Abstract
A unique bacterial GTPase, Der, containing two tandem GTP-binding domains, is essential for cell growth and plays a crucial role in a large ribosomal subunit in Escherichia coli. The depletion of Der resulted in accumulation of both large and small ribosomal subunits and also affected the stability of large ribosomal subunits. However, its exact cellular function still remains elusive. Previously, we have shown that two G domain mutants, DerN118D and DerN321D, cannot support cell growth at low temperatures, suggesting that both GTP-binding domains are indispensable. In this study, we show that both Der variants are defective in ribosome biogenesis. Genetic screening of an E. coli genomic library was performed to identify the genes which, when expressed from a multicopy plasmid, can restore the growth defect of the DerN321D mutant at restrictive temperatures. Among seven suppressors isolated, four were located at 62.7 min on the E. coli genomic map, and the gene responsible for the suppression of DerN321D was identified as the relA gene which encodes a ribosome-associated (p)ppGpp synthetase. The synthetic activity of RelA was found to be essential for its DerN321D suppressor activity. Overexpression of RelA in a suppressor strain did not affect the expression of DerN321D but suppressed the polysome defects caused by the DerN321D mutant. This is the first demonstration of suppression of impaired function of Der by a functional enzyme. A possible mechanism of the suppression of DerN321D by RelA overproduction is discussed.
GTP-binding proteins play critical roles in signal transduction, protein translation, and protein translocation, and the GTPase cycle, comprising a GTP-bound “on” state, a GDP-bound “off” state, and an “empty” state, is a key element which tightly regulates cellular events (2, 3). The subfamily of Der GTP-binding proteins is unique in that these proteins are highly conserved in eubacteria but not in eukaryotes or archaea and contain two consecutive GTP-binding domains that are highly homologous to each other (15). The Der proteins were found to be essential for cell growth in several bacteria, such as Escherichia coli, Neisseria gonorrhoeae, Bacillus subtilis, and Staphylococcus aureus (10, 16, 18, 20, 22). We previously demonstrated that Der cofractionates with 50S ribosomal subunits in a GTP-dependent manner and plays a critical role in 50S ribosome maturation at a later biogenesis step, although its precise role in large ribosomal subunits is still elusive (16). Based on these observations, it can be concluded that the essential function of E. coli Der is in 50S ribosome biogenesis. Interestingly, both GTP-binding domains (G domain) were essential for cell growth, suggesting that GTPase activity and GTP-induced conformational changes of Der are essential for its function as well as its viability (16).
Despite the lack of knowledge regarding the cellular role of Der, in recent years the X-ray crystal structures of two Der proteins were resolved from Thermotoga maritima and B. subtilis (23, 25). Both structures showed that the C-terminal domain shares similar topology with the KH domain. It does not have a consensus sequence motif (VIGXXGXXI) (12) and is flanked by two conventional GTP-binding domains. It was suggested that the GTP-bound form of YphC (a Der orthologue in B. subtilis) triggers a dramatic conformational change, which favors interaction with negatively charged ribonucleic acids by exposing a positively charged KH-like domain that has a high pI value (15), supporting the notion that Der associates with ribosomes (16, 23). However, the exact role of the C-terminal KH-like domain of Der remains to be determined.
Interestingly, the overexpression of E. coli Der was reported to suppress the slow-growth phenotype of an rrmJ mutant (6, 32). RrmJ is a methyltransferase that modifies the U2552 residue of 23S rRNA in an intact 50S ribosomal subunit. Even though this rrmJ mutant is viable, its growth is seriously affected due to the accumulation of 50S and 30S ribosomal subunits in the polysome profiles at the expense of 70S ribosomes. These results suggested that Der may play a critical role in 50S ribosome biogenesis. An interesting approach to further investigate the physiological role of the two GTP-binding domains of Der is to screen E. coli genomic libraries to identify a gene(s) to suppress the cell growth defect of mutant DerN118D (with a mutation of Asn to Asp at residue 118) and mutant DerN321D (with a mutation of Asn to Asp at residue 321). Here we report that the DerN118D and DerN321D mutants resulted in accumulation of both large and small ribosomal subunits, which led to severe growth and polysome defects. Screening of an E. coli genomic library revealed that the product of the relA gene suppresses the growth defect of both DerN118D and DerN321D.
When bacterial cells are exposed to nutritional deprivation, such as an amino acid or carbon source limitation, cell growth is regulated by a mechanism known as the “stringent response,” accumulating pppGpp or ppGpp, collectively termed (p)ppGpp (7). RelA is a ribosome-associated enzyme that is activated by amino acid starvation and synthesizes (p)ppGpp, using ATP and GDP(GTP) as substrates, in response to the uncharged tRNA in the ribosomal A site, the key event for the stringent response (7). In this study, we further demonstrate that both full-length and C-terminally truncated RelA proteins can suppress the growth defect of der deletion strains expressing DerN118D and DerN321D and that the point mutations in the catalytic domain of RelA cannot recover cell growth. This suggests that (p)ppGpp synthesis is essential for this suppression. Moreover, the polysome defect caused by the DerN321D mutant was suppressed by the overexpression of RelA. Our studies may provide a clue to understanding the critical cellular role of Der.
MATERIALS AND METHODS
Strains and plasmids.
