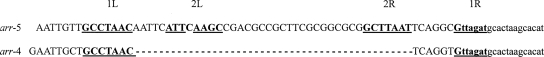Abstract
New arr alleles emerged in class 1 integrons from a clinical Pseudomonas aeruginosa strain (arr-4) and four Klebsiella pneumoniae strains (arr-5) in Brazil/American continent. arr-4 was preceded by aacA7-catB3, whereas arr-5 was the unique cassette. The putative proteins shared 75% (Arr-5) and 78% (Arr-4) identities with Arr-2.
Rifampin resistance is a notable global health problem concern (11), since it is the front line drug for treating tuberculosis (4) and preventing meningococcal diseases (13). More than 90% of rifampin resistance is due to mutational alterations in an 81-bp rifampin resistance determining region of the rpoB gene (9). However, resistance may also arise by horizontal acquisition of arr genes, which code for ADP-ribosyltransferases responsible for the drug inactivation. The arr-1 gene was first described in the Mycobacterium smegmatis chromosome (10). Subsequently, arr-2 and arr-3 alleles were described as gene cassettes in class 1 integrons present in gram-negative isolates from Europe and Asia (1, 6, 8, 14), showing high-level rifampin MICs ranging from 32 to >256 μg/ml.
We report here the identification and characterization of two new arr alleles, arr-4 and arr-5, and the emergence of this class of gene in the American continent as gene cassettes from class 1 integrons present in clinical Pseudomonas aeruginosa and Klebsiella pneumoniae strains.
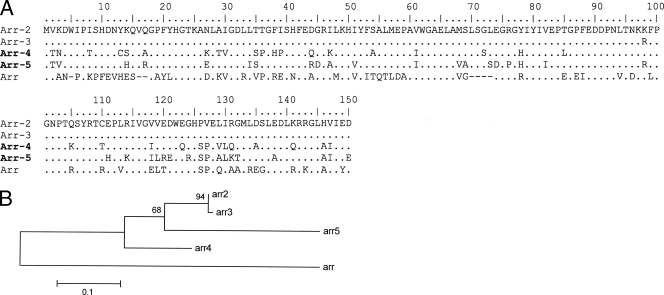
One P. aeruginosa (PS1111) and four K. pneumoniae (K56, K224, K688, and K830) isolates recovered from different inpatients of a Brazilian hospital (Rio de Janeiro city) in 2005 tested positive for class 1 integron signatures (3). DNA macrorestriction using SpeI, followed by pulsed-field gel electrophoresis, was performed as previously described (3) and demonstrated four distinct profiles among the K. pneumoniae strains. Sequence analysis of the class 1 integron variable regions showed the presence of a 453-bp open reading frame, encoding a predicted protein of 150 amino acid residues in all integrons. These sequences from the K. pneumoniae strains and from isolate PS1111 shared 78% amino acid identity, and 75 and 78% amino acid identity, respectively, with ADP-ribosyltransferase Arr-2 (Fig. 1) . Therefore, these open reading frames were named arr-4 (PS1111) and arr-5 (the K. pneumoniae strains). The genes were assigned to be parts of cassettes due to the presence of attC sites. The arr-4 gene cassette was preceded by a aacA7-catB3 array, which confer resistance to aminoglycosides and chloranphenicol, respectively, and arr-5 was the unique cassette inserted in all K. pneumoniae integrons. The arr-4 gene cassette is 516 bp long and presented an attC site only 22 bp in length due to the loss of 2L and 2R simple sites (Fig. 2). The arr-5 gene cassette is 555 bp, presenting a classical attC of 61 bp, encompassing the core site sequences that make up the left-hand and right-hand consensus sites composed of 1L, 2L, 1R, and 2R simple sites (Fig. 2) (12).
FIG. 1.
(A) Arr amino acid sequence alignment. The GenBank accession numbers for the putative proteins included in the comparison are as follows: Arr (AAC05822), Arr-2 (AAC64366), Arr-3 (AAP20922), Arr-4 (EF660562), and Arr-5 (EF660563). The sequences described here are in boldface. Identical amino acid residues are represented by dots. (B) Neighbor-joining genetic tree constructed with Arr predicted proteins (MEGA3).
FIG. 2.
arr-4 and arr-5 attC sites. The core sites representing 1L, 2L, 1R, and 2R simple sites are in boldface and underlined (12). Dashes indicate the partial deletion of arr-4 attC site. Bases in lowercase are part of the next cassette.
arr-2 and arr-3 gene cassettes share 99% nucleotide identity and, moreover, present identical attC site sequences. Considering that a gene cassette is characterized by a particular attC site sequence (12), it can be assumed that they were inappropriately designated as distinct cassettes, and it is more likely that arr-3 is a variant of the canonical arr-2, diverging by only two point mutations. Conversely, the arr-4 and arr-5 genes had 75 and 72% nucleotide identities, respectively, with arr-2 and were associated with unique attC sites, which presented no similarity to that from arr-2, characterizing three distinct arr gene cassettes. These findings altogether suggested that arr-2, arr-4, and arr-5 gene cassettes have evolved independently from each other.
