17617–17620. doi: 10.1074/jbc.271.30.17617. [DOI] [PubMed] [Google Scholar]
- 11.Hoodless P A, Haerry T, Abdollah S, Stapleton M, O’Connor M B, Attisano L, Wrana J L. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 12.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern S E. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 13.Savage C, Das P, Finelli A L, Townsend S R, Sun C Y, Baird S E, Padgett R W. Proc Natl Acad Sci USA. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker J C, Harland R M. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Rubock M J, Whitman M. Nature (London) 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Feng X, We R, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 17.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L C, Bapat B, Gallinger S, Andrulis I L, Thomsen G H, Wrana J L, Attisano L. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 18.Yingling J M, Das P, Savage C, Zhang M, Padgett R W, Wang X F. Proc Natl Acad Sci USA. 1996;93:8940–8944. doi: 10.1073/pnas.93.17.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomsen G H. Development (Cambridge, UK) 1996;122:2359–2366. doi: 10.1242/dev.122.8.2359. [DOI] [PubMed] [Google Scholar]
- 20.Graff J M, Bansal A, Melton D A. Cell. 1996;85:479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Hata A, Baker J C, Doody J, Carcamo J, Harland R M, Massague J. Nature (London) 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 22.Nakao A, Roijer E, Imamura T, Souchelnytskyi S, Stenman G, Heldin C, ten Dijke P. J Biol Chem. 1997;272:2896–2900. doi: 10.1074/jbc.272.5.2896. [DOI] [PubMed] [Google Scholar]
- 23.Derynck R, Gelbart W M, Harland R M, Heldin C H, Kern S E, Massague J, Melton D A, Mlodzik M, Padgett R W, Roberts A B, Smith J, Thomsen G H, Vogelstein B, Wang X F. Cell. 1996;87:173. doi: 10.1016/s0092-8674(00)81335-5. [DOI] [PubMed] [Google Scholar]
- 24.Riggins G J, Thiagalingam S, Rozenblum E, Weinstein C L, Kern S E, Hamilton S R, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi H, Abdollah S, Qui Y, Cai J, Xu Y-Y, Grinnell B W, Richardson M A, Topper J N, Gimbrone M A J, Wrana J L, Falb D. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 26.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Musci T, Derynck R. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Lebrun J J, Vale W. Proc Natl Acad Sci USA. 1996;93:12992–12997. doi: 10.1073/pnas.93.23.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 30.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker B W, Boettiger D, Spooncer E, Norton J D. Nucleic Acids Res. 1992;20:5234. doi: 10.1093/nar/20.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 33.Wrana J L, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X F, Massague J. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 34.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]