The der deletion strain (strain BK) and strain CF1944 (relA251::kan) were grown in Luria-Bertani (LB) or M9 minimal medium. pINEcDer (der+ lppp lacpo cat), pINEcDerGD-1 [der(N118D)], and pINEcDerGD-2 [der(N321D)] were used as described by Hwang and Inouye (16). Table 1 summarizes the strains, plasmids, and primers used in this study.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype, description, or sequence (5′-3′) | Reference |
|---|---|---|
| Strains | ||
| BW25113 | lacIp4000 (lacIq) rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 9 |
| BK | BW25113 der::kan pIEEcttg [der+bla ori(Ts)] | 16 |
| BD | BW25113 der::kan pINEcDer | This study |
| BD118 | BW25113 der::kan pINEcDerGD-1 | This study |
| BD321 | BW25113 der::kan pINEcDerGD-2 | This study |
| CF1944 | W3110 relA251::kan | 36 |
| DH5α | F−endA1 hsdR17 supE44 λ−thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 (φ80dlac)Δ(lacZ)M15 | Gibco-BRL |
| Plasmids | ||
| pACYC177 | bla aph(3′)-Ia ori p15A; cloning vector | New England Biolabs |
| pAC1 | relA+ pACYC177 (E. coli genomic library clone) | This study |
| pAC2 | Derivative of pAC1 | This study |
| pAC3 | Derivative of pAC1 | This study |
| pAC4 | Derivative of pAC1 | This study |
| pAC5 | Derivative of pAC3 | This study |
| pAC6 | relA(1-412) pACYC177 (E. coli genomic library clone) | This study |
| pAC7 | relA(1-522) pACYC177 (E. coli genomic library clone) | This study |
| pRelA | relA+ pACYC177 | This study |
| pRelAG251E | relA(G251E) pACYC177 | This study |
| pRelAH354Y | relA(H354Y) pACYC177 | This study |
| pRelAC638F | relA(C638F) pACYC177 | This study |
| pRelAG251EC638F | relA(G251E,C638F) pACYC177 | This study |
| pRelAH354YC638F | relA(H354Y,C638F) pACYC177 | This study |
| pIN | lppplacpocat | 11 |
| pINEcDer | der+ pIN | 16 |
| pINEcDerGD-1 | der(N118D) pIN | 16 |
| pINEcDerGD-2 | der(N321D) pIN | 16 |
| Primers | ||
| RelA5′ | ACGGTCACCGATGTCACAAAGCAGCCGTGG | This study |
| RelA3′ | ACGGATCCACCGCTATCATATGTAGATAC | This study |
| RelAG251E5′ | GCGGAAGTGTATGAGCGTCCGAAACAC | This study |
| RelAG251E3′ | GTGTTTCGGACGCTCATACACTTCCGC | This study |
| RelAH354Y5′ | GGTGTTGCTGCGTACTGGAAATATAAAGAGG | This study |
| RelAH354Y3′ | CCTCTTTATATTTCCAGTACGCAGCAACACC | This study |
| RelAC638F5′ | CACCGCGCCGATTTCGAACAACTGGCG | This study |
| RelAC638F3′ | CGCCAGTTGTTCGAAATCGGCGCGGTG | This study |
Growth conditions and media.
LB medium without isopropyl-β-d-thiogalactopyranoside (IPTG), unless indicated otherwise, was routinely used for cell growth, and ampicillin, kanamycin, and chloramphenicol were used at concentrations of 50, 35, and 50 μg/ml, respectively. Culture turbidity was measured at 600 nm. Sensitivity to 15 mM 3-amino-1,2,4-triazole (3-AT) was tested on M9 minimal plates containing 19 amino acids other than histidine, 1 mM adenine, and 1 mM thiamine, as described by Schreiber et al. (29) and Gropp et al. (13).
Construction of E. coli genomic library and screening for suppressor genes.
Genomic DNA from strain BW25113 (9) was prepared, and the genomic DNA was partially digested by using Sau3AI. Genomic DNA fragments of 2 to 5 kb were then ligated into the BamHI sites of pACYC177 (Ampr Kanr; New England Biolabs). The genomic library was transformed into strain BD321 (a der deletion strain complemented by pINEcDerGD-2), and transformants were plated on LB agar plates containing ampicillin, kanamycin, and chloramphenicol. The plates were incubated at 30°C for overnight. A total of 28 colonies formed at low temperature, and plasmids were extracted from these colonies. The extracted plasmids were transformed into DH5α cells, and transformed cells were plated on LB agar plates containing ampicillin to remove pINEcDerGD-2 (Cmr). Subsequently, a homogeneous library clone was purified from each ampicillin-resistant colony. To exclude clones containing the der gene, PCR was carried out using primers that amplify the full-length der gene. The library clones that did not contain the der gene were retransformed into BD321 cells to confirm their ability to suppress the growth defect of strain BD321. Clones that did not contain the der gene were sequenced from both ends to identify the inserts. To construct truncated library clones, a DNA blunting kit from Takara Mirus Bio was used per the manufacturer's guide. pAC1 was digested with BamHI to remove the mazEFG gene and was self-ligated to construct pAC2. The plasmids pAC3 and pAC4 were constructed by religation of the fragment after BstEII and DrdI digestion, respectively, followed by a blunt-end ligation reaction. The plasmid pAC4 was further digested with BstEII and ligated, yielding pAC5.
RelA cloning and site-directed mutagenesis.