The antimicrobial susceptibility was tested for β-lactams, aminoglycosides, and ciprofloxacin by the disk diffusion method (2), and MICs of imipenem and rifampin were determined by the E-test method. The K. pneumoniae strains were determined to be multiresistant, except to carbapenems. PS1111 was resistant only to ciprofloxacin and ticarcillin-clavulanic acid. The MIC of rifampin for K830 and PS1111 was increased (>32 μg/ml), as observed previously in arr-2-carrying strains (5), whereas the rifampin MICs for K56, K224, and K688 ranged from 8 to 16 μg/ml. However, for the K. pneumoniae ATCC 10031 control strain the rifampin MIC was 6 μg/ml. PCRs were carried out with primers flanking both arr-4 and arr-5 gene cassettes in order to obtain the entire arr coding regions to be cloned. arr-4, which was the last cassette, was amplified with primers annealing at the 3′ end of catB3 gene cassette (CATB3, F, 5′-CACTGGAGAAGATCAAAGCG-3′) and at the 3′CS (for 3′ conserved segment) from class 1 integrons (INB, 5′-GGGCAGACTTGACCTGAT-3′). arr-5, the unique cassette inserted in the integron variable region, was obtained with primers targeting the 5′CS (INF, 5′-GGCATCCAAGCAGCAAG-3′) and 3′CS (INB) from class 1 integrons. The resulted products were cloned into pGEM-T Easy vector according to the manufacturer's instructions (Promega). The ligation reaction was used to transform the rifampin-sensitive Escherichia coli DH5α, and transformants were selected on Luria-Bertani agar plates containing 100 μg of ampicillin and 50 μg of rifampin/ml. PCR and sequence analysis showed that all selected transformants harbored arr-4 or arr-5 genes. A fresh inoculum of transformants was prepared at a concentration of 0.5 on the McFarland scale (1.5 × 108 CFU/ml) and delivered onto Mueller-Hinton agar plates containing 100 μg of ampicillin/ml. The MICs were determined by the E-test method as the lowest drug concentration graded in the strip in which no growth was observed after 20 h of incubation at 37°C. The MICs of rifampin increased from 8 to >256 μg/ml for DH5α when transformed with both arr-4 and arr-5 genes. Therefore, the high-level rifampin MICs observed in transformed DH5α E. coli were due to arr-4 and arr-5 genes, proving the functionality of these new alleles in other bacterial species.
Sequence analysis revealed that the integron from PS1111 presented the strong version of cassette promoter (Pc), which is the most active configuration, whereas the K. pneumoniae integrons had the uncommon Pc configuration characterized by the weak Pc version, followed by the putative active second promoter (P2), usually responsible for 90% of the total promoter activity (7). These findings characterize the conditions for the expression of arr-4 and arr-5 genes found in these class 1 integrons.
Clinical P. aeruginosa and K. pneumoniae strains carrying new arr gene cassettes compose an ideal scenario for dissemination of these resistance determinants by horizontal gene transfer. In fact, the potential spread of arr gene is emphasized by our results, which showed four K. pneumoniae lineages carrying the same integron arrangement, composed by arr-5 gene cassette, circulating at the same moment in a unique clinical setting.
Nucleotide sequence accession numbers.
The nucleotide sequences of integrons harboring arr-4 and arr-5 have been submitted to GenBank under accession numbers EF660562 and EF660563, respectively.
Acknowledgments
We thank Anna Beatriz Robottom for English review of the manuscript and the PDTIS platform for sequencing.
This work was supported by a CNPq fellowship and an Oswaldo Cruz Institute grant.
Footnotes
Published ahead of print on 25 February 2008.
REFERENCES
- 1.Arlet, G., D. Nadjar, J.-L. Herrmann, J.-L. Donay, M. Rouveau, P. H. Lagrange, and A. Philippon. 2001. Plasmid-mediated rifampin resistance encoded by an arr-2-like gene cassette in Klebsiella pneumoniae producing an AAC-1 class C β-lactamase. Antimicrob. Agents Chemother. 45:2971-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. Document M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Fonseca, E. L., V. V. Vieira, R. Cipriano, and A. C. P. Vicente. 2006. Emergence of dhfrXVb and blaCARB-4 gene cassettes in class 1 integrons from clinical Pseudomonas aeruginosa isolated in Amazon region. Mem. Inst. Oswaldo Cruz 101:81-84. [DOI] [PubMed] [Google Scholar]
- 4.Grosset, J. H. 1989. Present status for chemotherapy for tuberculosis. Rev. Infect. Dis. 11(Suppl.):S347-352. [DOI] [PubMed] [Google Scholar]
- 5.Houang, E. T. S., Y.-W. Chu, W.-S. Lo, K.-Y. Chu, and A. F. B. Cheng. 2003. Epidemiology of rifampin ADP-ribosyltransferase (arr-2) and metallo-β-lactamase (blaIMP-4) gene cassettes in class 1 integrons in Acinetobacter strains isolated from blood cultures in 1997 to 2000. Antimicrob. Agents Chemother. 47:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, K., J. H. Yum, D. Yong, H. M. Lee, H. D. Kim, J.-D. Docquier, G. M. Rossolini, and Y. Chong. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lévesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 8.Mammeri, H., M. Van De Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolayevskyy, V. V., T. J. Brown, Y. I. Bazhora, A. A. Asmolov, Y. M. Balabanova, and F. A. Drobniewski. 2007. Molecular epidemiology and prevalence of mutations conferring rifampicin and isoniazid resistance in Mycobacterium tuberculosis strains from the southern Ukraine. Clin. Microbiol. Infect. 13:129-138. [DOI] [PubMed] [Google Scholar]
- 10.Quan, S., H. Venter, and E. R. Dabbs. 1997. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob. Agents Chemother. 41:2456-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sam, I. -C., F. Drobniewski, P. More, M. Kemp, and T. Brown. 2006. Mycobacterium tuberculosis and rifampin resistance, United Kingdom. Emerg. Infect. Dis. 12:752-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 13.Taha, M. K., M. L. Zarantonelli, C. Ruckly, D. Giorgini, and J. M. Alonso. 2006. Rifampin-resistance Neisseria meningitidis. Emerg. Infect. Dis. 12:859-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tribuddharat, C., and M. Fennewald. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960-962. [DOI] [PMC free article] [PubMed] [Google Scholar]