Primers RelA5′ (5′-ACGGTCACCGATGTCACAAAGCAGCCGTGG-3′ [BstEII site is underlined]) and RelA3′ (5′-ACGGATCCACCGCTATCATATGTAGATAC-3′ [BamHI site is underlined]) were used to amplify the coding sequence of the wild-type relA gene, including its native promoter. The PCR product was digested with BstEII-BamHI, and the digested DNA fragment was ligated into the BstEII-BamHI sites of pACYC177 (New England Biolabs), yielding pRelA. Primers RelA G251E5′ (5′-GCGGAAGTGTATGAGCGTCCGAAACAC-3′), RelA G251E3′ (5′-GTGTTTCGGACGCTCATACACTTCCGC-3′), RelA H354Y5′ (5′-GGTGTTGCTGCGTACTGGAAATATAAAGAGG-3′), RelA H354Y3′ (5′-CCTCTTTATATTTCCAGTACGCAGCAACACC-3′), RelA C638F5′ (5′-CACCGCGCCGATTTCGAACAACTGGCG-3′), and RelA C638F3′ (5′-CGCCAGTTGTTCGAAATCGGCGCGGTG-3′) (mutated sites are underlined) were used to make point mutations on pRelA as a template, yielding pRelAG251E, pRelAH354Y, and pRelAC638F (13). Plasmids pRelAG251EC638F and pRelAH354YC638F were constructed using pRelAG251E and pRelAH354Y as templates, with two primers, RelA C638F5′ and RelA C638F3′. Mutations were confirmed by sequence analysis.
Isolation of polysomes and sucrose density gradient sedimentation.
Polysomes were prepared and resolved as described previously (26), with minor modifications. E. coli strains were grown at either 42°C or 30°C in 100 ml of LB medium to log phase. Upon reaching an appropriate culture density, polysomes were trapped by the addition of hygromycin B to the culture at a final concentration of 0.1 mg/ml. After an additional 4 min of incubation, cells were harvested by centrifugation. The cell pellet was resuspended in 1 ml of buffer BP (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM NH4Cl, and 5 mM β-mercaptoethanol). Sample was mixed with a nonhydrolyzable GTP analog, GMPPNP, at a final concentration of 100 μM, and polysomes were resolved by applying 0.5 ml of the cell lysates to a 5 to 40% linear sucrose density gradient (10 ml) in buffer BP, with subsequent ultracentrifugation at 4°C in a Beckman SW41-Ti rotor for 2.5 h at 150,000 × g. The polysome profile was detected at 254 nm, and a total of 36 fractions of 0.4 ml were collected. For ribosomal subunit analysis, cell pellets were resuspended with buffer BP containing 0.25 mM MgCl2, and cell lysates were subjected to a 5 to 25% linear sucrose density gradient. Centrifugation was carried out at 4°C in a Beckman SW41-Ti rotor for 3.5 h at 170,000 × g as described by Hwang and Inouye (16).
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses were carried out as described previously (16). The immunoassay part of Western blot analyses utilized polyclonal anti-Der antiserum generated by Pocono Rabbit Farm and Laboratory, Inc., Canadensis, PA.
RESULTS
Growth defects and accumulation of ribosomal subunits caused by N118D and N321D mutations in Der.
Previously, we demonstrated that Der-depleted cells accumulated both 50S and 30S ribosomal subunits with a decreased amount of 70S monosomes and polysomes and that the DerN118D and DerN321D mutants could not complement the lethal phenotype of the der deletion strain at low temperatures; in addition, the der null strain expressing a double mutant DerN118/321D protein showed a complete loss of viability (16). Unlike E. coli Der, the mutation of Asn to Asp at the second NKXD motif of Thermotoga maritima Der did not show an inhibitory effect on its GTPase activity in vitro (25, 34). Thus, to further analyze the effect of G-domain mutations on cell growth and polysome profiles, the growth rate of the cells was measured in liquid culture, and sucrose density gradient experiments were carried out using der deletion strains harboring pINEcDer, pINEcDerGD-1 (an expression vector for DerN118D), or pINEcDerGD-2 (an expression vector for DerN321D) (16). Plasmid pINEcDer, pINEcDerGD-1, or pINEcDerGD-2 was transformed into strain BK (a der deletion strain) cells harboring a helper plasmid (pIEEcttg), which has a temperature-sensitive origin and contains the wild-type der gene.
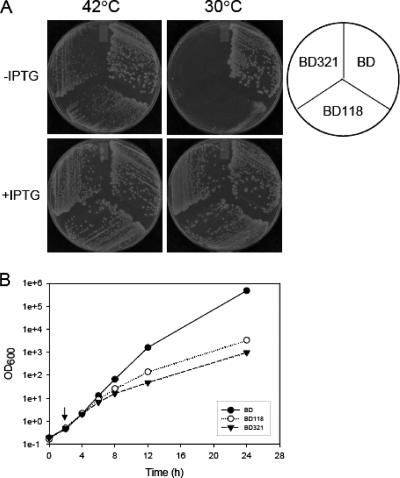
Transformants were first isolated on LB agar plates containing chloramphenicol (50 μg/ml; for the pIN vector), kanamycin (35 μg/ml; for the chromosomal der deletion), and ampicillin (50 μg/ml; for pIEEcttg) at 30°C. Single colonies were streaked on LB agar plates containing only chloramphenicol and kanamycin. Subsequently, the plates were incubated at 42°C in order to remove the Ampr helper plasmid as described previously (16). Colonies which were resistant to chloramphenicol and kanamycin but sensitive to ampicillin at 42°C were selected, and the final three strains, complemented with pINEcDer, pINEcDerGD-1, and pINEcDerGD-2, were designated BD, BD118, and BD321, respectively. All three strains were streaked on LB plates in the presence and absence of IPTG and grown at 42°C and 30°C. All three strains could form colonies at 42°C, as shown in Fig. 1A, but at 30°C, neither BD118 nor BD321 formed colonies in the absence of IPTG. This suggests that both DerN118D and DerN321D proteins are defective but that increased expression of these proteins can functionally compensate for the defective properties of the mutants. To further investigate strains BD118 and BD321, each complemented type of cells was grown overnight at 42°C in LB medium containing chloramphenicol and kanamycin, and the cell cultures were then diluted in the same medium. The diluted cultures were incubated at 42°C for 2 h, and the temperature was then shifted to 30°C, followed by further incubation. As shown in Fig. 1B, the isogenic wild-type strain BD grew exponentially at 30°C; however, the growth rates of BD118 and BD321 cells were significantly reduced. Note that the growth rate of BD321 was more severely reduced than that of the BD118 strain. These results indicate that either one of the intact GTP-binding domains of Der cannot complement the lethality of the der null mutant cells at 30°C and that a Der mutant in which the mutation of Asn to Asp at the GTP-binding domain is detrimental to the function of Der.
FIG. 1.
Complementation of the essential phenotype of the BK strain by wild-type or G-domain mutant Der proteins. (A) Strains BD, BD118, and BD321 were obtained as described in Materials and Methods. All three strains were streaked on LB plates containing chloramphenicol and kanamycin in the presence and absence of IPTG. IPTG was added at a concentration of 1 mM. The plates were incubated at 30°C or 42°C overnight. The pIN vector contains a chloramphenicol resistance gene. BD, der deletion cells containing pINEcDer; BD118, der deletion cells containing pINEcDerGD-1; BD321, der deletion cells containing pINEcDerGD-2. GD-1 and GD-2 correspond to the DerN118D and DerN321D mutations, respectively. (B) Growth curves for the BD, BD118, and BD321 strains. The der deletion strains expressing wild-type and mutant Der proteins were grown at 42°C in LB medium containing chloramphenicol and kanamycin. The cultures were shifted to 30°C at the time indicated by an arrow. During incubation at 30°C, the cultures were repeatedly diluted (1:10) in fresh medium. Closed circles, BD; open circles, BD118; closed triangles, BD321.
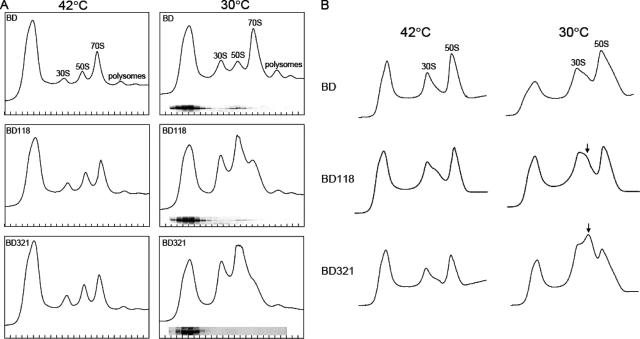
Next, we further investigated the effect of G-domain mutants on the ribosome profile. Polysome samples were prepared from cells grown at 42°C or 30°C for 6 h after temperature downshift, and cell lysates were applied to a 5 to 40% sucrose solution, followed by centrifugation as described in Materials and Methods. At 42°C, cells expressing wild-type or mutant Der proteins maintained normal ribosome profiles (Fig. 2A); however, both BD118 and BD321 grown at 30°C accumulated significant amounts of ribosomal subunits at the cost of 70S ribosomes, while the polysome pattern of the BD cells was not affected. Western blot analysis was then carried out to characterize the binding of Der mutants to 50S ribosomes. It was found that the two Der mutant proteins showed much reduced affinity in large ribosomal subunit fractions that may contain precursors of 50S ribosomes (Fig. 2A).
FIG. 2.
(A) Polysome defects of the BD118 and BD321 strains. The cells were grown as described in the legend to Fig. 1B. Samples were collected 0 and 6 h after temperature downshift. The cell pellets were resuspended with buffer BP containing 10 mM MgCl2. Polysomes were resolved by ultracentrifugation as described in Materials and Methods. Western blotting assays were carried out using anti-Der antiserum to detect the Der proteins in subsequent fractions from BD, BD18, and BD321 cells grown at 30°C. (B) Ribosomal subunit profiles of strains BD, BD118, and BD321. The same cell pellets as those used for panel A were resuspended with buffer BP containing 0.25 mM MgCl2. Ribosomal subunits were resolved by ultracentrifugation. An arrow indicates abnormal 50S ribosomal subunits observed in strains BD118 and BD321.
To further examine the effects of mutations in the Der protein, we carried out sucrose density gradient analysis at a lower concentration of Mg2+ (0.25 mM). The ribosome profiles of isogenic wild-type BD cells were normal at both 42°C and 30°C, but an unusual peak was observed between the 50S and 30S subunits for both strains BD321 and BD118 incubated at 30°C but not at 42°C (Fig. 2B). We previously observed that Der-depleted cells exhibit a similar ribosome pattern, suggesting that either the N118D or N321D mutation may dissociate Der from the 50S ribosome (16).
Isolation of multicopy suppressor for strain BD321.
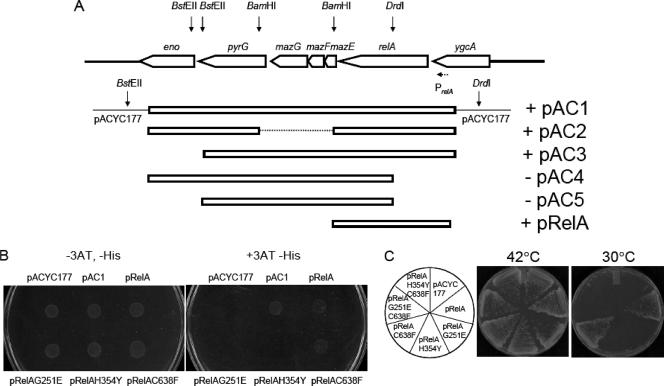
To search for a genetic element(s) in the E. coli genome which can restore the growth of strain BD321 at low temperature, BD321 (Kanr Cmr) cells were transformed with an E. coli genomic library constructed in the pACYC177 (Ampr) vector. The library contained partially Sau3AI-digested chromosomal DNA fragments from E. coli BW25113, which were ligated to BamHI-digested pACYC177. Note that the pACYC177-carried genomic library is compatible with pINEcDerGD-2, as they contain different replication origins, i.e., p15A and ColE1, respectively. Transformants were isolated by the ability to grow at 30°C on LB agar plates containing kanamycin, chloramphenicol, and ampicillin. Plasmids from those colonies that gained the ability to grow at 30°C were purified and retransformed into BD321 cells to confirm their ability to suppress the growth defect phenotype. Approximately 26,000 transformants were screened, and 28 library clones were isolated as possible candidates for suppression activity. Since the chromosomal library contained clones encoding the wild-type Der protein, PCR was carried out to identify the clones containing the full-length der gene. Among 28 clones, 21 clones carried the der gene. Among the remaining seven clones, three clones contained a genomic fragment of lacY, suggesting that overexpression of mutant Der protein by incorporating more inducer molecules rescues the growth defect of strain BD321. Interestingly, the remaining four clones contained the relA region (Fig. 3A).
FIG. 3.
(A) Identification of the gene responsible for suppression of the defective growth phenotype of strain BD321. pAC1-derived subclones and pRelA were constructed as described in Materials and Methods. Arrows indicate restriction enzyme sites. PrelA (dashed arrow) indicates the promoter of the relA gene (21). (B) Sensitivity of relA cells expressing mutant RelA proteins to 3-AT. Strain CF1944 (relA251::kan) was transformed with pACYC177, pAC1, pRelA, pRelAG251E, pRelAH354Y, or pRelAC638F as described above. Transformed cells were grown overnight in LB medium, and the culture was washed with M9 medium. Portions of cells were spotted onto M9 minimal plates containing ampicillin and all amino acids but histidine in the presence and absence of 15 mM 3-AT. The plates were incubated at 37°C for 18 h. (C) Strain BD321 was transformed with plasmids as indicated in the left circle, and cells harboring each plasmid were streaked on LB agar plates containing ampicillin, kanamycin, and chloramphenicol. The plates were incubated at 42°C and 30°C overnight.
Identification of a gene that suppresses the growth defect of strain BD321.
One of the suppressor clones was designated pAC1. The inserted genomic element from plasmid pAC1 was sequenced and found to contain seven genes, on a genomic fragment containing eno, pyrG, mazG, mazF, mazE, relA, and ygcA (Fig. 3A). To identify the exact gene that suppresses the growth defect of the BD321 strain, the pAC1 DNA was digested with BamHI, followed by self-ligation. The resulting plasmid, pAC2, retained the ability to suppress the defective growth phenotype of the BD321 strain, suggesting that the open reading frame of mazEFG is not responsible for the suppression observed. Likewise, cell growth was also supported by pAC3, which was constructed from pAC1 by BstEII digestion and blunt end cutting followed by self-ligation. However, the plasmids digested with DrdI followed by self-ligation after the blunt end cutting reaction (pAC4 and pAC5, respectively) lost the suppression activity. The relA gene was disrupted in both of these plasmids. These experiments suggest that the relA gene is responsible for suppression of the defective growth phenotype of the BD321 strain. This was further confirmed by transforming a plasmid that carries only the relA gene with its own promoter, PrelA (pRelA), as shown in Fig. 3A.
Another G-domain mutant, DerN118D, is also defective for cell growth (Fig. 1). Thus, we also tested whether the same suppressor clones are able to recover the defective growth phenotype of the BD118 strain expressing the DerN118D protein. Introduction of plasmids pAC1, pAC2, pAC3, and pRelA into the BD118 strain restored the ability of the BD118 strain to form colonies at 30°C (data not shown), suggesting that both the GTP-binding domains may associate directly with RelA or that overproduction of (p)ppGpp by RelA may indirectly suppress the Der-defective growth of the Der mutants. As described above, among four relA library clones, two clones encode only 412 and 522 residues of the N-terminal region, lacking 332 and 222 residues, respectively, from the C-terminal region (pAC6 and pAC7, respectively), indicating that the C-terminal region of RelA is dispensable for the suppression activity of the Der mutants. It is important that the relA gene could not suppress the null mutant of der.
(p)ppGpp synthetic activity is essential for suppression.
The RelA protein is made up of 744 residues and contains both catalytic (455 residues) and regulatory (289 residues) domains, in the N- and C-terminal regions, respectively. The N-terminal catalytic domain of RelA (amino acids 1 to 455) synthesizes (p)ppGpp by using ATP and GDP (or GTP), and it was suggested that the C-terminal domain (amino acids 456 to 744) is involved not only in the negative regulation of (p)ppGpp synthesis but also in RelA-RelA interaction (13, 37). Furthermore, the C-terminal region of RelA plays a role in its association with ribosomes (37). Previously, Gropp et al. (13) characterized the catalytic activities of three RelA mutant proteins, namely, RelAG251E, RelAH354Y, and RelAC638F. They showed that the G251E substitution dramatically inhibited (p)ppGpp synthesis and, on the other hand, that the H345Y substitution partially impairs the synthetic activity of RelA. In contrast, the RelAC638F mutant was shown to increase (p)ppGpp synthesis by eliminating the negative regulation of the C-terminal domain (13). Since the phenotype of the BD321 strain was suppressed by the full-length as well as C-terminally truncated RelA protein, it is likely that the direct association of RelA with ribosomes is not necessary for its suppressor activity.
To further characterize the role of RelA in suppression of the defective growth phenotype of strain BD321, site-directed mutagenesis of the relA gene was carried out as described by Gropp et al. (13). We first tested if mutations in RelA affect (p)ppGpp synthesis in relA mutant cells. Accumulation of (p)ppGpp can be tested in minimal medium devoid of histidine in the presence of a histidine analog, 3-AT. 3-AT induces His starvation, resulting in the synthesis of (p)ppGpp, which upregulates a biosynthesis pathway of the his operon (13, 14, 29, 30). Figure 3B shows that relA cells harboring pAC1, pRelA, or pRelAC638F were resistant to 3-AT, while transformants harboring pACYC177 (vector control), pRelAG251E, or pRelAH354Y were sensitive to 3-AT, confirming that RelAC638F, but not RelAG251E and RelAH354Y, is active.
Next, the BD321 cells were transformed with pRelA, pRelAG251E, pRelAH354Y, or pRelAC638F, and interestingly, cell growth was not supported by the pRelAG251E and pRelAH354Y plasmids, while pRelA and pRelAC638F were able to suppress the defective growth phenotype of the BD321 strain (Fig. 3C). This result suggests that (p)ppGpp synthesis is essential for the suppression effect. A C638F mutation in the RelA protein was proposed to disrupt the disulfide bond in the RelA protein, and therefore, in order to examine if inactive monomeric RelA can support the cell growth of the BD321 strain, we introduced the C638F mutation into the RelAG251E and RelAH354Y proteins. We transformed the pRelAG251EC638F and pRelAH354YC638F plasmids, carrying the respective mutant relA genes, into the BD321 strain. Neither of the mutant proteins was able to support the growth of the BD321 cells. Similar results were obtained with strain BD118 (data not shown). These results suggest that (p)ppGpp is directly involved in the suppression of the Der-defective mutant growth defect. It is important that although RelA overproduction causes overproduction of (p)ppGpp, resulting in cell growth inhibition, the suppressor plasmids pRelA, pAC1, pAC2, and pAC3 (Fig. 3) did not show any inhibiting effects on cell growth because the relA gene was cloned into pACYC, a low-copy-number plasmid.
Overexpression of RelA recovers the ribosome defect of the BD321 strain.
The RelA protein is both directly and indirectly involved in gene expression. It recognizes stalled ribosomes upon amino acid starvation and synthesizes (p)ppGpp, followed by release of reactivated ribosomes (4). Subsequently, (p)ppGpp affects gene expression by up- or downregulation of transcription (19). Therefore, we tested the possibility that overexpression of RelA may upregulate the expression of DerN321D in the suppressor strain. To examine this, Western blot analysis was carried out to compare the amounts of Der protein in BD321 and BD321 cells containing pRelA. The expression of DerN321D protein in the BD321 cells harboring pRelA was identical to the expression of DerN321D in the BD321 cells, indicating that RelA did not affect Der synthesis. In addition, cell lysates of these three strains were analyzed by SDS-PAGE, and the overall protein patterns were almost identical (data not shown).
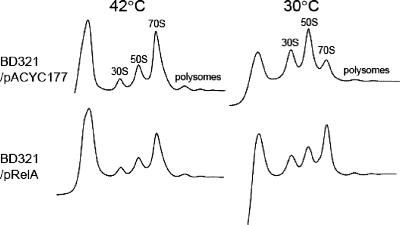
Next, we examined if the expression of RelA in the BD321 strain was able to result in a normal ribosome profile. First, samples from BD321 cells harboring pACYC177 or pRelA were prepared 0 and 6 h after the temperature shift, and cell lysates were subjected to a sucrose density gradient sedimentation experiment as shown in Fig. 4. Interestingly, for the BD321 cells expressing RelA, the polysome profile was normal and accumulation of ribosomal subunits was not observed. In contrast, for strain BD321 harboring the pACYC177 plasmid, ribosomal subunits were accumulated, similar to the case for strain BD321, as shown in Fig. 2A. Therefore, based on polysome analysis, it appears that overexpression of (p)ppGpp mediated by plasmid-encoded RelA may modulate ribosomal processing in the BD321 strain.
FIG. 4.
Recovery of polysome defects by expression of RelA. BD321 cells harboring pACYC177 or pRelA were grown in LB medium as described in the legend to Fig. 1B. Samples were collected 0 and 6 h after temperature downshift. The cell pellets were resuspended with buffer BP containing 10 mM MgCl2. Polysomes in sucrose gradient solution were resolved by ultracentrifugation as described in Materials and Methods.
DISCUSSION
Previously, we demonstrated that Der comigrated with 50S ribosomal subunits in a GTP-dependent manner and that, in Der-depleted cells, not only both ribosomal subunits but also rRNA precursors accumulated, suggesting that Der may be a novel ribosome assembly factor. In this report, we show that a single amino acid substitution of Asn to Asp in the GTP-binding domains inhibits the function of Der. Two mutations, N118D and N321D, in the Der protein resulted in severe impairment of cell growth and polysome profile when they were expressed in a der deletion strain. Total depletion of Der protein leads to a similar effect, suggesting that the two consecutive GTP-binding domains are critical to the cellular function of Der and that they may function cooperatively (16). Interestingly, we previously observed that the N300D mutation in T. maritima Der, which is equivalent to the N321D mutation in E. coli Der, did not affect GTPase activity in vitro (25). Thus, it is possible that an N300D mutation in T. maritima Der is unlikely to completely disrupt hydrogen bond formation in the guanine nucleotide or that an enzymatic action of T. maritima Der is different from that of E. coli Der. Recently, Bharat et al. (1) demonstrated that DerS16A or DerS216A protein could not support the growth of the der null strain (1). To further prove the essentiality of each GTP-binding domain in Der, we introduced S16A and S216A mutations, and both of them caused a lethal effect at 42°C (data not shown), suggesting that DerN118D and DerN118D are partially functional in the cell.
As reported previously, in BK (der deletion) cells cultured at 42°C, precursors of 23S rRNA and 50S ribosomal subunits were accumulated, and the accumulated 50S subunits were further dissociated into approximately 45S, 40S, and 35S subunits in vitro at Mg2+ concentrations of 1, 0.5, and 0.25 mM, respectively. The 40S subunit particles were shown to contain reduced amounts of L9 and L18 ribosomal proteins (16). In the present study, we observed that in BD321 cells, 50S ribosome biogenesis became defective at 30°C, accumulating abnormal, possibly destabilized, 50S ribosomal subunits at a low concentration of Mg2+ (0.25 mM). These data suggest that the essential role of Der in 50S ribosomal maturation at a later biogenesis step is critical for the integrity of the 50S subunit at all temperatures. Except for the ribosome-associated function of Der described above, its other cellular roles are not known. Therefore, in an attempt to elucidate a physiological role(s) of Der, we carried out screening of an E. coli genomic library, using strain BD321, which cannot grow at 30°C. We identified the relA gene as a multicopy suppressor for this strain. Interestingly, it was found that the lacY gene for β-galactoside permease was also able to suppress the growth defect of strain BD321 (data not shown). This is consistent with the fact that strains BD118 and BD321 were able to grow normally in the presence of 1 mM IPTG (Fig. 1A), possibly because the overexpression of DerN321D protein may overcome its weaker association activity with 50S ribosomes. Furthermore, Western blot assay revealed that the cold sensitivity of DerN321D was due to the lower expression of Der protein at 30°C than at 42°C (data not shown). Notably, however, the expression of RelA in strain BD321 did not upregulate the expression of DerN321D (data not shown).
From our genomic library screening and mutational analysis of RelA, we can rule out the possibility that interaction of RelA with ribosomes is critical for the suppression effect, since only the N-terminal domain of RelA, which is devoid of ribosome association, homodimerization activity, and negative regulation, could suppress the Der defect. It should be noted that RelA was not able to suppress the null phenotype of the der deletion mutant and did not interact with Der (data not shown). Thus, it appears that the suppression effect of RelA solely depends on the accumulation of (p)ppGpp. The stringent effector ppGpp was shown in crystal structures to associate with GTPases such as B. subtilis Obg and E. coli EF-Tu (5, 31). Our competition GTPase assay revealed that an excessive amount of ppGpp effectively inhibits the GTPase activity of E. coli Der, suggesting that ppGpp is able to bind to Der at the GTP-binding site (data not shown). However, an attempt to identify the association of Der in the presence of ppGpp in a ΔrelA ΔspoT mutant was not successful, implying that binding of Der to 50S ribosomes may depend on pppGpp (guanosine 3′-diphosphate, 5′-triphosphate) rather than ppGpp (guanosine 3′,5′-bispyrophosphate). Thus, it is possible that DerN118D and DerN321D mutant proteins associate only weakly with 50S ribosomal subunits, and this defect may be suppressed by association with (p)ppGpp. Our findings further suggest the possibility that Der may sense the total pool of guanine nucleotides, including the stringent effector (p)ppGpp, in the cell.
One of the features of the stringent response is the inhibition of biosynthesis of stable RNAs, rRNA, and tRNA. There have been several proposed models for direct negative regulation by ppGpp, including one to propose that ppGpp binds to the β and β′ subunits of RNA polymerase and that this complex destabilizes the RNA polymerase-promoter open structure to trigger a rapid downregulation of transcription initiation (8, 19, 33). During exponential growth, RNA and peptide chain elongation rates and the concentration of (p)ppGpp decrease with decreasing temperatures, although the concentrations of protein and RNA remain invariant between high and low temperatures (27). Based on the observations that the association of Der with 50S ribosomal subunits was pivotal for ribosome biogenesis (Fig. 2A) and that BD321 cells recovered the ability to form colonies in the presence of IPTG (Fig. 1A), it can be presumed that stable RNA synthesis in the BD321/pRelA strain is partially inhibited by (p)ppGpp, such that rRNA synthesis is downregulated and thus able to process properly. This may also support the notion that Der plays a critical role in ribosome assembly. However, we cannot rule out the possibility that the combination of (p)ppGpp-Der interaction and inhibition of RNA synthesis may work in concert to lead to suppression of the defective growth phenotype of the BD321 strain.
Der and ObgE have been identified as suppressors for an rrmJ null strain (32). Recent publications showed that the function of ObgE is implicated in late 50S ribosome assembly (28) and that Obg proteins regulate SopT (p)ppGpp degradation activity in E. coli and Vibrio cholerae (17, 24, 35). These GTPases appear to have overlapping functions in 50S ribosome assembly, but our genomic library screening revealed that the obgE gene was not isolated as a suppressor for strain BD321, implying a unique physiological function for the Der protein. These findings provide new insight into Der function in ribosome biogenesis and show that, clearly, further studies are necessary to understand the exact physiological role of Der.
Acknowledgments
We thank Koichi Inoue and Sangita Phadtare for critical readings of the manuscript and for scientific suggestions. The E. coli strain CF1944 (relA251::kan) was kindly provided by M. Cashel (Laboratory of Molecular Genetics, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD).
Footnotes
Published ahead of print on 22 February 2008.
REFERENCES
- 1.Bharat, A., M. Jiang, S. M. Sullivan, J. R. Maddock, and E. D. Brown. 2006. A cooperative and critical role for both G-domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 1887992-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourne, H. R., D. A. Sanders, and F. McCormick. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 8125-132. [DOI] [PubMed] [Google Scholar]
- 3.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 10117-127. [DOI] [PubMed] [Google Scholar]
- 4.Braeken, K., M. Moris, R. Daniels, J. Vanderleyden, and J. Michiels. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 1445-54. [DOI] [PubMed] [Google Scholar]
- 5.Buglino, J., V. Shen, P. Hakimian, and C. D. Lima. 2002. Structural and biochemical analysis of the Obg GTP binding protein. Structure 101581-1592. [DOI] [PubMed] [Google Scholar]
- 6.Caldas, T., E. Binet, P. Bouloc, A. Costa, J. Desgres, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23S ribosomal RNA methyltransferase. J. Biol. Chem. 2216414-16419. [DOI] [PubMed] [Google Scholar]
- 7.Cashel, M., D. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1488-1496. In F. C. Neidhardt, R. Curtis, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 8.Chatterji, D., N. Fujita, and A. Ishihama. 1998. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells 3279-287. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, K. G. C., P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Y. Zhu, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 431387-1400. [DOI] [PubMed] [Google Scholar]
- 11.Ghrayeb, J., H. Kimura, M. Takahara, H. Hsiung, Y. Masui, and M. Inouye. 1984. Secretion cloning vectors in Escherichia coli. EMBO J. 32437-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grishin, N. V. 2001. KH domain: one motif, two folds. Nucleic Acids Res. 29638-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gropp, M., Y. Strausz, M. Gross, and G. Glaser. 2001. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J. Bacteriol. 183570-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilton, J. L., P. C. Kearney, and B. N. Ames. 1965. Mode of action of the herbicide, 3-amino-1,2,4-triazole(amitrole): inhibition of an enzyme of histidine biosynthesis. Arch. Biochem. Biophys. 112544-547. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, J., and M. Inouye. 2001. An essential GTPase, der, containing double GTP-binding domains from Escherichia coli and Thermotoga maritima. J. Biol. Chem. 27631415-31421. [DOI] [PubMed] [Google Scholar]
- 16.Hwang, J., and M. Inouye. 2006. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 611660-1672. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, M., K. Datta, A. Walker, J. Strahler, P. Bagamasbad, P. C. Andrews, and J. R. Maddock. 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 1886757-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, G., S. Moriya, and C. Wada. 2001. Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol. Microbiol. 411037-1051. [DOI] [PubMed] [Google Scholar]
- 19.Magnusson, L. U., A. Farewell, and T. Nystrom. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13236-242. [DOI] [PubMed] [Google Scholar]
- 20.Mehr, I. J., C. D. Long, C. D. Serkin, and H. S. Seifert. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metzger, S., I. B. Dror, E. Aizenman, G. Schreiber, M. Toone, J. D. Friesen, M. Cashel, and G. Glaser. 1988. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J. Biol. Chem. 26315699-15704. [PubMed] [Google Scholar]
- 22.Morimoto, T., P. C. Loh, T. Hirai, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 1483539-3552. [DOI] [PubMed] [Google Scholar]
- 23.Muench, S. P., L. Xu, S. E. Sedelnikova, and D. W. Rice. 2006. The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding. Proc. Natl. Acad. Sci. USA 10312359-12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raskin, D. M., N. Judson, and J. J. Mekalanos. 2007. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 1044636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson, V. L., J. Hwang, E. Fox, M. Inouye, and A. M. Stock. 2002. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure 101649-1658. [DOI] [PubMed] [Google Scholar]
- 26.Ron, E. Z., R. E. Kohler, and B. D. Davis. 1966. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science 1531119-1120. [DOI] [PubMed] [Google Scholar]
- 27.Ryals, J., R. Little, and H. Bremer. 1982. Temperature dependence of RNA synthesis parameters in Escherichia coli. J. Bacteriol. 151879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato, A., G. Kobayashi, H. Hayashi, H. Yoshida, A. Wada, M. Maeda, S. Hiraga, K. Takeyasu, and C. Wada. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10393-408. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber, G., S. Metzger, E. Aizenman, S. Roza, M. Cashel, and G. Glaser. 1991. Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 2663760-3767. [PubMed] [Google Scholar]
- 30.Stephens, J. C., S. W. Artz, and B. N. Ames. 1975. Guanosine 5′-diphosphate 3′-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino-acid deficiency. Proc. Natl. Acad. Sci. USA 724389-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suck, D., and W. Kabsch. 1981. X-ray determination of the GDP-binding site of Escherichia coli elongation factor Tu by substitution with ppGpp. FEBS Lett. 126120-122. [DOI] [PubMed] [Google Scholar]
- 32.Tan, J., U. Jakob, and J. C. Bardwell. 2002. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 1842692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toulokhonov, I. I., I. Shulgina, and V. J. Hernandez. 2001. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta′-subunit. J. Biol. Chem. 2761220-1225. [DOI] [PubMed] [Google Scholar]
- 34.Walter, M., S. G. Clark, and A. D. Levinson. 1986. The oncogenic activation of human p21ras by a novel mechanism. Science 233649-652. [DOI] [PubMed] [Google Scholar]
- 35.Wout, P., K. Pu, S. M. Sullivan, V. Reese, S. Zhou, B. Lin, and J. R. Maddock. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 1865249-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 2665980-5990. [PubMed] [Google Scholar]
- 37.Yang, X., and E. E. Ishiguro. 2001. Involvement of the N terminus of ribosomal protein L11 in regulation of the RelA protein of Escherichia coli. J. Bacteriol. 1836532-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]